Abstract
One important function of the human adenovirus E1B 55-kDa protein is induction of selective nuclear export of viral late mRNAs. This protein interacts with the viral E4 Orf6 and four cellular proteins to form an infected-cell-specific E3 ubiquitin ligase. The assembly of this enzyme is required for efficient viral late mRNA export, but neither the relevant substrates nor the cellular pathway that exports viral late mRNAs has been identified. We therefore examined the effects on viral late gene expression of inhibition of the synthesis or activity of the mRNA export receptor Nxf1, which was observed to colocalize with the E1B 55-kDa protein in infected cells. When production of Nxf1 was impaired by using RNA interference, the efficiency of viral late mRNA export was reduced to a corresponding degree. Furthermore, synthesis of a dominant-negative derivative of Nxf1 during the late phase of infection interfered with production of a late structural protein. These observations indicate that the Nxf1 pathway is responsible for export of viral late mRNAs. As the infected-cell-specific E3 ubiquitin ligase targets its known substrates for proteasomal degradation, we compared the concentrations of several components of this pathway (Nxf1, Thox1, and Thoc4) in infected cells that did or did not contain this enzyme. Although the concentration of a well-established substrate, Mre11, decreased significantly in cells infected by adenovirus type 5 (Ad5), but not in those infected by the E1B 55-kDa protein-null mutant Hr6, no E1B 55-kDa protein-dependent degradation of the Nxf1 pathway proteins was observed.
During the late phase of productive infection by human species C adenovirus, such as adenovirus type 5 (Ad5), viral late mRNAs are exported selectively from the nucleus to the cytoplasm, with concomitant inhibition of export of the majority of cellular mRNAs (see references 6, 28, and 34 for reviews). Such regulation of mRNA export requires the viral E1B 55-kDa protein (3, 67, 71) and the complex it forms with the E4 Orf 6 protein (13, 23, 87). In both transformed and normal human cells, efficient export of viral late mRNAs correlates with the E4 Orf6 protein-dependent recruitment of the E1B protein to the peripheral zones of viral replication centers (40, 41, 66), which are the sites of synthesis and at least initial processing of viral late pre-mRNAs (2, 14, 68, 69). In infected cells, the E1B 55-kDa and E4 Orf6 proteins associate with the cellular proteins cullin 5, elongins B and C, and Rbx to form an E3 ubiquitin ligase (46). Substrates of the infected-cell-specific ubiquitin ligase include the cellular p53 protein (21, 46, 61, 71), the Mre11, Rad50, and Nbs1 components of the MRN complex (85), DNA ligase IV (5), and integrins α3 (25), which are targeted for proteasomal degradation. The infected-cell-specific ligase has also been implicated in regulation of mRNA export during the late phase of infection. Synthesis in infected cells of a dominant-negative derivative of cullin 5 that stabilized p53 and Mre11 resulted in decreases in viral L3 and L5 mRNA export and in synthesis of late protein similar to those observed in cells infected by an E1B 55-kDa protein-null mutant (88). Furthermore, a mutation in the E4 Orf6 protein-coding sequence that blocks formation of the infected-cell-specific ligase caused the same defects in export of viral late L5 mRNA, late protein synthesis, and viral replication as in an E4 Orf6-null mutant (7). Inhibition of proteasome activity in infected cells has also been reported to impair viral late gene expression (22). These observations indicate that ubiquitinylation and perhaps degradation of one or more proteins contribute directly or indirectly to regulation of mRNA export during the late phase of Ad5 infection. However, the identity of such substrates of the E1B 55-kDa and E4 Orf6 protein-containing E3 ligase is not yet known, nor has the cellular pathway by which viral late mRNAs are exported from the nucleus been identified.
Initial efforts to address the latter issue focused on the cellular export receptor exportin 1 (Crm-1). This receptor binds specifically to leucine-rich nuclear export signals (NES), like the first such signal to be identified in the human immunodeficiency type 1 (HIV-1) Rev protein (32, 36, 37, 63). The E1B 55-kDa protein contains an autonomous NES that is necessary and sufficient to direct export of the protein in both uninfected and Ad5-infected cells (27, 29, 57). Such transport of the E1B protein and its shuttling between nucleus and cytoplasm are inhibited by the HIV-1 Rev protein and the exportin 1 inhibitor, leptomycin B (29, 57), indicating that these processes are mediated by exportin 1. The E1B 55-kDa protein has also been observed to associate with the pp32 protein (46). This cellular protein contains an NES that is recognized by exportin 1 and binds to the human HuR protein, which has been identified as an export adaptor for certain unstable cellular mRNAs (12, 39). Nevertheless, exportin 1 is not responsible for export of viral late mRNAs: treatment of infected cells with leptomycin B under conditions that blocked accumulation of the E1B 55-kDa protein in the cytoplasm had no impact on synthesis of viral late proteins (17, 72). Furthermore, a cell-permeative peptide containing the HIV-1 Rev NES, but not an inactive derivative, significantly reduced the cytoplasmic accumulation of the E1B 55-kDa protein and inhibited its shuttling between the nucleus and cytoplasm (35), but it did not impair the efficiency with which processed major late (ML) penton or fiber mRNAs were exported from the nucleus (35).
The export receptor for the majority of processed eukaryotic mRNAs is the conserved protein termed Nxf1(Tap) in metazoans (Mex67p in Saccharomyces cerevisiae), which functions as an heterodimer with Nxt1 (reviewed in references 31, 51, 56, 75, and 86). Although Nxf1 binds specifically to the constitutive transport element present in unspliced and partially spliced RNAs of some retroviruses, such as Mason-Pfizer monkey virus (MPMV) (11, 43, 54), it initially recognizes most mRNA substrates indirectly, via adaptor proteins such as Thoc4 (Aly/Ref) and certain SR (Ser/Arg-rich) proteins (31, 50, 56). The E1B 55-kDa protein has not been reported to interact with these components of the Nxf1 export pathway. However, it does bind directly to a member of the RNP family termed E1B-Ap5 (38), which was subsequently reported to bind to glutathione S-transferase (GST)-Nxf1 (4). Redirection of one or more components necessary for Nxf1-dependent export from the normal cellular mRNA substrates to mature viral late mRNAs could account for the selectivity of mRNA export during the late phase of Ad5 infection. We therefore examined the contributions of the Nxf1 export pathway to viral late mRNA export by inhibiting the production or function of this protein in Ad5-infected cells.
MATERIALS AND METHODS
Cells and virus.
HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM) with 5% (vol/vol) fetal bovine serum and 5% (vol/vol) calf serum (Gibco-Invitrogen Corporation). Adenovirus type 5 and Hr6 were propagated in HeLa and 293 cells, respectively, and titers measured by plaque assay on 293 cells, as described previously (33, 35). HeLa Tet-ON cells (Clontech) were maintained in DMEM containing 10% fetal bovine serum lacking tetracycline (Clontech).
DNA constructs.
Expression constructs for chloramphenicol acetyltransferase (CAT) under the control of the cytomegalovirus (CMV) promoter that contain or lack the Mason-Pfizer monkey virus (MMPV) constitutive transport element (CTE) (8, 54) were provided by B. Cullen. Plasmids that carry the coding sequences for full-length Nxf1 or for a dominant-negative derivative lacking amino acids 518 to 619 fused to an N-terminal FLAG tag (19) were provided by R. Sandri-Goldin. The FLAG-Nxf1-coding sequences were subcloned by standard procedures into the multiple-cloning site of pTRE2hygro2-HA (Clontech) to create the plasmids pTRE2FLAGNxf1 and pTRE2FLAGNxf1ΔC. After amplification, plasmid DNA was purified by using the Qiagen plasmid purification kit according to the manufacturer's instructions.
RNAi.
Nxf1 Stealth RNA interference (RNAi) or universal control (low-GC) Stealth RNAi (Invitrogen) was introduced into the HeLa cells at 60 to 70% confluence in six-well dishes by using the Lipofectamine (Invitrogen) or Turbofect (Fermentas) reagent according to the manufacturers' protocols. Unless otherwise stated, 300 pmol of the double-stranded interfering RNAs was applied per well, either after introduction of reporter plasmids or prior to infection with Ad5.
Reporter assay for Nxf1-dependent export.
The CAT reporter plasmids (2.0 μg) containing or lacking the MMPV CTE were introduced in HeLa cells at 60 to 70% confluence by using Lipofectamine (Invitrogen). At 24 h thereafter, the anti-Nxf1 or control interfering RNAs were introduced as described above for 24 h, or cells were mock treated. Cells were then harvested and washed in cold phosphate-buffered saline (Gibco-BRL), and cytoplasmic extracts were prepared as described previously (41). Protein concentrations were determined by the method of Bradford (9) and 250 μg each sample assayed for CAT activity using the phase extraction method (83). Samples were incubated for 3 h at 37°C with 0.25 μCi [3H]chloramphenicol (30 to 60 Ci/nmol; Perkin-Elmer Life Sciences) and 40 μg/ml n-butyryl coenzyme A (CoA) and extracted with mixed xylenes (Sigma-Aldrich). After centrifugation for 3 min at 10,000 × g, the organic phase was removed and back extracted with 1 volume of 0.25 M Tris-HCl, pH 8.0. The phases were again separated, and the tritiated n-butyryl chloramphenicol recovered in the organic phase was measured by liquid scintillation counting in Econoscint (National Diagnostics).
Export of viral late mRNA.
Export of viral late mRNA was measured as the ratio of cytoplasmic to nuclear concentrations of the viral L5 fiber mRNA a few hours after the onset of the late phase of infection (41). Cytoplasmic and nuclear fractions of Ad5-infected cells were separated by two extractions with 0.01 M Tris-HCl (pH 7.5) containing 0.15 M NaCl, 1 mM EDTA, and 0.065% (vol/vol) NP-40. RNA was purified from both fractions using the Qiagen RNA isolation kit according to the manufacturer's instructions. For complete extraction of RNA from cytoplasmic and nuclear fractions from equal numbers of cells, the lysates were passed through Qiashredder columns (Qiagen). The concentrations of specific mRNAs were measured by using real-time reverse transcription-PCR (RT-PCR). Reverse transcription was performed manually as described previously (35, 41). Real-time PCR was carried out using the ABI Prism 7700 sequence detection system and TaqMan (Applied Biosystems) probes of an amplicon within the ML tripartite leader sequence described previously (35) and a human β-actin amplicon (Applied Biosystems) labeled with VIC and 6-carboxyfluorescein (FAM), respectively.
Immunoblotting.
Total cell extracts were prepared at the indicated times after infection or introduction of interfering RNAs as described previously (41). Proteins were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, transferred to Immobilon (Millipore Incorporated), and blocked as described previously (76). The viral E1B 55-kDa protein was detected with the monoclonal antibody 2A6 (80). The cellular Nxf1, Mre11, Thoc4 (Aly/Ref), and Thocl proteins were detected with mouse monoclonal antibody 31 (BD Biosciences), mouse monoclonal antibody ab397 (Abcam), rabbit polyclonal antibody from Sigma-Aldrich, and rabbit polyclonal antibody from Protein Tech Group, respectively. Primary antibodies were visualized with horseradish peroxidase conjugated to anti-mouse or anti-rabbit IgG (Millipore Corporation) and enhanced chemiluminescence (Amersham Biosciences). Cellular β-actin was visualized with a monoclonal antibody conjugated to horseradish peroxidase (Abcam). To estimate protein concentrations, enhanced chemiluminescence images were scanned and the areas under the proteins peaks of interest measured using the gel analysis function of Image J (73).
Immunofluorescence.
HeLa Tet-ON cells were seeded onto glass coverslips in six-well tissue culture dishes. When cells were 60 to 70% confluent, 4 μg pTRE2FlagNxf1 or pTRE2FlagNxf1ΔC DNA per well was introduced by using Lipofectamine (Invitrogen) according to the manufacturer's instructions. After 24 h, the cells were infected with 10 PFU/cell Ad5. Twelve hours after infection, doxycycline was added to the medium to a final concentration of 10 μg/ml. Twelve hours after induction (24 h after infection), cells were fixed with 4% (vol/vol) paraformaldehyde in phosphate-buffered saline (Gibco-Invitrogen Corp.), permeabilized with 0.5% (vol/vol) Triton X-100 in the same buffer, and blocked as described by Ornelles and Shenk (66). FLAG-Nxf1 was detected using rabbit anti-FLAG polyclonal antibody (Sigma-Aldrich) and Cy5-conjugated anti-rabbit donkey IgG (Jackson ImmunoResearch Labs). The viral E1B 55-kDa protein and protein V were visualized with mouse monoclonal antibody 2A6 and F58#1 (60), respectively, and fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG. Cells were stained with DAPI (4′,6′-diamidino-2-phenylindole) (Jackson ImmunoResearch Labs) and mounted, and single sections were visualized using a Zeiss LSM 510 confocal microscope with a C-Apochromat 40× 1.2-numerical-aperture (NA) water immersion objective, as described previously (41).
The endogenous Nxf1 protein was visualized in Ad5-infected HeLa cells processed for immunofluorescence as described above by using mouse monoclonal anti-Nxf1 antibody clone 31 IgG (BD Biosciences) and Cy5-labeled anti-mouse IgG (Jackson ImmunoResearch Labs). Subsequently, cells were incubated with 2A6 IgG, which had been conjugated to Alexa Fluor 488 (Molecular Probes) as described previously (41).
RESULTS
Colocalization of the Nxf1/Tap export receptor with the E1B 55-kDa protein in Ad5-infected cells.
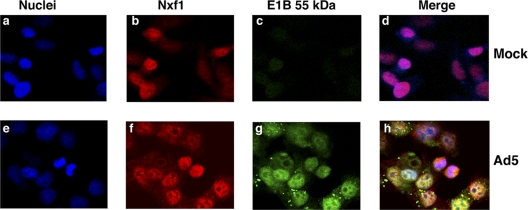
No components of the Nxf1 mRNA export pathway were recovered in experiments to identify E1B 55-kDa-interacting protein by immunoprecipitation and mass spectrometry (46, 71). However, screening of a HeLa cDNA phage display expression library for proteins that bound specifically to a GST-E1B55-kDa fusion protein identified a member of the RNP family termed E1B-AP5 (38). Subsequently, the E1B-AP5 protein, which exhibits RNA-binding activity in vitro (38), was reported to interact with the N-terminal domain of Nxf1 (4). These properties suggested that E1B-Ap5 might mediate an interaction between the E1B 55-kDa protein and Nxf1 in Ad5-infected cells. To begin to assess this possibility, HeLa cells were infected with 10 PFU/cell Ad5 and the localization of these viral and cellular proteins examined during the late phase of infection by immunofluorescence. The Nxf1 protein was visualized by using a mouse monoclonal anti-Nxf1 antibody and Cy5-conjugated anti-mouse IgG (see Materials and Methods). Subsequently the cells were incubated with anti-E1B 55-kDa monoclonal antibody 2A6 IgG, which had been labeled with Alexa Fluor 488. In uninfected cells, Nxf1 was observed to be located largely in the nucleus (Fig. 1 b and d). Infection with Ad5 (for 18 h in the example shown in Fig. 1) did not reduce the concentration of Nxf1. However, the protein was observed to be recruited to foci and ring-like structures (Fig. 1f), where it was extensively colocalized with the E1B 55-kDa protein (Fig. 1g). Such structures closely resemble those formed by accumulation of the E1B 55-kDa protein in the peripheral zones of viral replication centers (41, 66).
FIG. 1.
Colocalization of the E1B 55-kDa protein and Nxf1 in Ad5-infected cells. HeLa cells were mock infected (a to d) or infected with 10 PFU/cell Ad5 for 18 h (e to h) and the E1B 55-kDa and Nxf1 proteins examined by immunofluorescence, as described in Materials and Methods.
Nxf1-dependent export can be inhibited by RNAi.
The colocalization of the Nxf1 and E1B 55-kDa proteins during the late phase of Ad5 infection suggested that Nxf1 might serve as the export receptor for viral late mRNAs. We therefore wished to investigate whether inhibition of Nxf1 synthesis by RNA interference (RNAi) affected export of viral late mRNAs. However, the majority of processed eukaryotic mRNAs appear to be exported from the nucleus to the cytoplasm via the Nxf1 export receptor (see the introduction), suggesting that this protein is essential. It was therefore necessary to determine whether the concentration of Nxf1 and mRNA export could be reduced effectively by RNAi under conditions in which cells remained viable. To address these issues, we examined the effects of such RNAi on the intracellular concentrations and function of Nxf1.
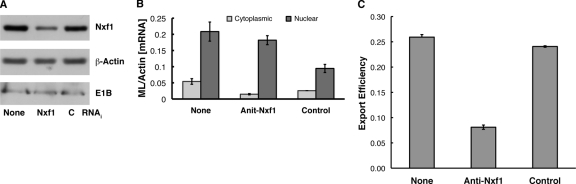
Short, double-stranded RNAs specific for Nxf1 mRNA (anti-Nxf1 RNAi) or a nonspecific control (control RNAi) were introduced into HeLa cells as described in Materials and Methods, and the concentration of Nxf1 was examined by immunoblotting at various times thereafter. The total steady-state concentration of this protein was reduced significantly by 24 and 48 h after addition of anti-Nxf1 RNAi (compare anti-Nxf1 and control samples in Fig. 2 A). Comparison of the Nxf1 signals observed in cells treated with the two interfering RNAs indicated that the concentration of Nxf1 was decreased by 76% by 24 h after addition of the anti-Nxf1 RNAi and further reduced by 48 h, with no corresponding changes in the concentrations of β-actin. However, at 48 h and later times after introduction of anti-Nxf1 RNAi, a significant number of cells became detached from the substratum and did not appear to be viable. To investigate whether the degree of inhibition of synthesis of Nxf1 achieved before this time was sufficient to impair export, we exploited a reporter assay developed by Kang and Cullen (54), in which efficient synthesis of chloramphenicol acetyltransferase (CAT) depends on export from the nucleus of an intronless reporter mRNA that contains the MMPV constitutive transport element (CTE). Nxf1 binds specifically to this RNA sequence to direct export of RNAs containing it from the nucleus to the cytoplasm (11, 43, 54). Plasmids carrying the CTE-containing CAT reporter (CTE+ CAT) or a control reporter gene lacking the CTE (CTE− CAT) were introduced in HeLa cells 24 h prior to addition of anti-Nxf1 or control RNAi, as described in Materials and Methods. Cytoplasmic extracts were isolated from cells harvested at various times after addition of interfering RNAs, and CAT activity was measured as described in Materials and Methods. The results of one such experiment, in which cells were harvested 24 h after addition of interfering RNA, are shown in Fig. 2B. As expected (54), much more CAT was made in cells that received the CTE+ CAT reporter than in those receiving the CTE− CAT reporter and the control RNAi, as was also observed in cells that were not exposed to any interfering RNA (data not shown). Addition of anti-Nxf1 RNAi led to a greater-than-3-fold decrease in CAT activity in CTE+ CAT-containing cells but did not inhibit synthesis of CAT from the gene lacking the CTE (Fig. 2B). These data indicate that an RNAi-induced reduction of the Nxf1 concentration in HeLa cells of some 4-fold results in specific and significant inhibition of Nxf1-dependent export of mRNA from the nucleus.
FIG. 2.
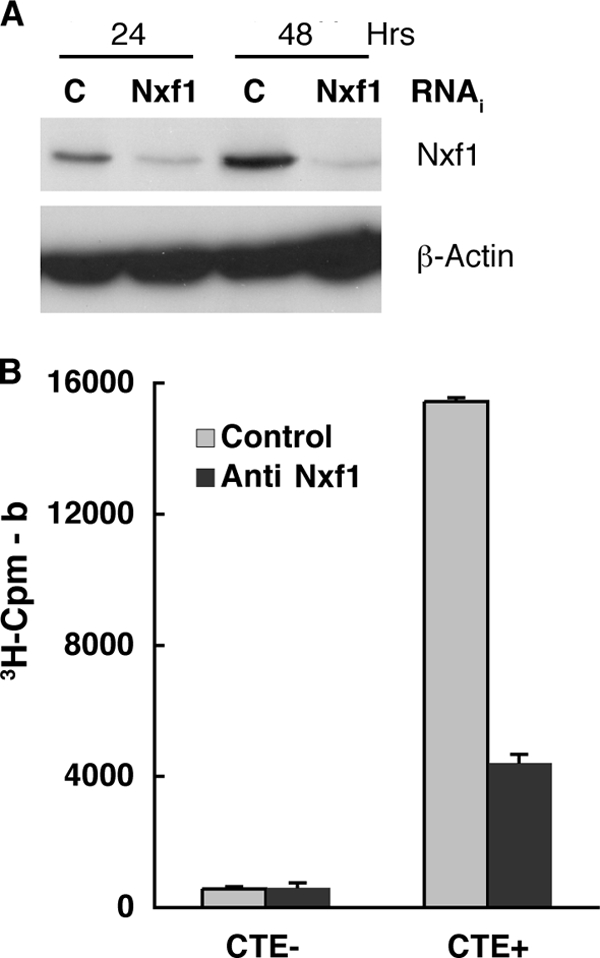
Inhibition of Nxf1 production and function by RNAi. (A) Non targeting control (C) or anti-Nxf1 (Nxf1) interfering RNAs were introduced into HeLa cells as described in Materials and Methods. After the periods indicated, cells were harvested and the concentrations of Nxf1 and β-actin in total cell extracts examined by immunoblotting. (B) Plasmids carrying a CAT reporter gene lacking (CTE−) or containing (CTE+) the MPMV constitutive transport element and control or anti-Nxf1 interfering RNAs were introduced into HeLa cells as described in Materials and Methods. At 24 h after introduction of interfering RNAs, CAT activity was assayed in cytoplasmic extracts by measurement of synthesis of tritiated n-butyryl chloramphenicol. All samples were assayed in duplicate, and the cpm values corrected for background (b) in reactions that contained no cytoplasmic cell extracts. Error bars indicate average deviations.
Inhibition of Nxf1 synthesis results in impaired export of viral late mRNAs.
To investigate the role of the Nxf1 pathway in export of adenoviral late mRNAs, Nxf1-dependent mRNA export was inhibited by RNAi in Ad5-infected cells. An important consideration in the design of these experiments was to minimize effects of inhibition of Nxf1-dependent export on entry into the late phase of infection or synthesis of the viral early proteins required for regulation of mRNA export (see the introduction). Addition of interfering RNAs and Ad5 infection were therefore carried out sequentially, with a interval designed to allow the infection to enter the late phase before production of the interfering RNAs at a concentration sufficient to impair synthesis of Nxf1. The concentrations of the control or anti-Nxf1 interfering RNAs used in the experiments described in the previous section were introduced into HeLa cells 12 h prior to Ad5 infection. Cells were harvested 12 to 14 h after infection, and one sample was processed for immunoblotting and a second separated into nuclear and cytoplasmic fractions, from which RNA was purified as described in Materials and Methods. Typical results of these experiments are shown in Fig. 3.
FIG. 3.
Anti-Nxf1 RNAi reduces the efficiency of export of viral ML mRNA. HeLa cells were sequentially transduced with anti-Nxf1 (Nxf1) or control (C) interfering RNAs, or were not treated (None), and infected with Ad5 as described in Materials and Methods. At 12 h postinfection (p.i.), cells were harvested and divided into two samples. (A) Total protein extracts were prepared from one set of samples and the concentrations of Nxf1, β-actin, and the E1B 55-kDa protein examined by immunoblotting. (B) Nuclear and cytoplasmic fractions were isolated from the second set of samples and RNA purified from each fraction. The concentrations of ML fiber and β-actin mRNAs were measured by quantitative RT-PCR as described in Materials and Methods. The values measured for ML fiber mRNA were expressed relative to those for internal control β-actin mRNA. (C) The results shown in panel B were used to calculate export efficiency, i.e., the ratio of cytoplasmic to nuclear concentrations of ML fiber mRNA. Error bars indicate average deviations.
To monitor the efficacy of inhibition of Nxf1 production, the total steady-state concentrations of the protein were compared by immunoblotting in cells that received the anti-Nxf1 or control interfering RNAs or that were not treated prior to infection. A significantly lower concentration of Nxf1 was observed in cells containing the anti-Nxf1 RNAi than in cells that received the control or that were not exposed to any interfering RNA (Fig. 3A). Despite the reduction in Nxf1 concentration, no significant decrease in the steady-state concentration of the E1B 55-kDa protein was detected in infected cells exposed to anti-Nxf1 RNAi (Fig. 3A). The efficiency of export of viral late mRNA was examined by measurement of the ratios of the nuclear to cytoplasmic concentrations of processed ML fiber mRNA (41). Quantitative RT-PCR was used to measure the concentrations of this mRNA in nuclear and cytoplasmic fractions isolated from the same number of cells, relative to those of cellular β-actin mRNA as an internal control: as β-actin mRNA is abundant and stable in HeLa cells, the inhibition of export of newly synthesized actin mRNA when Ad5-infected cells enter the late phase of infection has no detectable impact on the steady-state concentration (3, 58, 67, 87, 89). Exposure of cells to anti-Nxf1 RNAi reduced the concentration of cytoplasmic ML mRNA but not the nuclear concentration (Fig. 3B), and the efficiency of export of the viral late mRNA was reduced by some 3-fold when the anti-Nxf1 RNAi was introduced prior to Ad5 infection (Fig. 3C). In contrast, this parameter was very similar in infected cells that were not exposed to interfering RNA or that received the control RNAi (Fig. 3C). Under the conditions used in these experiments to avoid both loss of viability of cells exposed to anti-Nxf1 short hairpin RNA (shRNA) and inhibition of expression of viral early genes required for selective export of viral late mRNAs, the production of Nxf1 was reduced but not eliminated (Fig. 2A and 3A). This property seems likely to account for the incomplete inhibition of viral late mRNA export observed.
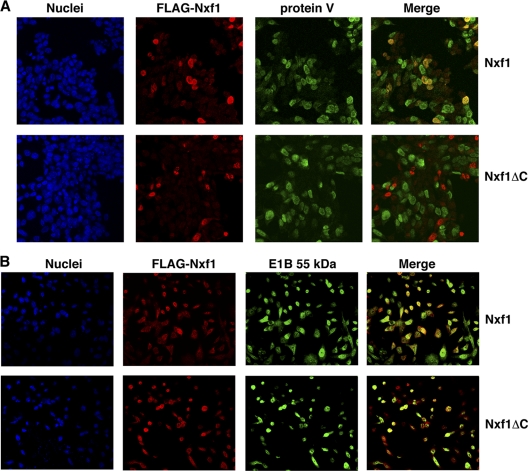
In an alternative approach to inhibit Nxf1-dependent mRNA export, we exploited a dominant-negative derivative of human Nxf1 that lacks residues 518 to 619 (19). This C-terminal domain mediates interaction of the export receptor with nucleoporins and is essential for Nxf1-dependent export (4, 10, 19, 59). The coding sequences for this truncated Nxf1 and the full-length protein fused to that for an N-terminal FLAG tag were placed under the control of the Tet-ON promoter as described in Materials and Methods, so that synthesis of these protein could be induced specifically during the late phase of the Ad5 infectious cycle. The expression plasmids were introduced as described in Materials and Methods into HeLa Tet-ON cells, which were infected with Ad5 24 h thereafter. Expression of the FLAG-Nxf1-coding sequences was induced by addition of doxycycline to the medium 10 to 12 h later, during the initial period of the late phase of infection. Because the efficiency with which plasmid DNAs entered the cells was much less than 100%, it was not possible to examine the effects of synthesis of the FLAG-Nxf1 proteins on viral late mRNA export. Rather, this parameter was monitored indirectly, by taking advantage of the large decreases in synthesis of viral late proteins observed when late mRNAs are not exported selectively (42, 46, 67, 87). The accumulation of protein V, a late structural protein (6), as well as of the exogenous, FLAG-tagged Nxf1 proteins, was therefore examined by immunofluorescence, as described in Materials and Methods. As illustrated in Fig. 4 A, the majority of cells in which full-length FLAG-Nxf1 was made also contained protein V. In contrast, fewer cells that produced the dominant-negative FLAG-Nxf1 protein also contained high concentrations of protein V. In several experiments of this kind, we consistently observed that protein V was synthesized in a significantly smaller percentage of the cells that contained dominant-negative FLAG-Nxf1 than in the cells in which full-length FLAG-Nxf1 was made (Table 1) The reduction in viral late gene expression induced by this dominant-negative derivative of Nxf1 was not an indirect result of effects on synthesis of the E1B 55-kDa protein, as E1B 55-kDa protein staining was observed in the majority of cells that produced full-length or dominant-negative FLAG-Nxf1 (Fig. 4B).
FIG. 4.
Production of dominant-negative Nxf1 in Ad5-infeced cells inhibits synthesis of viral protein V. Plasmids directing synthesis of FLAG-Nxf1 or a C-terminal domain-deleted dominant-negative derivative, Nxf1ΔC, were introduced into HeLa Tet-ON cells as described in Materials and Methods. Cells were infected with 10 PFU/cell Ad5 24 h later, and expression of the exogenous Nxf1 coding sequences was induced 12 h thereafter by addition of doxycycline to the medium. Cells were processed for immunofluorescence 12 h after addition of the drug and examined by confocal microscopy, as described in Materials and Methods. FLAG-Nxf1 proteins were detected by indirect staining with Cy5 (red), and viral protein V (A) and the E1B 55-kDa protein (panel B) were detected by indirect staining with FITC (green). To facilitate visualization of costaining, the images of DAPI stained nuclei were omitted from the merged images.
TABLE 1.
Dominant-negative Nxf1 inhibits production of protein V in Ad5-infected cells
| Expt | Exogenous FLAG-Nxf1a | No. of cellsb |
% FLAG-Nxf1-producing cells that synthesize V | |
|---|---|---|---|---|
| FLAG positive | FLAG and V positive | |||
| 1 | WT | 44 | 40 | 90.9 |
| DN | 44 | 3 | 6.8 | |
| 2 | WT | 86 | 56 | 65.1 |
| DN | 44 | 8 | 18.1 | |
| 3 | WT | 136 | 67 | 49.3 |
| DN | 86 | 8 | 9.3 | |
WT, wild type; DN, dominant negative.
The numbers of cells exhibiting nuclear FLAG-Nxf1 signals in individual fields such as those shown in Fig. 4A (experiment 1) and in two additional experiments were counted by using the cell-counting function of Image J (73). This same function was then used to determine the numbers of such cells that also exhibited nuclear protein V staining.
Effects of Ad5 infection on components of the Nxf1 export pathway.
The results described in the previous section indicate that the Nxf1 export receptor is responsible for transport of viral late mRNAs from the nucleus to the cytoplasm. This finding suggested that one or more components of the Nxf1 export pathway might be substrates of the infected-cell-specific E3 ubiquitin ligase, which is required for efficient export of these viral mRNAs (7, 88). Ubiquitinylation of all known substrates of this enzyme targets the modified protein for degradation by the proteasome, and it has been suggested that degradation of a cellular protein necessary for export of cellular mRNAs, but dispensable for viral mRNA export, might result in selective export of viral mRNAs (88). It is also possible that this characteristic feature of the late phase of infection is the result of the regulation of the activity of a cellular export protein, or proteins, as a result of ubiquitinylation by the Ad E3 ligase (see Discussion). We therefore compared the properties of Nxf1 and other proteins that participate in Nxf1-dependent mRNA export in infected cells in which the E1B 55-kDa and E4 Orf6 protein-containing E3 ubiquitin ligase was present or absent.
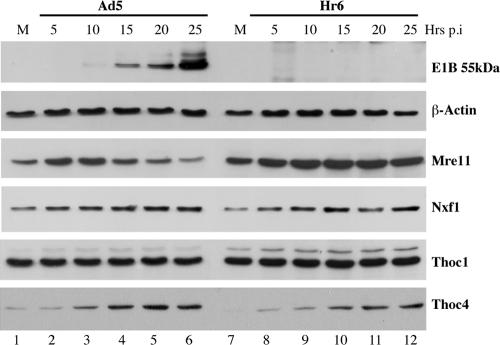
HeLa cells were harvested at regular intervals after infection with Ad5 or the E1B 55-kDa-null mutant Hr6, and the total steady-state concentrations of proteins of interest were examined by immunoblotting. The changes in concentration of a bona fide substrate of the infected-cell-specific E3 ubiquitin ligase, the Mre11 protein (16, 85), were first compared to those of the E1B 55-kDa protein and of β-actin as an internal control. This E1B protein was first detected at 10 h after Ad5 infection and accumulated to a much higher concentration by 25 h, but, as expected, it was not present in Hr6-infected cells (Fig. 5). The concentration of Mre11 initially increased following Ad5 infection but declined continuously from 10 h after infection as the E1B 55-kDa protein accumulated, in agreement with previous studies (25, 82). Also as expected, no such reduction in Mre11 concentration was observed in the absence of the E1B 55-kDa protein in Hr6-infected cells (Fig. 5). Having demonstrated E1B 55-kDa protein-dependent turnover of Mre11, we examined the concentrations of Nxf1 in Ad5- and Hr6-infected cells. Consistent with the immunofluorescence results described above (Fig. 1), no decrease in Nxf1 concentration was observed in Ad5-infected cells (Fig. 5). Rather, this protein appeared to accumulate slightly as the infectious cycle progressed, as it did in Hr6-infected cells (Fig. 5).
FIG. 5.
Comparison of the concentrations of Nxf1 pathway components in the presence and absence of the E1B 55-kDa protein. HeLa cells were mock infected (M) or infected with 20 PFU/cell Ad5 or Hr6 for the periods indicated. Total protein extracts were prepared and the proteins listed at the right examined by immunoblotting.
We next assessed the effects of infection on the Nxf1 pathway component Thoc1 (hHpr1). This protein, which is a component of the TREX complex (56, 74), was chosen because in S. cerevisiae, Hpr1 is polyubiquitinylated by the E3 ubiquitin ligase Rsp5 and thus targeted for proteasomal degradation (44). Both stabilization of Hpr1 by inactivation of Rsp5 and deletion of the HPR1 gene result in defects in mRNA export (45, 64, 78), indicating that precise control of Hpr1 concentration is necessary for mRNA export. These properties suggested that targeting for proteasomal degradation of the human homologue Thoc1 by the Ad5-infected-cell-specific E3 ligase might contribute to regulation of mRNA export during the late phase of infection. However, no change in the concentration of Thoc1 was observed during the late phase of Ad5 infection (Fig. 5), when the E1B 55-kDa protein accumulated and Mre11 was degraded, nor was this protein stabilized in Hr6-infected cells (Fig. 5).
A second component of the metazoan TREX complex, Thoc4 (Aly/Ref), has been shown to be limiting for the export of mRNA from the nucleus in Xenopus oocytes (62, 77, 91). Furthermore, inhibition of expression of the single human THOC4 gene in HeLa cells impaired export of poly(A)-containing mRNA (55, 65), indicating that Thoc4 is necessary for mRNA export in human cells. These properties suggested that targeting of Thoc4 for proteasomal degradation by the infected-cell-specific E3 ubiquitin ligase could account for inhibition of cellular mRNA export in Ad5-infected cells. We therefore compared the effects of Ad5 and Hr6 infection on the concentration of this protein. Thoc4 was observed to be present at only low concentrations in mock-infected cells (Fig. 5, lanes 1 and 7) but accumulated to substantially higher concentrations from 10 h after either Ad5 or Hr6 infection (Fig. 5).
DISCUSSION
Two receptors for export of mRNA from the nucleus to the cytoplasm in metazoan cells have been indentified, Nxf1 (Tap) and exportin 1 (Crml) (see references 15, 31, 56, and 74 for reviews). The former mediates export of the majority of processed cellular mRNAs via interactions with both F/G repeat-containing nucleoporins and various adaptor proteins that bind to mRNA. A subset of the short-lived cellular mRNAs characterized by the presence of AU-rich sequences in their 3′ untranslated regions have been reported to leave the nucleus by the exportin 1 pathway (39). Previous studies have demonstrated that inhibition of exportin 1 does not impair synthesis of viral late protein (2, 72) or of export of viral late mRNAs (35), despite the exportin 1-dependent shuttling of the E1B protein between the nucleus and cytoplasm (27, 29, 57). Rather, data reported here implicate Nxf1 in export of these viral mRNAs from the nucleus to the cytoplasm: synthesis in Ad5-infected cells of an interfering RNA that reduced the concentration of Nxf1, but not of a control nontargeting interfering RNA, results in decreased efficiency of export of viral late mRNA (Fig. 3). Furthermore, synthesis of a dominant derivative of Nxf1 during the late phase of infection impaired viral late protein synthesis (Fig. 4; Table 1).
Perhaps the most enigmatic aspect of selective export of viral late mRNAs, which possess all the hallmarks of cellular mRNAs and are made by cellular transcription and processing systems (6), is the mechanism(s) that discriminates viral from cellular mRNAs. Early studies established that the E4 Orf6 protein is required for localization of the E1B 55-kDa protein to the peripheral zones of replication centers within infected-cell nuclei (66). Such colocalization is necessary for selective export of viral late mRNAs: a 4-amino-acid insertion in the E1B 55-kDa protein that impairs interaction with the E4 Orf6 protein (79) prevents localization of the E1B protein to these intranuclear sites and substantially reduces the efficiency of viral late mRNA export (41). The peripheral zones of viral replication centers are the sites of transcription and initial processing of viral late pre-mRNAs (2, 14, 68-70). It was therefore proposed that recruitment of a cellular protein necessary for mRNA export to such sites by association with the E1B 55-kDa-E4 Orf6 protein complex could prevent transport of cellular mRNA to the cytoplasm, while promoting viral late mRNA export (66). As efficient viral late mRNA export depends on the assembly of the Ad E3 ubiquitin ligase (7, 88), it has been suggested that ubiquitinylation and subsequent degradation of a cellular protein necessary for export of cellular, but not of viral, mRNAs might stimulate viral late mRNA export (88). One possibility is that degradation of such a cellular protein reduces competition between viral and cellular mRNAs for one or more proteins that mediate mRNA export and facilitates recruitment of such a protein to the peripheral zones of viral replication centers. In fact, we observed that the export receptor Nxf1 colocalizes with the E1B 55-kDa protein at these sites during the late phase of infection (Fig. 1), consistent with the initial model summarized above. It will be of particular interest to determine whether such colocalization of the E1B protein and Nxf1 depends on the Ad E3 ubiquitin ligase, as well as the mechanism by which Nxf1 is recruited to these sites. It seems unlikely that Nxf1 binds directly to the E1B 55-kDa and/or the E4 Orf6 protein, as this protein was not identified when proteins associated with the E1B 55-kDa-E4 Orf6 protein complex were analyzed by mass spectrometry (46, 71). The hnRNP protein E1B-AP5 was identified by virtue of its binding to the E1B 55-kDa protein (38) and subsequently was reported to interact with Nxf1 in vitro (4). Whether this interaction contributes to regulation of mRNA export in Ad5-infected cells is not yet clear: the effects of a number of amino acid insertions in the E1B 55-kDa protein on both binding to E1B-AP5 (38) and viral late mRNA export (41) have been examined, but all insertions that both impaired binding to E1B-AP5 and led to reduced efficiency of viral late mRNA export also destabilized and/or resulted in mislocalization of the E1B protein (38, 41, 90).
The conclusion that the Nxf1 export receptor is responsible for the export of viral late mRNAs raises the obvious question of which components of this export pathway are exploited or targeted by viral gene products to allow selective export of viral late mRNAs. The F/G repeat nucleoporins with which Nxf1 must interact during transport of mRNA from the nucleus via nuclear pore complexes (4, 53) can be excluded: these proteins mediate export of all classes of RNA, and previous studies have demonstrated that export of cellular rRNAs, tRNAs and other classes of small RNAs is not perturbed in Ad5-infected cells (18, 84). The simplest interpretation of the stimulation of viral late mRNA export by the Ad E3 ligase (7, 88) and the results reported here is that ubiquitinylation of one or more proteins that participate in Nxf1-dependent export is necessary for selective export of viral late mRNAs. Clear precedent for the view that this modification plays an important role in mRNA export has been provided by studies in yeast. As described previously (see Results), regulation of the concentration of S. cerevisiae Hrp1, which requires ubiquitinylation of the protein by the E3 ubiquitin ligase Rsp5p, is crucial for mRNA export (44, 45, 64). In addition, the S. cerevisiae Tom1p E3 ligase and its fission yeast homologue Ptr1-1p are required for export of poly(A)-containing RNA from the nucleus (1, 30). One substrate of this enzyme has recently been identified as the export adaptor Yral/Ref (52). We therefore sought to identify Nxf1 pathway components that, like all known substrates of the infected-cell-specific E3 ubiquitin ligase (5, 25, 61, 71), are targeted for degradation. However, none of the proteins examined, i.e., Nxf1, Thoc1, and Thoc4, exhibited E1B protein-dependent decreases in concentration as the infectious cycle progressed (Fig. 5). Indeed, we unexpectedly observed E1B 55-kDa protein-independent increases in the concentrations of Nxf1 and Thoc4 as the infectious cycle progressed (Fig. 5). The effect of infection on Thoc4 was particularly striking. Quantification of data such as those shown in Fig. 5 indicated that this protein increased in concentration relative to that of the β-actin control by more than 10-fold. Furthermore, it continued to accumulate during the late phase of infection (Fig. 5), when translation of the great majority of cellular mRNA is inhibited (6). The mechanism(s) by which one or more viral gene products induce increased expression of the Thoc4 gene remains to be determined. Nevertheless, it is tempting to speculate that this increase in the concentration of Thoc4, which may be limiting for mRNA export (62, 77, 91), facilitates transport of the large quantities of viral mRNA made during the late phase of infection to the cytoplasm.
A substantial number of other proteins have been implicated in Nxf1-dependent export of mRNA in metazoan cells. These proteins include the several additional components of the TREX complex, the spliceosome component Uap56, certain serine-arginine-rich splicing proteins (SR proteins) (reviewed in references 15, 31, 56, and 74), and the cap-binding protein Cpb80, which recruits the TREX complex to the 5′ ends of human mRNAs (21). Additional proteins, such as the ATP-dependent helicase Dbp5, Gle1 and Gle2, and some hnRNP proteins, are also important in this process (15, 51, 56). Furthermore, as has been suggested previously (88), inhibition of mRNA export upon induction of an Ad E3 ubiquitin ligase-dependent antiviral response might favor export of viral late mRNAs, which are produced in very large quantities. These considerations emphasize the likely value of more global approaches to identify the substrate(s) of the Ad E3 ligase that is responsible for regulation of mRNA export. In fact, the results of a proteomic approach to identify substrates of this ligase by exploiting two-dimensional (2D) differential imaging gel electrophoresis to detect protein with decreased concentration in cells that contain the ligase have been reported recently (24, 25). The studies clearly established integrin α3 as a substrate and identified several other potential substrates, but none that was reported to participate in Nxf1-dependent mRNA export or in nuclear export more generally (24, 25). Technical limitations of the methods employed in these experiments, such as failure to separate proteins closely similar in mass and pI during 2D electrophoresis and, importantly, screening of only proteins present at relatively high concentrations (24), may well have precluded detection of such proteins.
Addition of polyubiquitin chains to lysine residues in proteins has long been known to target proteins for degradation by the proteasome (reviewed in references 47 and 49). More recently, ubiquitinylation, as well as addition of ubiquitin-related proteins such as Sumo-1, has been recognized as an important device for regulation of the activity of many proteins and cellular processes (20, 26, 48, 81). Assembly of the E1B 55-kDa and E4 Orf6 protein-containing E3 ubiquitin ligase is necessary for efficient export of viral late mRNAs (7, 88), but whether all (or any) relevant substrates are targeted for proteasomal degradation is not clear. It has been reported that exposure of infected cells to proteasome inhibitors during the early phase of infection impaired expression of viral genes (22). However, viral DNA synthesis, upon which expression of late genes depends (6), was reduced and delayed under such conditions (22), to a degree apparently sufficient to account for the reduced and delayed accumulation of viral late proteins. Consequently, it is possible that regulation of mRNA export in adenovirus-infected cells is mediated, at least in part, by the ubiquitinylation-dependent modulation of the activity of one or more cellular proteins. No such substrates of the E1B 55-kDa and E4 Orf6 protein-containing E3 ubiquitin ligase have been identified, but on the other hand, all substrates identified to date were recognized by virtue of their decreased concentrations in cells in which this enzyme was present compared to those in which it was absent. Development of methods for identification of substrates of the Ad E3 ligase that do not rely on this criterion seems likely to provide a valuable complement to the approaches applied previously.
Acknowledgments
We thank Brian Cullen and Rozanne Sandri-Goldin for gifts of CTE-CAT reporter plasmids and plasmids carrying human Nxf1 coding sequences, respectively, and Ellen Brindle-Clark for assistance with preparation of the manuscript.
This work was supported by a grant (R01AI058172) to S.J.F. from the National Institute of Allergy and Infectious Disease, National Institutes of Health.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1.Andoh, T., A. K. Azad, A. Shigematsu, Y. Ohshima, and T. Tani. 2004. The fission yeast ptr1+ gene involved in nuclear mRNA export encodes a putative ubiquitin ligase. Biochem. Biophys. Res. Commun. 317:1138-1143. [DOI] [PubMed] [Google Scholar]
- 2.Aspegren, A., C. Rabino, and E. Bridge. 1998. Organization of splicing factors in adenovirus-infected cells reflects changes in gene expression during the early to late phase transition. Exp. Cell Res. 245:203-213. [DOI] [PubMed] [Google Scholar]
- 3.Babich, A., L. T. Feldman, J. R. Nevins, J. E. Darnell, Jr., and C. Weinberger. 1983. Effect of adenovirus on metabolism of specific host mRNAs: transport control and specific translational discrimination. Mol. Cell. Biol. 3:1212-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachi, A., et al. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6:136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34K and E1b 55K oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berk, A. J. 2007. Adenoviridae: the viruses and their replication, p. 2355-2394. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5 ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Blanchette, P., et al. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 82:2642-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogerd, H. P., A. Echarri, T. M. Ross, and B. R. Cullen. 1998. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol. 72:8627-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the detection of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Braun, I. C., A. Herold, M. Rode, and E. Izaurralde. 2002. Nuclear export of mRNA by TAP/NXF1 requires two nucleoporin-binding sites but not p15. Mol. Cell. Biol. 22:5405-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun, I. C., E. Rohrbach, C. Schmitt, and E. Izaurralde. 1999. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 18:1953-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 14.Bridge, E., and U. Pettersson. 1996. Nuclear organization of adenovirus RNA biogenesis. Exp. Cell Res. 229:233-239. [DOI] [PubMed] [Google Scholar]
- 15.Carmody, S. R., and S. R. Wente. 2009. mRNA nuclear export at a glance. J. Cell Sci. 122:1933-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carson, C. T., et al. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter, C. C., R. Izadpanah, and E. Bridge. 2003. Evaluating the role of CRM1-mediated export for adenovirus gene expression. Virology 315:224-233. [DOI] [PubMed] [Google Scholar]
- 18.Castiglia, C. L., and S. J. Flint. 1983. Effects of adenovirus infection on rRNA synthesis and maturation in HeLa cells. Mol. Cell. Biol. 3:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, Z. J. 2005. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 7:758-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng, C. Y., P. Blanchette, and P. E. Branton. 2007. The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion. Virology 364:36-44. [DOI] [PubMed] [Google Scholar]
- 22.Corbin-Lickfett, K. A., and E. Bridge. 2003. Adenovirus E4-34kDa requires active proteasomes to promote late gene expression. Virology 315:234-244. [DOI] [PubMed] [Google Scholar]
- 23.Cutt, J. R., T. Shenk, and P. Hearing. 1987. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J. Virol. 61:543-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dallaire, F., P. Blanchette, and P. E. Branton. 2009. A proteomic approach to identify candidate substrates of human adenovirus E4orf6-E1B55K and other viral cullin-based E3 ubiquitin ligases. J. Virol. 83:12172-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dallaire, F., P. Blanchette, P. Groitl, T. Dobner, and P. E. Branton. 2009. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J. Virol. 83:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Fiore, P. P., S. Polo, and K. Hofmann. 2003. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat. Rev. Mol. Cell Biol. 4:491-497. [DOI] [PubMed] [Google Scholar]
- 27.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobner, T., and J. Kzhyshkowska. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 259:25-34. [DOI] [PubMed] [Google Scholar]
- 29.Dosch, T., et al. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4Orf6 is mediated by a CRM1-independent mechanism. J. Virol. 75:5677-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duncan, K., J. G. Umen, and C. Guthrie. 2000. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 10:687-696. [DOI] [PubMed] [Google Scholar]
- 31.Erkmann, J. A., and U. Kutay. 2004. Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp. Cell Res. 296:12-20. [DOI] [PubMed] [Google Scholar]
- 32.Fischer, A., J. Huber, W. C. Boulens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 33.Flint, S. J., P. H. Gallimore, and P. A. Sharp. 1975. Comparison of viral RNA sequences in adenovirus 2-transformed and lytically infected cells. J. Mol. Biol. 96:47-68. [DOI] [PubMed] [Google Scholar]
- 34.Flint, S. J., and R. A. Gonzalez. 2003. Regulation of mRNA production by the adenoviral E1B 55kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272:287-330. [DOI] [PubMed] [Google Scholar]
- 35.Flint, S. J., W. Huang, J. Goodhouse, and S. Kyin. 2005. A peptide inhibitor of exportin1 blocks shuttling of the adenoviral E1B 55 kDa protein but not export of viral late mRNAs. Virology 337:7-17. [DOI] [PubMed] [Google Scholar]
- 36.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda, M., et al. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 38.Gabler, S., et al. 1998. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 72:7960-7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallouzi, I. E., and J. A. Steitz. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294:1895-1901. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez, R., W. Huang, R. Finnen, C. Bragg, and S. J. Flint. 2006. Adenovirus E1B 55-kilodalton protein is required for both regulation of mRNA export and efficient entry into the late phase of infection in normal human fibroblasts. J. Virol. 80:964-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez, R. A., and S. J. Flint. 2002. Effects of mutations in the adenoviral E1B 55-kDa protein coding sequence on viral late mRNA metabolism. J. Virol. 76:4507-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodrum, F. D., and D. A. Ornelles. 1997. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J. Virol. 71:548-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grüter, P., et al. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1:649-659. [DOI] [PubMed] [Google Scholar]
- 44.Gwizdek, C., et al. 2005. The mRNA nuclear export factor Hpr1 is regulated by Rsp5-mediated ubiquitylation. J. Biol. Chem. 280:13401-13405. [DOI] [PubMed] [Google Scholar]
- 45.Gwizdek, C., et al. 2006. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc. Natl. Acad. Sci. U. S. A. 103:16376-16381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 48.Hicke, L. 2001. A new ticket for entry into budding vesicles-ubiquitin. Cell 106:527-530. [DOI] [PubMed] [Google Scholar]
- 49.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 50.Huang, Y., and J. A. Steitz. 2005. SRprises along a messenger's journey. Mol. Cell 17:613-615. [DOI] [PubMed] [Google Scholar]
- 51.Iglesias, N., and F. Stutz. 2008. Regulation of mRNP dynamics along the export pathway. FEBS Lett. 582:1987-1996. [DOI] [PubMed] [Google Scholar]
- 52.Iglesias, N., et al. 2010. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 24:1927-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang, Y., H. P. Bogerd, and B. R. Cullen. 2000. Analysis of cellular factors that mediate nuclear export of RNAs bearing the Mason-Pfizer monkey virus constitutive transport element. J. Virol. 74:5863-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang, Y., and B. R. Cullen. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 13:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katahira, J., H. Inoue, E. Hurt, and Y. Yoneda. 2009. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J. 28:556-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kohler, A., and E. Hurt. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761-773. [DOI] [PubMed] [Google Scholar]
- 57.Kratzer, F., et al. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19:850-857. [DOI] [PubMed] [Google Scholar]
- 58.Leppard, K. N., and T. Shenk. 1989. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 8:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levesque, L., et al. 2001. RNA export mediated by tap involves NXT1-dependent interactions with the nuclear pore complex. J. Biol. Chem. 276:44953-44962. [DOI] [PubMed] [Google Scholar]
- 60.Lunt, R., M. E. Vayda, M. Young, and S. J. Flint. 1988. Isolation and characterization of monoclonal antibodies against the adenovirus core proteins. Virology 164:275-279. [DOI] [PubMed] [Google Scholar]
- 61.Luo, K., et al. 2007. Adenovirus E4orf6 assembles with Cullin5-ElonginB-ElonginC E3 ubiquitin ligase through an HIV/SIV Vif-like BC-box to regulate p53. FASEB J. 21:1742-1750. [DOI] [PubMed] [Google Scholar]
- 62.Luo, M. L., et al. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413:644-647. [DOI] [PubMed] [Google Scholar]
- 63.Malim, M. H., D. F. McCarn, L. S. Tiley, and B. R. Cullen. 1991. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J. Virol. 65:4248-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neumann, S., et al. 2003. Formation and nuclear export of tRNA, rRNA and mRNA is regulated by the ubiquitin ligase Rsp5p. EMBO Rep. 4:1156-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okada, M., S. W. Jang, and K. Ye. 2008. Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc. Natl. Acad. Sci. U. S. A. 105:8649-8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ornelles, D., and T. Shenk. 1991. Location of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55kd transforming polypeptide modulates transport or cytoplasmic stablization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pombo, A., J. Ferreira, E. Bridge, and M. Carmo-Fonseca. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13:5075-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Puvion-Dutilleul, F., J. P. Bachellerie, N. Visa, and E. Puvion. 1994. Rearrangements of intranuclear structures involved in RNA processing in response to adenovirus infection. J. Cell Sci. 107:1457-1468. [DOI] [PubMed] [Google Scholar]
- 70.Puvion-Dutilleul, F., and E. Puvion. 1991. Sites of transcription of adenovirus type 5 genomes in relation to early viral DNA replication in infected HeLa cells. A high resolution in situ hybridization and autoradiographical study. Biol. Cell 71:135-147. [DOI] [PubMed] [Google Scholar]
- 71.Querido, E., et al. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rabino, C., A. Aspegren, K. Corbin-Lickfett, and E. Bridge. 2000. Adenovirus late gene expression does not require a Rev-like nuclear RNA export pathway. J. Virol. 74:6684-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rasband, W. S. 1997-2009. ImageJ. National Institutes of Health, Bethesda, MD.
- 74.Reed, R., and H. Cheng. 2005. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 17:269-273. [DOI] [PubMed] [Google Scholar]
- 75.Reed, R., and E. Hurt. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108:523-531. [DOI] [PubMed] [Google Scholar]
- 76.Rickards, B., S. J. Flint, M. D. Cole, and G. LeRoy. 2007. Nucleolin is required for RNA polymerase I transcription in vivo. Mol. Cell. Biol. 27:937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodrigues, J. P., et al. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. U. S. A. 98:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodriguez, M. S., C. Gwizdek, R. Haguenauer-Tsapis, and C. Dargemont. 2003. The HECT ubiquitin ligase Rsp5p is required for proper nuclear export of mRNA in Saccharomyces cerevisiae. Traffic 4:566-575. [DOI] [PubMed] [Google Scholar]
- 79.Rubenwolf, S., H. Schutt, M. Nevels, H. Wolf, and T. Dobner. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 71:1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the Ad5 E1B-58K tumor antigen in adenovirus-infected and transformed cells. Virology 120:387-394. [DOI] [PubMed] [Google Scholar]
- 81.Schnell, J. D., and L. Hicke. 2003. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278:35857-35860. [DOI] [PubMed] [Google Scholar]
- 82.Schwartz, R. A., et al. 2008. Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J. Virol. 82:9043-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seed, B., and J. Y. Sheen. 1988. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene 67:271-277. [DOI] [PubMed] [Google Scholar]
- 84.Smiley, J. K., M. A. Young, C. L. Bansbach, and S. J. Flint. 1995. The metabolism of small cellular RNA species during productive subgroup C adenovirus infection. Virology 206:100-107. [DOI] [PubMed] [Google Scholar]
- 85.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 86.Stutz, F., and E. Izaurralde. 2003. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 13:319-327. [DOI] [PubMed] [Google Scholar]
- 87.Williams, J., et al. 1986. The adenovirus E1B 495R protein plays a role in regulating the transport and stability of the viral late messages. Cancer Cells. 4:275-284. [Google Scholar]
- 88.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81:575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang, U.-C., W. Huang, and S. J. Flint. 1996. mRNA export correlates with activation of transcription in human subgroup C adenovirus-infected cells. J. Virol. 70:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8:190-202. [DOI] [PubMed] [Google Scholar]
- 91.Zhou, Z., et al. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407:401-405. [DOI] [PubMed] [Google Scholar]