Abstract
HIV replication is suppressed in vitro by a CD8+ cell noncytotoxic antiviral response (CNAR). This activity directly correlates with an asymptomatic clinical state. The objective of this study was to identify the phenotype of CD8+ cell subsets having strong CNAR activity. CD8+ cell subset frequencies and CNAR levels were measured for human immunodeficiency virus (HIV)-uninfected individuals and three groups of HIV type 1 (HIV-1)-infected individuals: asymptomatic individuals with low-level viremia (vHIV), antiretroviral-drug-treated subjects with undetectable virus levels (TxHIV), and therapy-naïve aviremic elite controllers (EC). CD8+ cells from the vHIV individuals exhibited the highest HIV-suppressing activity and had elevated frequencies of CD45RA− CD27+ and PD-1+ (CD279+) cells. Functional assessments of CD8+ cells sorted into distinct subsets established that maximal CNAR activity was mediated by CD45RA− CCR7− CD27+ and PD-1+ CD8+ cells. T cell receptor (TCR) repertoire profiles of CD8+ cell subsets having strong CNAR activity exhibited increased perturbations in comparison to those of inactive subsets. Together, these studies suggest that CNAR is driven by HIV replication and that this antiviral activity is associated with oligoclonally expanded activated CD8+ cells expressing PD-1 and having a transitional memory cell phenotype. The findings better describe the identity of CD8+ cells showing CNAR and should facilitate the evaluation of this important immune response in studies of HIV pathogenesis, resistance to infection, and vaccine development.
CD8+ cells from human immunodeficiency virus (HIV)-infected individuals potently suppress the in vitro replication of HIV in primary CD4+ cells without eliminating the infected cells (24, 32, 50, 51, 54). This CD8+ cell noncytotoxic antiviral response (CNAR) becomes evident during the acute stage of infection (22, 38), varies in magnitude among HIV-infected persons (21, 35, 52), and directly correlates with a healthy clinical state (3, 6, 7, 14, 25, 35). Strong CNAR activity is a feature of long-term survivors (LTS) of HIV infection (3, 14). CNAR activity is also associated with resistance to HIV infection among exposed seronegative individuals (27, 45). CD8+ cells from uninfected persons, individuals with AIDS, and HIV-infected subjects receiving long-term antiretroviral therapy typically exhibit little or no CNAR activity (21, 22, 44).
CNAR is associated with the production of a soluble CD8+ cell antiviral factor (CAF) (26) that suppresses HIV replication by blocking transcription from the virus promoter (9, 32). CAF is not present in cytolytic granules (37), and CNAR does not involve apoptosis (36). CNAR activity is effective against all HIV and simian immunodeficiency virus (SIV) isolates and is not virus type specific (5, 53). In addition, CD8+ cells are able to suppress HIV replication in major histocompatibility complex (MHC)-mismatched CD4+ cells (28, 34, 51).
CNAR has been found to be associated with an activated CD8+ cell phenotype (25) and with vascular cell adhesion molecule 1 (VCAM-1)-expressing CD8+ cells (11). To further characterize the CD8+ cells that mediate CNAR, we evaluated this activity in phenotypically distinct CD8+ cell subsets obtained directly from peripheral blood without in vitro stimulation. Here we report that the natural suppression of HIV type 1 (HIV-1) replication is mediated by memory CD8+ T cells, particularly those that express PD-1 and exhibit a transitional memory cell phenotype.
MATERIALS AND METHODS
Human subjects.
HIV-1-infected (n = 100) and uninfected (HIV−) (n = 19) subjects were selected from participants in ongoing studies at the University of California San Francisco (UCSF). Among the HIV-1-infected subjects, all of whom had been infected for more than 5 years, three groups were studied: (i) individuals on antiretroviral therapy with very low viral loads (TxHIV+) (n = 44), (ii) elite controllers (EC) (n = 15) who had been infected with HIV-1 for at least 10 years without exhibiting AIDS-defining symptoms and had undetectable plasma viral loads (<50 copies HIV RNA/ml) and normal CD4+ T cell counts (>400 CD4+ T cells/μl) in the absence of antiretroviral therapy, and (iii) viremic individuals (vHIV+) (n = 41) who were asymptomatic, had viral loads ranging from 3.6- to 4.8-log RNA copies per ml, and were not receiving antiretroviral therapy. Each subject signed informed consent forms, and the study received approval from the Committee for Human Research at UCSF. Salient features of the study population are provided in Table 1 .
TABLE 1.
Demographic and immunologic characteristics of the study subjectsa
| Status of study subjects | Gender (no. of males/females) | Age (yr) | No. of lymphocytes/μl (× 1,000) | No. of CD4+ T cells/μl | % CD4+ T cells | No. of CD8+ T cells/μl | % CD8+ T cells | CD4/CD8 ratio | HIV-1 RNA log no. of copies/ml |
|---|---|---|---|---|---|---|---|---|---|
| HIV− | 16/3 | 43 (33-56) | 1.7 (1.4-2.0) | 835 (650-1,209) | 40 (37-48) | 580 (513-647) | 26 (23-30) | 1.6 (1.2-1.8) | |
| TxHIV+ | 44/0 | 50 (40-56) | 2.1 (1.8-2.4) | 497 (312-662) | 29 (20-35) | 802 (588-1,160) | 47 (39-55) | 0.6 (0.4-0.9) | <1.7 |
| EC | 15/0 | 51 (48-53) | 2.1 (1.7-2.4) | 690 (638-830) | 43 (35-49) | 693 (554-890) | 37 (32-42) | 1.1 (0.9-1.5) | <1.7 |
| vHIV+ | 39/2 | 48 (39-56) | 2.3 (1.6-2.8) | 447 (357-633) | 27 (20-33) | 901 (687-1,350) | 52 (44-60) | 0.5 (0.4-0.8) | 3.9 (3.6-4.8) |
Except for gender, numbers provided are median values, with interquartile ranges in parentheses. Tx, treated.
Clinical measures.
Complete differential blood cell counts (CBCs) to determine erythrocyte numbers, hemoglobin levels, and levels of total leukocytes, granulocytes, lymphocytes, monocytes, platelets, and T cell subsets were determined by the UCSF clinical laboratories. Measurements of plasma HIV RNA levels were performed using a branched-DNA (bDNA) assay (Siemens Diagnostics, Emeryville, CA) or were self-reported.
Cell specimens.
Whole blood was collected in evacuated tubes (BD) containing EDTA and sodium heparin for immunophenotyping and functional studies, respectively. Peripheral blood mononuclear cells (PBMC) were isolated from whole blood collected in evacuated tubes containing EDTA (BD Biosciences) by density gradient separation over Ficoll (Sigma). From the PBMC of each study subject, CD4+ and CD8+ cells were serially isolated by immunomagnetic bead separation (Miltenyi) prior to cell sorting. Purities of the isolated cells were >95%, as measured by flow cytometry.
CNAR assays.
To determine the respective levels of CNAR activity, incremental numbers of CD8+ cells (without in vitro stimulation) were cocultured with acutely HIV-infected autologous or heterologous CD4+ cells and the ensuing level of HIV replication were measured. Briefly, CD4+ cells were resuspended (3 × 106 cells/ml) in growth medium (RPMI 1640 medium supplemented with fetal calf serum [heat inactivated at 56°C for 30 min; 10%, vol/vol], penicillin [100 U/ml], streptomycin [100 μg/ml], l-glutamine [2 mM], and recombinant interleukin 2 [IL-2; 100 U/ml; Invitrogen]) and stimulated for 3 days in the presence of phytohemagglutinin-leucoagglutinin (PHA-L; 3 μg/ml; Sigma) in a 37°C humidified incubator. Subsequently, 10 × 106 cells were treated with Polybrene (2 μg/ml; Sigma) for 20 min at 37°C and pelleted. The pelleted cells were resuspended in 1 ml of HIV-1SF33 (10,000 50% tissue culture infective doses [TCID50]/ml in PBMC) for 1 h at 37°C with periodic mixing. HIV-1SF33, a syncytium-inducing (SI), CXCR4-tropic (X4) strain, has been maintained in primary cells since its isolation, exhibits rapid replication kinetics with a high degree of cytopathicity in cell culture, and is not sensitive to β-chemokine-mediated antiviral effects (31). The acutely infected CD4+ cells were then washed and resuspended at 106 cells/ml of growth medium, and 200-μl aliquots were placed into a flat-bottom 96-well tissue culture plate (Falcon 3072; BD) in triplicate. Cocultures were established by adding CD8+ cells to wells containing acutely infected CD4+ cells at 1:1, 0.5:1, and 0.25:1 CD8+ cell-to-CD4+ cell input ratios.
Measurement of CNAR activity.
To measure HIV replication levels in the cultures, 100-μl aliquots of the culture supernatant from each well were collected on days 3 and 6 of culture and centrifuged at 12,000 × g for 1 h at 4°C, and the resulting virus pellets were assayed for reverse transcriptase (RT) activity as described previously (17). In ongoing cultures, the supernatant removed for measurement of HIV levels was replaced with an equal volume of fresh growth medium. In the assays for RT activity, 2.5 units (U) of purified avian myeloblastosis virus (AMV) reverse transcriptase (Roche) was used as a positive control. The extent of CNAR activity in each culture, assessed as percent suppression, was calculated based on the magnitude of HIV replication in the cocultures in comparison to replication levels in CD4+ cells cultured alone as follows: percent suppression = (1 − [HIV level in coculture/HIV level in CD4+ cells]) × 100%. Differential CNAR activity was defined to be present when the CD8+ cell subsets compared were discordant (e.g., >60% versus <40% suppression) at various CD8+ cell-to-CD4+ cell input ratios.
Conditioned medium and transwell assays.
All experiments were performed without the in vitro stimulation of CD8+ cells, except those involving conditioned medium from cultured CD8+ cells or the cells used in transwell inserts. For such experiments, the CD8+ cells were stimulated with anti-CD3 beads following their separation into distinct subsets. Conditioned medium was generated by culturing the stimulated CD8+ cells of HIV-infected individuals in serum-free F12 medium, and its anti-HIV activity was measured as previously described (33). For the transwell assays, the CD4+ cells and CD8+ cells were placed into a 24-well plate, where they were physically separated by a semipermeable insert (0.45 μm; BD). These cultures were established with the upper chamber containing 1 × 105 HIV-infected CD4+ cells and the lower reservoir having 4 × 105 CD8+ cells.
Immunophenotyping and cell sorting.
To enumerate the frequencies of distinct CD8+ cell subsets, fresh whole blood was stained with various combinations of fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)-, and allophycocyanin (APC)-conjugated monoclonal antibodies. Antibodies to CD3 (clone UCHT1), CD7 (M-T701), CD11b (D12), CD25 (2A3), CD27 (L128), CD28 (L293), CD38 (HB7), CD45RA (L48), CD57 (HNK-1), CD62L (SK11), CD95 (DX2), PD-1 (MIH4), HLA-DR (L243), CD122 (TU27), CD127 (M21), and CCR7 (3D12) were obtained from BD, whereas anti-CD8β (IM2217U) and -2B4 (C1.7.1) were purchased from Beckman Coulter. Negative cell populations and cursor settings for each specimen were established using the appropriate isotype control reagents (BD). Four-color flow cytometric analyses were performed on a FACSort (BD) using CellQuest (BD). Seven-color analyses were performed using an LSRII cytometer (BD) and FlowJo (TreeStar) software. To isolate subsets within the CD8+ cell compartment, CD8+ cells were first separated from PBMC using immunomagnetic beads (see above). The CD8+ cells were stained with various antibodies (described above) and then sorted into distinct subsets using a FACSDiva (BD) or FACSAria (BD) instrument (UCSF core facilities). Postsort cell populations were analyzed and confirmed to exceed 90% purity. Where presented in figures, dot plots have been gated on CD8+ cells that fall within a lymphocyte region of characteristic size and complexity.
CD107 degranulation assay.
To evaluate cytotoxic potential (i.e., the release of lytic granules), CD107 degranulation assays were performed similarly to those described previously (4). Briefly, CD8+ cells alone or mixed with HIV-infected heterologous CD4+ cells were cultured in complete medium (106 cells/ml) in the presence of anti-CD107a and anti-CD107b FITC-conjugated monoclonal antibodies (BD; 50 μl/ml each) for 1 h at 37°C. After 1 h, monensin (BD; 1 μl/ml) was added and the cells were cultured for another 4 to 5 h at 37°C. Then, the cells were collected, stained with anti-CD8-APC (BD), and analyzed for CD107 surface expression. For use as positive controls in the CD107 degranulation assay, primary CD8+ cells from HLA-A0201+ HIV-infected individuals were stimulated for 3 days in complete medium containing the HIV-1 gag consensus peptide SLYNTVATL (1 μg/ml) or a cocktail of cytomegalovirus (CMV)-Epstein-Barr virus (EBV)-influenza virus (CEF; 10 μg/ml) peptides (NIH AIDS Research and Reference Reagent Program) (23). The peptide-stimulated CD8+ cells were then cultured for an additional 11 days to generate appreciable frequencies of the antigen-specific cells. These HIV-specific and CMV-specific CD8+ cells were then used in CD107 degranulation assays in the absence or presence of SLYNTVATL and CEF peptides. Flow cytometric analyses were performed following staining of the cells with CMV- and HIV-specific tetramer reagents (Beckman Coulter).
T cell receptor repertoire analysis.
In evaluating the clonal diversity of the T cells mediating strong CNAR activity, T cell receptor (TCR) repertoire analysis was performed as previously described (19, 20, 23). Historical nomenclature for the TCRVβ families is used in this text in order to maintain consistency with those prior reports. Briefly, RNA was extracted from cell lysates using RNeasy columns (Qiagen) and reverse transcribed using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and a TCRβ chain constant region primer (CTCAGCTCCAGTG). Specific combinations of one, two, or three TCRVβ-specific forward primers (0.1 μM each) were then used for each multiplex PCR on the resulting cDNA. Each reaction mixture also contained a 6-carboxyfluorescein (FAM) fluorescently labeled reverse primer (0.1 μM) specific for the TCRβ constant region. The nucleotide lengths and fluorescent intensities of the resulting amplicons were measured using an ABI PRISM 3730xl DNA analyzer (Applied Biosystems). Fluorescence data were collected and processed using GeneScan and Genotyper software packages (ABI).
Statistical analyses.
All data were compiled in an Access database (Microsoft). Group comparisons were performed using the nonparametric Mann-Whitney test with Splus 6.1 (Insightful). Graphs were prepared using SigmaPlot 11.2 (Systat).
RESULTS
CNAR activity is associated with persistent low-level HIV replication in vivo.
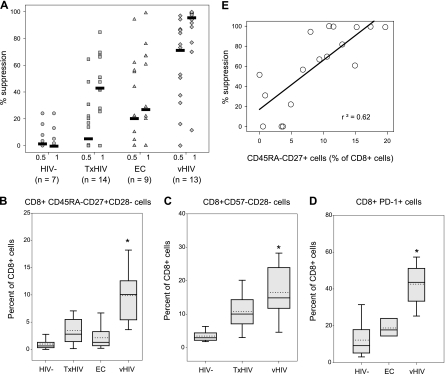
In previous studies, we have consistently observed that mitogen-stimulated CD8+ cells from HIV-uninfected individuals, HIV-infected individuals with advanced disease, and those receiving antiretroviral therapy exhibit reduced or no CNAR activity in comparison to the CNAR activity in asymptomatic HIV-infected individuals (see the introduction). In the present study, we evaluated unstimulated (ex vivo) CD8+ cells from healthy uninfected blood donors (HIV−), elite controllers (EC), viremic HIV-infected individuals (vHIV), and HIV-infected subjects receiving antiretroviral therapy (TxHIV) (Fig. 1 and Table 1). We observed the following trend of CNAR activity among CD8+ cells from individuals in these groups: vHIV ≫ EC ≈ TxHIV > HIV− (Fig. 1A).
FIG. 1.
CNAR activities and cell subset frequencies of CD8+ cells from viremic and aviremic individuals. (A) Shown are the relative abilities of primary CD8+ cells from HIV-uninfected (HIV−) individuals, HIV-infected subjects receiving antiretroviral therapy (TxHIV+), elite controllers (EC), and low-viremia HIV-infected individuals (vHIV+) to suppress HIV replication in heterologous primary CD4+ cells. Results are provided for day 6 cultures containing CD8+ cells and acutely HIV-infected CD4+ cells that were plated at 0.5:1 and 1:1 input ratios, respectively. HIV replication levels consistently peaked 6 days postinfection in the cultures of CD4+ cells alone, with RT activity exceeding 106 cpm/100 μl of cell culture supernatant. Bars show median values. Box plots detail whole-blood levels of CD45RA− CD27+ CD28− (B), CD57− CD28− (C), and PD-1+ CD8+ (D) cells among HIV-infected and uninfected individuals. The lower, central, and upper lines of the boxes identify quartiles; dotted lines demark mean values; and whiskers and dots mark the 5th, 10th, 90th, and 95th percentiles. *, significantly different from aviremic groups (P < 0.05). (E) Correlation between CNAR activity (y axis) (1:1 CD8+ cell/CD4+ cell ratio) and the frequency of CD45RA− CD27+ cells within the CD8+ cell compartment (x axis).
To identify potential subsets of cells that mediate strong CNAR activity, we measured the whole-blood frequencies of CD8+ cell subsets in the aforementioned groups (Table 2). In comparison to the HIV− and EC subjects, vHIV individuals exhibited elevated frequencies (P < 0.05) of CD45RA−, CD11b−, HLA-DR+, and CD27+ CD8+ cells. vHIV subjects also exhibited the highest frequencies of CD45RA− CD27+, CD57− CD28−, and PD-1+ CD8+ cells (Fig. 1B, C, and D, respectively). A direct correlation (r2 = 0.62) was observed between CNAR activity and the frequency of CD45RA− CD27+ cells within the CD8+ cell compartment (Fig. 1D). These cross-sectional comparisons of CD8+ cells from aviremic and viremic individuals demonstrate an association between low-level HIV replication, frequencies of distinct CD8+ cell subsets, and CNAR activity.
TABLE 2.
CD8+ cell subset frequencies in HIV-infected and uninfected individuals
| Cell subset | % CD8+ cells among subjects who weree: |
|||
|---|---|---|---|---|
| HIV− (n = 19) | TxHIV+ (n = 12) | EC (n = 15) | vHIV+ (n = 31) | |
| CD38+ | 64 (50-69) | 62 (49-69) | 60 (55-70) | 70 (59-76) |
| HLA-DR+ | 15 (10-32) | 31 (22-46) | 33 (25-46) | 49d (36-62) |
| CD27+ | 22 (19-26) | 26 (20-31) | 21 (17-23) | 31d (25-37) |
| CD28+ | 57 (49-71) | 47 (38-53) | 40 (33-56) | 35a,c (27-46) |
| CD25+ | 4 (3-6) | 5 (3-6) | 3 (2-4) | 2a,c (1-4) |
| CD122+ | 31 (19-44) | 16 (11-25) | 21 (18-29) | 17a (13-26) |
| CD45RO+ | 35 (28-46) | 50 (43-54) | 42 (34-48) | 47a (38-58) |
| CD45RA+ | 69 (59-83) | 67 (57-72) | 73 (64-78) | 59a,b (52-71) |
| CD57+ | 19 (12-22) | 43 (30-54) | 36 (32-41) | 41a (34-55) |
| CD62L+ | 62 (54-71) | 50 (45-57) | 52 (43-58) | 42d (36-49) |
| CD11b+ | 31 (19-41) | 15 (7-27) | 17 (11-21) | 10a,b (6-16) |
| PD-1+ | 9 (6-15) | ND | 18 (15-23) | 44a,b (34-51) |
Significantly different from the value for HIV− subjects (P < 0.05).
Significantly different from the value for EC subjects (P < 0.05).
Significantly different from the value for TxHIV+ subjects (P < 0.05).
Signficantly different from the values for HIV−, EC, and TxHIV+ subjects (P < 0.05).
Values are medians (interquartile ranges). ND, no data.
Maximal CNAR activity is mediated by activated CD8+ T cells.
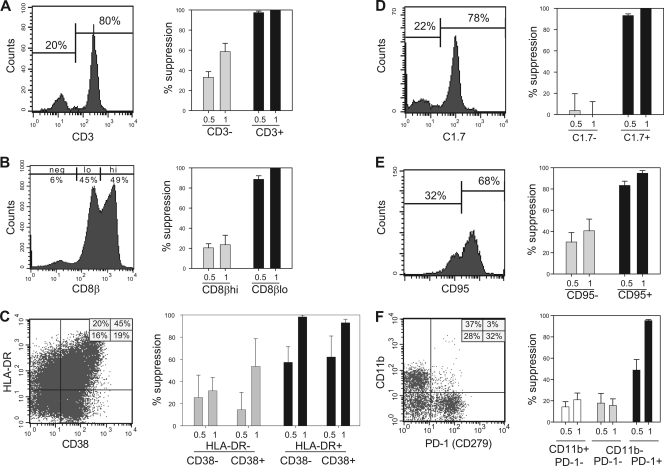
Having identified correlations between the heightened CNAR activity of bulk CD8+ cells and the frequencies of selected CD8+ cell subsets (e.g., CD45RA− CD27+ cells) measured by fluorescence-activated cell sorting (FACS), we next evaluated the functional ability of those subsets (without in vitro stimulation procedures) to suppress HIV-1SF33 (X4) replication in primary CD4+ cell cultures (Fig. 2). Toward this objective, more than 50 independent cell sorting experiments were performed. First, to determine whether or not CNAR activity is a characteristic of all CD8+ lymphocytes or restricted to CD8+ T cells, CD3+ and CD3− CD8+ cells were evaluated for their CNAR activity (Fig. 2A). As shown in Fig. 2, most CD8+ cells express CD3. Note that this and all other flow cytometry plots presented in Fig. 2 and 3 are gated on CD8+ lymphocytes. The CD8+ CD3+ (T) cells exhibited substantially greater CNAR activity than did the CD8+ CD3− (NK) cells (95% versus 35% suppression of HIV replication at a 0.5:1 input ratio). Thus, with respect to CD8+ cells having distinct hematopoietic lineages, the CNAR is mediated by CD8+ T cells.
FIG. 2.
CNAR activity is mediated by CD8+ T cells that are activated in vivo. Primary CD8+ cells from asymptomatic, low-viremia HIV-1-infected individuals were sorted into distinctive subsets and then cocultured with heterologous HIV-infected primary CD4+ cells at 0.5:1 and 1:1 CD8+ cell-to-CD4+ cell input ratios. Shown are representative staining profiles (left) and antiviral activities (right) for CD8+ cell subsets differing in expression of CD3 (A), CD8β (B), HLA-DR and CD38 (C), C1.7 (D), CD95 (E), and CD11b and PD-1 (F). All FACS plots shown are gated on live CD8+ lymphocytes. Results are representative of at least 2 independent experiments with different CD8+ cell sources. neg, negative for marker; lo, low levels of the marker; hi, high levels of the marker.
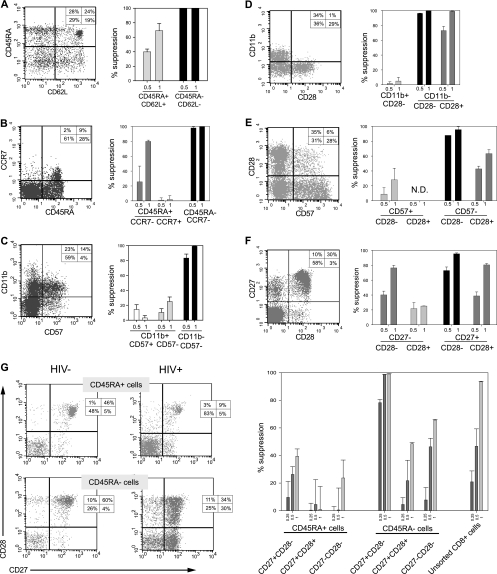
FIG. 3.
Transitional memory CD8+ cells exhibit maximal CNAR activity. Shown are staining profiles (left) and anti-HIV activities (right) of CD8+ cells that were separated into distinct populations based on the expression of CD45RA and CD62L (A), CD45RA and CCR7 (B), CD57 and CD11b (C), CD28 and CD11b (D), CD57 and CD28 (E), CD27 and CD28 (F), and CD45RA, CD27, and CD28 (G). Suppression data are shown for CD8+ cell/infected CD4+ cell coculture ratios of 0.25:1 (G only), 0.5:1, and 1:1 (left to right). Results are representative of at least 2 independent experiments with different CD8+ cell sources.
CD8+ T cells, as they encounter environmental stimuli, become “activated” and newly express a variety of surface antigens. To investigate their association with CNAR, cells differing in their levels of expression of several activation markers (e.g., the CD8 beta chain [CD8β], HLA-DR, and CD38, IL-25, IL-122, C1.7, CD95, CD11b, and PD-1) were evaluated. Among the CD8+ T cells, two distinct populations were apparent: dimly fluorescent CD8β (CD8βdim) and brightly fluorescent CD8β (CD8βbright) cells (Fig. 2B). In cocultures with HIV-1-infected cells, the CD8βdim subset consistently exhibited greater suppression of virus replication than did the CD8βbright subset (90% versus 20% at a 0.5:1 input ratio). Noteworthy is the fact that CD8+ NK cells do not express CD8β (39). Upon evaluation of CD8+ cells differing in their levels of expression of HLA-DR and CD38, HLA-DR+ cells had the strongest CNAR activity, regardless of CD38 expression levels (Fig. 2C). We observed that maximal CNAR activity was mediated by C1.7+, CD95+, and CD11b− PD-1+ CD8+ cells (Fig. 2D-F), in addition to CD8βdim and HLA-DR+ cells. Notably, CD8βdim cells and C1.7+ cells were primarily CD95+ cells and CD28− cells, respectively. These results for unstimulated CD8+ cells, with respect to HLA-DR, CD38, and CD11b, confirm previous studies with mitogen-stimulated CD8+ cells (25) and further establish that the CNAR is mediated by activated CD8+ cells.
We also investigated CD8+ cells differing in expression of the IL-2 receptors CD122 and CD25. Differential expression of the intermediate-affinity IL-2 receptor CD122 was not observed to be associated with CNAR activity (data not shown). Moreover, depletion of the minor population of CD8+ cells that express the low-affinity IL-2 receptor CD25 had no appreciable effect on CNAR levels (data not shown). Thus, IL-2 receptor expression does not distinguish ex vivo CD8+ cells with high and low CNAR activity.
CD8+ cells with a memory phenotype exhibit maximal CNAR activity.
To investigate the differentiation state of CNAR-mediating cells, CD8+ cells differing in their levels of coexpression of CD45RA CD62L, CD45RA CCR7, CD11b CD57, CD11b CD28, CD57 CD28, and CD45RA CD27 CD28 were assessed (Fig. 3). First, we compared the CNAR activities of CD8+ cells that differed in their levels of coexpression of CD45RA and CD62L or CCR7 (Fig. 3A and B). In comparison to naïve (CD45RA+ CD62L+ or CD45RA+ CCR7+) cells, the more immunologically mature CD8+ cells (CD45RA− CD62L− or CD45RA− CCR7+/−) exhibited superior CNAR activity (e.g., >90% suppression versus <50% suppression when the 0.5:1 cell input values were compared). Next, we evaluated the CNAR activity of CD8+ cells that differed in their levels of coexpression of CD11b and CD57 or CD28 (Fig. 3C and D). CD11b− cells were observed to be mostly CD57 negative, whereas these cells were heterogeneous for expression of CD28. Maximal suppression was associated with a CD57− phenotype, while both CD11b− CD28− and CD11b− CD28+ cells exhibited strong CNAR activity. Then, we evaluated the CNAR activity of CD8+ cells that varied in their levels of coexpression of CD28 and CD57 or CD27 (Fig. 3E and F). Maximal suppression was exhibited by CD57− CD28− and CD27+ CD28− cells. Finally, among six CD8+ cell subsets that differentially express CD45RA, CD27, and CD28, the two CD45RA− CD27+ subsets were found to most potently suppress HIV replication (Fig. 3G). Therefore, the CD8+ cells that exhibited maximal suppression of HIV replication were CD45RA− CD27+ CD28− cells, although appreciable anti-HIV activity was also mediated by CD45RA− CD27+ CD28+ cells. Comparisons of bulk CD8+ cells, which were unstained or stained with antibodies, revealed no substantial differences in CNAR activity (data not shown). Thus, activation of the cells and/or blocking of the surface antigens due to antibody binding was not involved. These results show that CNAR is associated with memory CD8+ cells, particularly those with a transitional memory phenotype.
CNAR is associated with oligoclonal CD8+ cell populations.
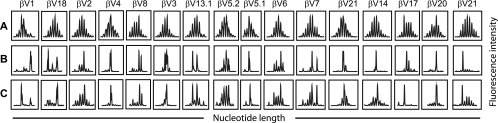
To investigate the diversity of T cell receptor (TCR) usage among CD8+ cell subsets having strong CNAR activity, TCR profiling was performed (Fig. 4). Specifically, bulk CD8+ cells (Fig. 4C), CD45RA− CD27− CD28−, CD45RA− CD27+ CD28+, CD45RA− CD27+ CD28− (Fig. 4B), CD45RA+ CD27+ CD28+ (Fig. 4A), CD45RA+ CD27− CD28−, and CD45RA+ CD27+ CD28− CD8+ cells were analyzed from 3 HIV-1-infected (vHIV+) individuals. Among the subsets evaluated, the CD45RA+ CD27+ CD28+ CD8+ cells exhibited Gaussian-like distributions of CDR3 lengths in each TCRVβ family, indicating a lack of clonal dominance within this subset. As described above (for Fig. 3G), this population exhibited poor CNAR activity. In contrast, TCRVβ families within the CD45RA− CD27+ CD28− CD8+ cell subset, a population with robust CNAR activity (Fig. 3G), exhibited a striking degree of perturbation (Fig. 4B). These data provide evidence that the CNAR is associated with CD8+ cell subsets having a biased (i.e., a non-Gaussian-like distribution) T cell receptor usage.
FIG. 4.
HIV-suppressing CD8+ cells are increased in frequency among cells that exhibit biased T cell receptor repertoires. T cell receptor diversity profiling was performed on CD45RA+ CD27+ CD28+ (naïve) (A), CD45RA− CD27+ CD28− (differentiated) (B), and bulk CD8+ (C) cells that had been freshly isolated from the blood of an HIV-infected individual. Elevated CNAR activity was exhibited by the CD45RA− CD27+ CD28− CD8+ cells in parallel assays. Shown are the resulting spectratypes for 16 of the 24 TCRVβ families evaluated. Within each TCRVβ family, peaks are separated by 3 nucleotides (1 amino acid). Similar results were observed among 3 HIV-infected individuals.
CD8+ cells with strong CNAR activity do not exhibit classical cytotoxic T lymphocyte (CTL) activity.
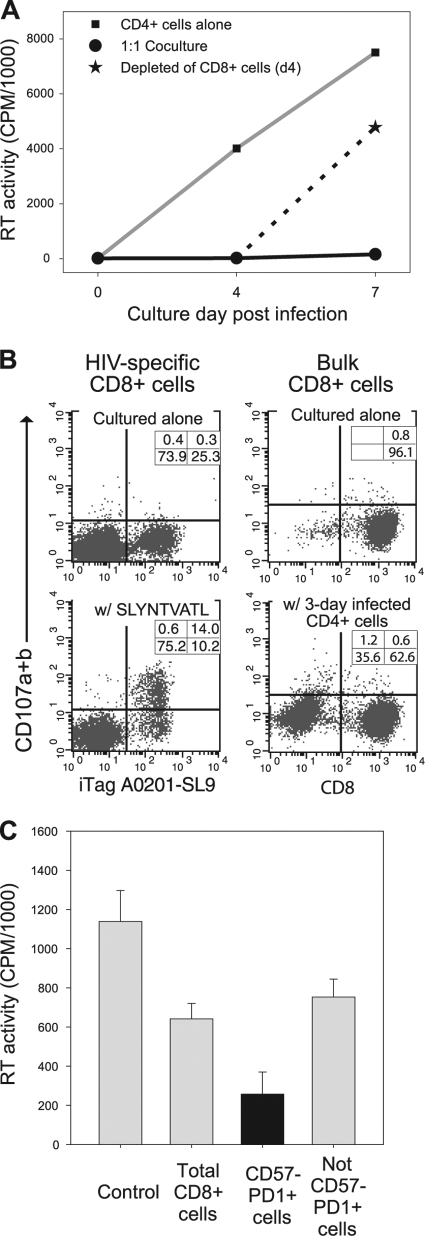
Past studies have shown that CNAR does not involve cell killing (28). To confirm those findings in the present studies, the CD8+ cells were removed after 3 days of cocultivation with the HIV-infected CD4+ cells (Fig. 5). As shown previously (50, 51), HIV-infected cells persist in the presence of CNAR activity. HIV levels rapidly increased in the cell culture supernatants upon removal of the CD8+ cells (Fig. 5A).
FIG. 5.
Noncytotoxic features of HIV-suppressing CD8+ cells. (A) HIV replication levels were evaluated in cultures from which CD8+ cells were removed following suppression of HIV replication. Shown are reverse transcriptase (RT) levels in the supernatants of HIV-infected cells cultured alone (▪), in the presence of HIV-suppressing CD8+ cells (•), and upon removal of the HIV-suppressing CD8+ cells after 4 days of coculture (★). (B) CD107a/CD107b levels were measured in HIV-specific CD8+ cells (left) and bulk CD8+ cells (right) upon exposure to cognate antigen and heterologous HIV-infected CD4+ cells, respectively. (C) CD8+ cells from a viremic HIV-infected individual were sorted into 2 subsets: CD57− PD1+ cells and those lacking this phenotype (not CD57− PD1+). Conditioned medium collected from cultures containing these CD8+ cell subsets was placed onto CD4+ cells that were acutely infected with HIV. Shown are the RT levels in each culture at the time of peak virus replication in the control. Data in each panel are representative of at least 2 separate experiments.
In separate experiments, CD8+ cells were evaluated for degranulation upon their exposure to HIV peptides or HIV-infected heterologous CD4+ cells in CD107 mobilization assays. The mobilization of lysosome-associated membrane glycoproteins (LAMPs), including CD107a (LAMP-1) and CD107b (LAMP-2), to the cell surfaces of CD8+ cells is directly associated with CTL activity (4). CD8+ cells that were expanded in vitro with HIV-specific (Fig. 5B, left) or CMV-specific (data not shown) antigens and then incubated in the presence of their cognate peptides underwent marked degranulation. In comparison, CD8+ cells did not degranulate when placed into culture with heterologous CD4+ cells that had been infected for 3 days and were producing substantial levels of HIV (Fig. 5B, right). Also, the levels of CD107 expression on CD8+ cells were not found to differ between CD8+ cells cultured alone and those cocultured with acutely infected CD4+ cells (data not shown). Notably, the CD8+ cells that were stimulated with HIV peptides were able to suppress HIV replication in acutely infected heterologous CD4+ cells poststimulation.
In another series of experiments, we assessed the ability of various subsets of CD8+ cells to produce soluble factors having anti-HIV activity. In two independent experiments (Fig. 5C), conditioned medium from the CD57− PD-1+ CD8+ cells exhibited increased anti-HIV activity in comparison to conditioned medium from bulk CD8+ cells or those lacking a CD57− PD-1+ cell phenotype. Similarly, CD57− PD-1+ cells suppressed HIV replication by >50% when assessed in transwell assays (data not shown). These results demonstrate that HIV-suppressing CD8+ cells do not eliminate HIV-infected cells and do not exhibit detectable degranulation in the presence of HIV-infected CD4+ cells. Moreover, CD8+ cell subsets with strong CNAR activity do suppress HIV replication via the secretion of a soluble factor(s). Thus, they are unlike classical CTL in function.
DISCUSSION
In previous studies, we observed that CD8+ cells from HIV-infected individuals vary in their abilities to suppress HIV replication in primary CD4+ cells; CD8+ cells from asymptomatic persons have the highest CD8+ cell noncytotoxic antiviral response (CNAR) (3, 7, 14, 21, 25, 35, 52). To further characterize this anti-HIV activity, we systematically compared the whole-blood frequencies (Table 1) and HIV-suppressing activities of various CD8+ cell subsets (Fig. 2 and 3) among HIV-infected and uninfected individuals.
In comparison to CD8+ cells from aviremic (EC and TxHIV) HIV-infected individuals, those from asymptomatic subjects with low-level viremia (vHIV) were found to exhibit the strongest CNAR activity (Fig. 1). These findings support past results showing that when viral loads are below detectable levels, as is characteristic of elite controllers and subjects treated with antiretroviral therapy, the CNAR is generally low or not detectable (44). This observation indicates that CNAR is activated upon HIV replication. Those elite controllers who exhibited some CNAR activity (Fig. 1) most likely had blips of virus replication that were sufficient to sustain this response (16). Importantly, the viremic individuals in this study were healthy long-term survivors of HIV infection and exhibited low viral loads (median, 3.9 logs) (Table 1). Indeed, our findings are consistent with those of previous studies establishing this anti-HIV response as a characteristic of CD8+ cells from asymptomatic long-term survivors with low-level viremia (3, 7, 14, 25, 35).
Cross-sectional analyses of whole blood revealed increased frequencies of CD45RA− CD27+ and CD57− CD28− CD8+ cells in the vHIV group (Fig. 1). In addition, we observed a direct correlation between CNAR activity and the frequency of CD45RA− CD27+ CD8+ cells. These observations link CNAR activity with transitional memory cells (see below). The finding of decreased frequencies of CD45RA− CD27+ CD8+ cells in patients receiving antiretroviral therapy (Fig. 1A and B) provides an explanation for the previously observed loss of CNAR activity in CD8+ cells from these subjects (22, 44). Still, differences in CD8+ cell subset frequencies, as measured by flow cytometry, do not necessarily account for a loss or gain of antiviral function. Therefore, cell sorting experiments were performed to evaluate directly the antiviral activities of CD8+ cell subsets that change in frequency with HIV infection.
In assessing cell function, our experiments demonstrate that CD8+ cell subsets (without prior in vitro stimulation) exhibit differential abilities to suppress HIV replication. With respect to CD8+ cells of distinct hematopoietic lineages, CD3+ (T) cells expressing CD8β have strong CNAR activity, whereas CD3+ (NK) cells do not (Fig. 2). Of note, CD8β is not expressed by circulating CD8+ γ/δ T cells (30), thus excluding γ/δ T cells from being part of CNAR.
In previous investigations with mitogen-stimulated CD8+ cells, we observed that noncytotoxic anti-HIV activity was highest among HLA-DR+, CD11b−, and VCAM+ cells (11, 25). Similarly, in this study of peripheral blood CD8+ cells that were not mitogen stimulated, CNAR activity was found to be mediated by CD8+ cells having activated phenotypes (Fig. 2). Specifically, CD8βdim, HLA-DR+, CD95+, C1.7+, and PD-1+ CD8+ cells exhibited maximal CNAR activity. CD8β is downmodulated upon activation, and the frequency of this downmodulation in CD8βdim cells is increased in HIV-infected individuals (43). CD8βdim cells also suppress virus replication in feline immunodeficiency virus (FIV)-infected cats (13).
Moreover, as noted in other studies of CD8+ cells that were activated in vitro, CNAR was mediated chiefly by CD57− CD8+ cells (2). In the present studies of CD8+ cells not stimulated in vitro, both CD57− CD28+ and CD57− CD28− subsets were able to suppress HIV replication, although the CD28− cells showed superior CNAR activity. At the time of the earlier study (2), CD28 and CD57 were believed to be mutually exclusive antigens on CD8+ cells and the depletion of CD57+ cells was thought to yield relatively pure populations of CD28+ cells. However, as described in this study (Fig. 1C), HIV-infected persons can harbor appreciable numbers of CD57− CD28− CD8+ cells. Furthermore, CD8+ cells have now been shown to downmodulate CD28 expression during the process of immunologic maturation (46).
Further phenotypic analyses of CD8+ cells that potently suppress HIV replication provided insight into the differentiation state of cells mediating CNAR. Circulating CD8+ cells can be classified into the following subsets: naïve cells (CD45RA+ CD27+ CD28+), central memory cells (CD45RA− CCR7+ CD62L+), transitional memory cells (CD45RA− CD27+ CCR7−), and effector cells (CD45RA+ CD27− CCR7−) (15, 42). Memory cells express high surface levels of CD95 and exhibit little cytolytic activity in the absence of in vitro prestimulation. Effector cells express high levels of CD11b (10) and have high cytolytic activity without in vitro prestimulation. As noted above, we found that CNAR activity is mediated by CD8+ memory cells (Fig. 3), chiefly of the transitional memory (CD45RA− CD27+ CCR7−) phenotype. Supportive evidence that CNAR activity is mediated by memory CD8+ cells is our observation that the CD8+ cell population having strong CNAR activity exhibits skewed T cell receptor usage (Fig. 4). Additional studies are needed to establish the overall contribution of clonally expanded CD8+ cells to CNAR. In this regard, CD8+ cell clones isolated from HIV-infected individuals can exhibit CNAR activity without HIV-specific CTL activity (18, 47). Also, our findings are consistent with the very recent report that memory CD8+ cells, particularly CD45RA− CD27+ cells, effectively suppress HIV replication (12). However, our studies used a primary HIV-1 isolate and included biologic assessments of the antiviral function of CD8+ cells from HIV-1-infected individuals. Thus, we were able to distinguish the CNAR from classical CTL activity (see below).
HIV-specific (tetramer-positive) CD8+ cells have been described to predominantly exhibit a CD45RA− CD27+ CD57− CCR7− phenotype characteristic of transitional memory cells (8). In contrast, strong CTL responses have traditionally been associated with a CD45RA− CD27− CD28− effector cell phenotype (46). In comparison to bulk CD8+ cells or those that are specific for other viruses (e.g., CMV), HIV-specific CD8+ cells contain substantially lower levels of perforin, a protein required for granule-mediated cytolysis (1). These observations have led to speculation that HIV-specific CD8+ cells are defective killers (29, 49). Indeed, CD8+ CD45RA− CD27+ cells exhibit very little lytic activity in CD3 monoclonal antibody (MAb)-mediated redirected cytotoxicity assays (15). This finding is consistent with our past and present observations that HIV-suppressing CD8+ cells do not eliminate HIV-infected cells and that their antiviral effect is rapidly reversible (Fig. 5A) (50, 51). Moreover, CD8+ cells exhibiting strong CNAR activity do not degranulate in the presence of HIV-infected CD4+ cells (Fig. 5B) yet do secrete a soluble antiviral factor (Fig. 5C). Therefore, we propose that CD8+ cells having a CD45RA− CD27+ phenotype, likely including some that are HIV specific, are noncytotoxic HIV-suppressing cells (8).
Considerable attention has been given to the role of PD-1-expressing CD8+ cells in HIV infection. PD-1 is a member of the CD28 family, which has immunoregulatory functions (40), and this antigen is frequently expressed on HIV-specific CD8+ cells having a transitional memory phenotype (41). In agreement with other studies (48), we observed that PD-1 expression on CD8+ cells is increased in the context of HIV infection (Table 2). However, in those studies the HIV-specific CD8+ cells exhibiting high levels of PD-1 expression were found to be functionally defective (48). In contrast, our studies demonstrate that PD-1+ CD8+ cells have a previously unappreciated anti-HIV activity (Fig. 2F).
In summary, maximal CNAR activity is associated with CD8+ T cells having a CD3+ CD8βdim CD11b− CD57− CD95+ C1.7+ PD-1+ cell phenotype. These markers, along with HLA-DR (25), indicate that the CNAR is mediated by activated CD8+ cells. Furthermore, strong suppression of HIV replication is associated with CD8+ cells having a CD45RA− CD27+ CD28− CCR7− phenotype that is characteristic of transitional memory cells. Notably as well, PD-1+ CD8+ cells, previously considered dysfunctional, exhibit strong CNAR activity. Overall, our immunophenotyping and functional studies indicate that fewer than 50% of CD8+ cells mediate greater than 90% of CNAR activity. These studies better distinguish the ex vivo phenotypes of CD8+ T cells that mediate CNAR and provide insight for why HIV-infected subjects can differ in their levels of this important antiviral activity. This information can be helpful in developing novel immunotherapeutic strategies and directing vaccine design.
Acknowledgments
This research was supported by NIH grant 5R01AI056992-07 and California HIV/AIDS Research Program grant F05-SF-218.
We thank Vernon Maino (BD Biosciences) for advice and some reagents used for this study.
Footnotes
Published ahead of print on 8 December 2010.
REFERENCES
- 1.Appay, V., et al. 2000. HIV-specific CD8(+) T cells produce antiviral cytokines but are impaired in cytolytic function. J. Exp. Med. 192:63-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, E., K. N. Bossart, S. H. Fujimura, and J. A. Levy. 1997. CD28 costimulation increases CD8+ cell suppression of HIV replication. J. Immunol. 159:5123-5131. [PubMed] [Google Scholar]
- 3.Barker, E., et al. 1998. Virological and immunological features of long-term human immunodeficiency virus-infected individuals who have remained asymptomatic compared with those who have progressed to acquired immunodeficiency syndrome. Blood 92:3105-3114. [PubMed] [Google Scholar]
- 4.Betts, M. R., et al. 2003. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods 281:65-78. [DOI] [PubMed] [Google Scholar]
- 5.Blackbourn, D. J., et al. 1997. CD8+ cells from HIV-2-infected baboons control HIV replication. AIDS 11:737-746. [DOI] [PubMed] [Google Scholar]
- 6.Blackbourn, D. J., et al. 1996. Suppression of HIV replication by lymphoid tissue CD8+ cells correlates with the clinical state of HIV-infected individuals. Proc. Natl. Acad. Sci. U. S. A. 93:13125-13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castelli, J. C., S. G. Deeks, S. Shiboski, and J. A. Levy. 2002. Relationship of CD8(+) T cell noncytotoxic anti-HIV response to CD4(+) T cell number in untreated asymptomatic HIV-infected individuals. Blood 99:4225-4227. [DOI] [PubMed] [Google Scholar]
- 8.Champagne, P., et al. 2001. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature 410:106-111. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. H., K. J. Weinhold, J. A. Bartlett, D. P. Bolognesi, and M. L. Greenberg. 1993. CD8+ T lymphocyte-mediated inhibition of HIV-1 long terminal repeat transcription: a novel antiviral mechanism. AIDS Res. Hum. Retroviruses 9:1079-1086. [DOI] [PubMed] [Google Scholar]
- 10.Christensen, J. E., S. O. Andreasen, J. P. Christensen, and A. R. Thomsen. 2001. CD11b expression as a marker to distinguish between recently activated effector CD8(+) T cells and memory cells. Int. Immunol. 13:593-600. [DOI] [PubMed] [Google Scholar]
- 11.Diaz, L. S., et al. 2005. VCAM-1 expression on CD8+ cells correlates with enhanced anti-HIV suppressing activity. J. Immunol. 174:1574-1579. [DOI] [PubMed] [Google Scholar]
- 12.Freel, S. A., et al. 2010. Phenotypic and functional profile of HIV-inhibitory CD8 T cells elicited by natural infection and heterologous prime/boost vaccination. J. Virol. 84:4998-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebhard, D. H., et al. 1999. Progressive expansion of an L-selectin-negative CD8 cell with anti-feline immunodeficiency virus (FIV) suppressor function in the circulation of FIV-infected cats. J. Infect. Dis. 180:1503-1513. [DOI] [PubMed] [Google Scholar]
- 14.Gomez, A. M., F. M. Smaill, and K. L. Rosenthal. 1994. Inhibition of HIV replication by CD8+ T cells correlates with CD4 counts and clinical stage of disease. Clin. Exp. Immunol. 97:68-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamann, D., et al. 1997. Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 186:1407-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatano, H., et al. 2009. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J. Virol. 83:329-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman, A. D., B. Banapour, and J. A. Levy. 1985. Characterization of the AIDS-associated retrovirus reverse transcriptase and optimal conditions for its detection in virions. Virology 147:326-335. [DOI] [PubMed] [Google Scholar]
- 18.Hsueh, F. W., C. M. Walker, D. J. Blackbourn, and J. A. Levy. 1994. Suppression of HIV replication by CD8+ cell clones derived from HIV-infected and uninfected individuals. Cell. Immunol. 159:271-279. [DOI] [PubMed] [Google Scholar]
- 19.Killian, M. S., J. Matud, R. Detels, J. V. Giorgi, and B. D. Jamieson. 2002. MaGiK method of T-cell receptor repertoire analysis. Clin. Diagn. Lab. Immunol. 9:858-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Killian, M. S., et al. 2004. Persistent alterations in the T-cell repertoires of HIV-1-infected and at-risk uninfected men. AIDS 18:161-170. [DOI] [PubMed] [Google Scholar]
- 21.Killian, M. S., S. Ng, C. E. Mackewicz, and J. A. Levy. 2005. A screening assay for detecting CD8(+) cell non-cytotoxic anti-HIV responses. J. Immunol. Methods 304:137-150. [DOI] [PubMed] [Google Scholar]
- 22.Killian, M. S., J. Roop, S. Ng, F. M. Hecht, and J. A. Levy. 2009. CD8+ cell anti-HIV activity rapidly increases upon discontinuation of early antiretroviral therapy. J. Clin. Immunol. 29:311-318. [DOI] [PubMed] [Google Scholar]
- 23.Killian, M. S., et al. 2005. Clonal breadth of the HIV-1-specific T-cell receptor repertoire in vivo as determined by subtractive analysis. AIDS 19:887-896. [DOI] [PubMed] [Google Scholar]
- 24.Kinter, A. L., S. M. Bende, E. C. Hardy, R. Jackson, and A. S. Fauci. 1995. Interleukin 2 induces CD8+ T cell-mediated suppression of human immunodeficiency virus replication in CD4+ T cells and this effect overrides its ability to stimulate virus expression. Proc. Natl. Acad. Sci. U. S. A. 92:10985-10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landay, A. L., C. E. Mackewicz, and J. A. Levy. 1993. An activated CD8+ T cell phenotype correlates with anti-HIV activity and asymptomatic clinical status. Clin. Immunol. Immunopathol. 69:106-116. [DOI] [PubMed] [Google Scholar]
- 26.Levy, J. A. 2003. The search for the CD8+ cell anti-HIV factor (CAF). Trends Immunol. 24:628-632. [DOI] [PubMed] [Google Scholar]
- 27.Levy, J. A., F. Hsueh, D. J. Blackbourn, D. Wara, and P. S. Weintrub. 1998. CD8 cell noncytotoxic antiviral activity in human immunodeficiency virus-infected and -uninfected children. J. Infect. Dis. 177:470-472. [DOI] [PubMed] [Google Scholar]
- 28.Levy, J. A., C. E. Mackewicz, and E. Barker. 1996. Controlling HIV pathogenesis: the role of the noncytotoxic anti-HIV response of CD8+ T cells. Immunol. Today 17:217-224. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman, J., P. Shankar, N. Manjunath, and J. Andersson. 2001. Dressed to kill? A review of why antiviral CD8 T lymphocytes fail to prevent progressive immunodeficiency in HIV-1 infection. Blood 98:1667-1677. [DOI] [PubMed] [Google Scholar]
- 30.MacDonald, H. R., M. Schreyer, R. C. Howe, and C. Bron. 1990. Selective expression of CD8 alpha (Ly-2) subunit on activated thymic gamma/delta cells. Eur. J. Immunol. 20:927-930. [DOI] [PubMed] [Google Scholar]
- 31.Mackewicz, C. E., E. Barker, G. Greco, G. Reyes-Teran, and J. A. Levy. 1997. Do beta-chemokines have clinical relevance in HIV infection? J. Clin. Invest. 100:921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackewicz, C. E., D. J. Blackbourn, and J. A. Levy. 1995. CD8+ T cells suppress human immunodeficiency virus replication by inhibiting viral transcription. Proc. Natl. Acad. Sci. U. S. A. 92:2308-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackewicz, C. E., C. S. Craik, and J. A. Levy. 2003. The CD8+ cell noncytotoxic anti-HIV response can be blocked by protease inhibitors. Proc. Natl. Acad. Sci. U. S. A. 100:3433-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackewicz, C. E., M. R. Garovoy, and J. A. Levy. 1998. HLA compatibility requirements for CD8(+)-T-cell-mediated suppression of human immunodeficiency virus replication. J. Virol. 72:10165-10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackewicz, C. E., H. W. Ortega, and J. A. Levy. 1991. CD8+ cell anti-HIV activity correlates with the clinical state of the infected individual. J. Clin. Invest. 87:1462-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackewicz, C. E., S. Ridha, and J. A. Levy. 2000. Fas and Fas ligand are not involved in the suppression of HIV replication by CD8 cells. AIDS 14:204-205. [DOI] [PubMed] [Google Scholar]
- 37.Mackewicz, C. E., et al. 2003. Lack of the CD8+ cell anti-HIV factor in CD8+ cell granules. Blood 102:180-183. [DOI] [PubMed] [Google Scholar]
- 38.Mackewicz, C. E., L. C. Yang, J. D. Lifson, and J. A. Levy. 1994. Non-cytolytic CD8 T-cell anti-HIV responses in primary HIV-1 infection. Lancet 344:1671-1673. [DOI] [PubMed] [Google Scholar]
- 39.Moebius, U., G. Kober, A. L. Griscelli, T. Hercend, and S. C. Meuer. 1991. Expression of different CD8 isoforms on distinct human lymphocyte subpopulations. Eur. J. Immunol. 21:1793-1800. [DOI] [PubMed] [Google Scholar]
- 40.Okazaki, T., and T. Honjo. 2006. The PD-1-PD-L pathway in immunological tolerance. Trends Immunol. 27:195-201. [DOI] [PubMed] [Google Scholar]
- 41.Petrovas, C., et al. 2006. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J. Exp. Med. 203:2281-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708-712. [DOI] [PubMed] [Google Scholar]
- 43.Schmitz, J. E., et al. 1998. Expression of the CD8alpha beta-heterodimer on CD8(+) T lymphocytes in peripheral blood lymphocytes of human immunodeficiency virus− and human immunodeficiency virus+ individuals. Blood 92:198-206. [PubMed] [Google Scholar]
- 44.Stranford, S. A., et al. 2001. Reduction in CD8+ cell noncytotoxic anti-HIV activity in individuals receiving highly active antiretroviral therapy during primary infection. Proc. Natl. Acad. Sci. U. S. A. 98:597-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stranford, S. A., et al. 1999. Lack of infection in HIV-exposed individuals is associated with a strong CD8(+) cell noncytotoxic anti-HIV response. Proc. Natl. Acad. Sci. U. S. A. 96:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomiyama, H., T. Matsuda, and M. Takiguchi. 2002. Differentiation of human CD8(+) T cells from a memory to memory/effector phenotype. J. Immunol. 168:5538-5550. [DOI] [PubMed] [Google Scholar]
- 47.Toso, J. F., et al. 1995. Oligoclonal CD8 lymphocytes from persons with asymptomatic human immunodeficiency virus (HIV) type 1 infection inhibit HIV-1 replication. J. Infect. Dis. 172:964-973. [DOI] [PubMed] [Google Scholar]
- 48.Trautmann, L., et al. 2006. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 12:1198-1202. [DOI] [PubMed] [Google Scholar]
- 49.van Baarle, D., S. Kostense, M. H. van Oers, D. Hamann, and F. Miedema. 2002. Failing immune control as a result of impaired CD8+ T-cell maturation: CD27 might provide a clue. Trends Immunol. 23:586-591. [DOI] [PubMed] [Google Scholar]
- 50.Walker, C. M., A. L. Erickson, F. C. Hsueh, and J. A. Levy. 1991. Inhibition of human immunodeficiency virus replication in acutely infected CD4+ cells by CD8+ cells involves a noncytotoxic mechanism. J. Virol. 65:5921-5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker, C. M., D. J. Moody, D. P. Stites, and J. A. Levy. 1986. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science 234:1563-1566. [DOI] [PubMed] [Google Scholar]
- 52.Walker, C. M., D. J. Moody, D. P. Stites, and J. A. Levy. 1989. CD8+ T lymphocyte control of HIV replication in cultured CD4+ cells varies among infected individuals. Cell. Immunol. 119:470-475. [DOI] [PubMed] [Google Scholar]
- 53.Walker, C. M., et al. 1991. CD8+ T cells from HIV-1-infected individuals inhibit acute infection by human and primate immunodeficiency viruses. Cell. Immunol. 137:420-428. [DOI] [PubMed] [Google Scholar]
- 54.Wiviott, L. D., C. M. Walker, and J. A. Levy. 1990. CD8+ lymphocytes suppress HIV production by autologous CD4+ cells without eliminating the infected cells from culture. Cell. Immunol. 128:628-634. [DOI] [PubMed] [Google Scholar]