Abstract
The molecular basis of pathogenicity of H5N1 highly pathogenic avian influenza (HPAI) viruses in chickens remains largely unknown. H5N1 A/chicken/Yamaguchi/7/2004 virus (CkYM7) replicates rapidly in macrophages and vascular endothelial cells in chickens, causing sudden death without fever or gross lesions, while H5N1 A/duck/Yokohama/aq10/2003 virus (DkYK10) induces high fever, severe gross lesions, and a prolonged time to death, despite the 98% amino acid identity between the two viruses. To explore the molecular basis of this difference in pathogenicity, a series of eight single-gene reassortant viruses from these HPAI viruses were compared for pathogenicity in chickens. Two reassortants possessing the NP or PB2 gene from DkYK10 in the CkYM7 background reduced pathogenicity compared to other reassortants or CkYM7. Inversely, reassortants possessing the NP or PB2 gene of CkYM7 in the DkYK10 background (rgDkYK-PB2Ck, rgDkYK-NPCk) replicated quickly and reached higher titers than DkYK10, accompanied by more rapid and frequent apoptosis of macrophages. The rgDkYK-NPCk and rgDkYK-PB2Ck reassortants also replicated more rapidly in chicken embryo fibroblasts (CEFs) than did rgDkYK10, but replication of these viruses was similar to that of CkYM7 and DkYK10 in duck embryo fibroblasts. A comparison of pathogenicities of seven rgDkYK10 mutants with a single amino acid substitution in NPDk demonstrated that valine at position 105 in the NPCk was responsible for the increased pathogenicity in chickens. NPCk, NP105V, and PB2Ck enhanced the polymerase activity of DkYK10 in CEFs. These results indicate that both NP and PB2 contribute to the high pathogenicity of the H5N1 HPAI viruses in chickens, and valine at position 105 of NP may be one of the determinants for adaptation of avian influenza viruses from ducks to chickens.
Avian influenza (AI) virus belongs to the family Orthomyxoviridae and is classified into low-pathogenicity (LP) and high-pathogenicity (HP) pathotypes based on a pathogenicity test for chickens. The HPAI virus has over 75% mortality in chickens and can have devastating economic consequences, which can be controlled by a World Organization for Animal Health stamping-out policy, in the poultry industry. The HPAI viruses identified to date are only of the H5 and H7 subtypes and have multiple basic amino acid residues at the hemagglutinin (HA) cleavage site. Of the HPAI viruses, the H5N1 HPAI virus that has continued to circulate in poultry in East Asia since 1996 has been shown to be extremely virulent in chickens (47) and a serious threat to human health. The H5N1 virus has caused over 400 human infections in 15 countries and has a mortality rate of more than 50%.
Molecular mechanisms for adaptation of AI viruses from natural reservoirs to new hosts are important for understanding the evolution of influenza viruses. The binding property of hemagglutinin (HA) proteins to avian or mammalian sialic acid receptors (α2-3 or α2-6, respectively) is a first step in overcoming the interspecies barrier. The NS1 protein plays an important role in countering host cell antiviral cytokines or the initial host immune responses of chickens (22, 43). Recently, it was shown that amino acids at position 627 (8, 11) and 701 (21, 46) in polymerase subunit PB2 and 97, 349, and 550 in polymerase subunit PA (40, 45) may play important roles in the adaptation of H5N1 HPAI viruses from birds to mammals. In contrast, the molecular basis of the pathogenicity of AI viruses in chickens on the particle surface HA and neuraminidase (NA) proteins has been intensely investigated. The acquisition of polybasic amino acids at the HA cleavage site is a main determinant permitting the systemic replication of AI viruses in chickens, and the glycosylation patterns of HA molecules influence the accessibility of proteinases to the HA cleavage site (15, 35, 42). Amino acids in proximity to the HA receptor binding site affect the pathogenicity of the virus in chickens (17). The NA protein is implicated in the release of influenza virus from cells by removing sialic acid residues from the cell-derived glycoproteins (33, 47); virus particles with low NA activity cannot be released efficiently from infected cells (23, 27). A functional association between the HA and NA proteins has been suggested to directly affect pathogenicity (17, 28). Interestingly, recent studies have shown that polymerase subunits (PB1, PB2, and PA) and nucleoprotein (NP) also contribute to pathogenicity in avian species, as has been shown for mammals. The amino acids at positions 515 in PA and 436 in PB1 of H5N1 virus are associated with lethality in ducks (16), and NP, PB1, and PB2 genes (51, 52) of H5N1 virus contribute to the enhanced replication in chickens. These studies suggest that viral polymerase subunits and NP may also contribute to the adaptation or pathogenicity of AI viruses, even from ducks to chickens.
We previously identified two H5N1 HPAI viruses with different pathogenicities in chickens despite having 98% amino acid identity between the two genomes. The H5N1 A/chicken/Yamaguchi/7/2004 virus (CkYM7) replicates rapidly and quickly and kills chickens without clinical signs or gross lesions, whereas H5N1 A/duck/Yokohama/aq10/2003 virus (DkYK10) induces severe clinical signs and gross lesions in chickens, as well as a high fever, with a prolonged time to death (47). Both HPAI viruses have multiple basic amino acids at the HA cleavage site (14, 15, 48), and five amino acids surrounding the HA receptor binding site that are involved in the pathogenicity of H5N1 viruses in chickens are identical between the two strains (17). Both strains have glutamic acid at position 92 in NS1, which confers resistance to antiviral cytokines in pigs (43, 44), and alanine at position 149 of NS1, which is critical to high pathogenicity in chickens (22). The amino acid at position 627 in PB2 is glutamic acid in both strains (11), and the amino acids at position 515 in PA and 436 in PB1, which are associated with lethality in ducks, are the same in these strains (16), as is lysine at position 184 of NP, which is related to high pathogenicity in chickens (52). Therefore, these HPAI viruses are useful for deepening our knowledge of the molecular basis of the extreme virulence of H5N1 HPAI viruses in chickens.
We explored the molecular basis of the difference in pathogenicity between two H5N1 HPAI viruses in chickens by comparing the pathogenicities of several reassortant viruses prepared using a reverse genetics system. Our investigation demonstrates that NP and PB2 are associated with an increased pathogenicity of H5N1 HPAI viruses in chickens, and the valine at position 105 of the NP body domain may play critical roles in the high polymerase activity and host range restriction between ducks and chickens.
MATERIALS AND METHODS
Viruses.
Two HPAI viruses were used. CkYM7 (genotype V, clade 2-5) isolated from a chicken in the outbreak in Japan (25) has extreme virulence in chickens; it does not cause clinical signs or gross lesions, and there is a short mean death time (MDT) of 34 ± 2.2 h for animals infected with this virus. DkYK10 (clade 6) isolated from duck meat imported from China during Japanese quarantine service (24) induces severe clinical signs and gross lesions, with a long MDT of 87 ± 2.2 h (47). The amino acid sequences of the HA cleavage site of CkYM7 and DkYK10 are PQRERRKKR and PQRERRRKKR, respectively.
Generation of reassortant viruses.
CkYM7 and DkYK10 RNAs were extracted using a viral RNA purification kit (Qiagen) and transcribed into cDNA using PrimeScript reverse transcriptase (Takara) and random 6-mer primers. Each of eight full-length gene segments of CkYM7 and DkYK10 were amplified using gene-specific primer sets as previously described (12) and cloned into the pCR2.1 TOPO cloning vector (Invitrogen). After sequencing verification, the gene segments were subcloned into the pHW2000 expression vector (kindly provided by R.G. Webster, St. Jude Children's Research Hospital, Memphis, TN). To generate reassortant viruses using the reverse genetics method, MDCK and 293T cells were cocultured and transfected with 1 μg of each of the eight plasmids and 16 μl of Trans-IT-LT1 (Mirus Bio) in 200 μl of Opti-MEM (Gibco). After 48 h of incubation at 37°C, 100 μl of the supernatant was injected into the allantoic cavity of 10-day-old embryonated eggs. The allantoic fluid was harvested after 24 h of cultivation, and the viral RNA was used in sequencing confirmation to verify the gene constellation.
Comparison of pathogenicities in chickens.
In order to determine the pathogenicities of the reassortant viruses, 4-week-old specific-pathogen-free (SPF) White Leghorn chickens purchased from the Nippon Institute of Biological Science (Kobuchizawa, Yamanashi, Japan) were inoculated intranasally with one of the reassortant viruses (106.0 50% embryo infectious doses [EID50] in 0.1 ml). All of the chickens were housed in negative-pressure isolators with HEPA filters in the biosafety level 3 (BSL 3) animal facility at the NIAH in accordance with the institution's biosafety manual. The kinetics of the intranasal pathogenicity index (INPI) were determined every 12 h based on the following scores: 0, no clinical sign; 1, ruffled feathers and/or weakness; 2, other clinical signs; 3, death. An accurate time of death and the temperature of each chicken were determined using a wireless thermosensor attached to the abdominal skin surface of each chicken as previously described (47).
Virus replication kinetics in chickens.
To compare the replication speed of each reassortant virus in chickens, 4-week-old SPF chickens were inoculated intranasally with 106.0 EID50 of one of the reassortant viruses, three chickens per group at each sampling point were euthanized by ether, and the lungs, kidneys, spleens, and brains were harvested and stored at −80°C until use. For the viral titration, 0.1 g of tissue was mixed with 0.9 ml phosphate-buffered saline (PBS), and the 10% tissue homogenate was prepared with a Multi-Beads Shocker (Yasui Kikai, Osaka, Japan). After centrifugation at 10,000 rpm for 5 min at 4°C, the supernatant was used for virus titration with 10-day-old embryonated chicken eggs. The EID50/g of tissue was calculated by the Reed and Muench method (39).
Immunohistochemistry.
Lungs, spleens, brains, livers, and hearts were sequentially harvested from the same chickens used for the virus replication experiment and processed for the histopathological study and immunohistochemical staining. The collected tissues were fixed in 10% neutral buffered formalin, dehydrated, and embedded in paraffin, and tissue sections (4 μm thick) were prepared and stained with hematoxylin and eosin (HE) solution (Sigma). Immunohistochemical staining using a monoclonal antibody against the matrix (M) protein of influenza A virus was performed as described previously (49) to compare the sequential distributions of AI virus antigens in tissues.
Detection of apoptotic cells.
In order to compare the distribution of apoptotic cells in tissues, lungs and livers were harvested sequentially from the same chickens as for the virus replication experiment, and within 7 days after sacrifice, the tissue sections were stained using an ApopTag peroxidase in situ apoptosis detection kit (S7100; Chemicon International, Inc.) according to the manufacturer's protocol.
Viral growth curve kinetics in cells.
The growth kinetic curves of the reassortant viruses in chicken embryo fibroblasts (CEFs) were compared with those in duck embryonic fibroblasts (DEFs) to determine the adaptation of each reassortant virus from ducks to chickens. Briefly, 2.5 × 105 CEFs or DEFs were cultured in a culture dish and inoculated with each reassortant virus at a multiplicity of infection (MOI) of 0.001. After cultivation at 37°C in 5% CO2, culture supernatants were harvested at 0, 12, 24, 36, and 48 h postinoculation and stored at −80°C until titration. Virus titration was conducted with CEFs and determined by the cytopathic effect and hemagglutination with chicken red blood cells. The 50% tissue culture infectious dose (TCID50)/ml was calculated using the Reed and Muench method (39).
Luciferase assay.
The luciferase assay was conducted as previously described (41). The luciferase gene flanked by the noncoding region of the M segment was inserted into pHH21 (kindly provided by Yoshihiro Kawaoka, University of Wisconsin, Madison, WI) between the human RNA polymerase I promoter and mouse RNA polymerase I terminator regions to express the minus-sense gene (pHH21-lucR). Also, a chicken RNA polymerase I promoter (26) was amplified from CEF DNA and replaced with the human RNA polymerase I promoter in pHH21-lucR, and the resultant plasmid was designated pCk-polI-lucR. 293T cells or DF-1 cells in a six-well tissue culture plate (Falcon) were transfected at 60% confluence with 2 μg pHH21-lucR or pCk-polI-lucR, 1 μg pHW2000-PB2, pHW2000-PB1, and pHW2000-PA, and 2 μg pHW2000-NP using Trans-IT-LT1 (Mirus Bio). After 24 h of transfection, cell extracts were prepared in 250 μl passive lysis buffer, and the luciferase levels were assayed with a luciferase assay system (Promega) using an Arvo MX/Light (PerkinElmer, Japan) instrument. The experiment was performed using triplicate wells, and the average result was determined.
Identification of the critical amino acid of NP for high pathogenicity.
There were seven different amino acids in NP between CkYM7 and DkYK10, at positions 34, 67, 77, 105, 373, 377, and 482. To identify the critical amino acids in NP for high pathogenicity in chickens, seven amino acid mutant NPDk genes were prepared by PCR and cloned into pHW2000. Each of these mutant NPDk genes was cotransfected with the remaining seven genes derived from DkYK10 in 293T cells to generate seven NP-mutant viruses with a single amino acid substitution of NP. These NP mutant viruses (106.0 EID50) were inoculated intranasally into chickens, and the INPIs of the viruses and MDTs of the chickens were compared with those for rgDkYK-NPCk and rgDKYK10 as described above.
Molecular epidemiological analysis.
Involvement of NP105V identified in this study and NP184K identified previously (52) was determined for the adaptation of 1,025 duck viruses to chickens, amino acid sequences of NP genes of 1,025 AI viruses of all subtypes isolated from ducks and chickens were downloaded from Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html), and the numbers of amino acids at positions 105 and 184 of NP genes of duck or chicken strains were compared.
Statistical analysis.
Student's t test was used to determine the significance of fever and MDT in chickens infected with CkYM7, DkYK10, or their reassortant viruses. Significance was set at a P value of <0.05.
RESULTS
Screening of genes responsible for high pathogenicity in chickens.
To identify the gene segment responsible for the high virulence of H5N1 HPAI virus CkYM7 in chickens, pathogenicity of the two parent viruses, CkYM7 and DkYK10, and their single-gene reassortants, with one of eight segments from DkYK10 in the CkYM7 background, were compared by monitoring the body temperature of the chickens using a wireless thermosensor system.
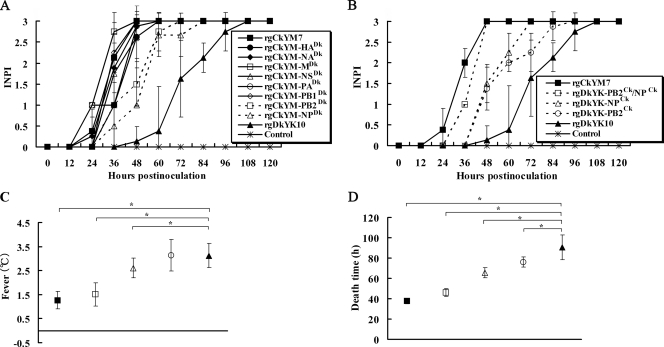
The INPI test indicated that the kinetic curves of rgCkYM-PB2Dk and rgCkYM-NPDk were delayed 1 day from those of the other six reassortant viruses or rgCkYM7 but were 1 day earlier than that of rgDkYK10. The INPIs of rgCkYM-PB2Dk and rgCkYM-NPDk slowly increased and reached a plateau 72 h postinfection (hpi) and 84 hpi, respectively, whereas the INPIs of the other six reassortant viruses increased quickly and reached a plateau within 60 hpi, similar to results for rgCkYM7 (Fig. 1 A).
FIG. 1.
Screening and identification of genes responsible for high pathogenicity of H5N1 HPAI virus CkYM7 in chickens. (A) Each of eight 4-week-old chickens were inoculated intranasally with 106 EID50 of each single-gene reassortant virus based on the CkYM7 background (rgCkYM7, rgCkYM-HA, rgCkYM-NA, rgCkYM-M, rgCkYM-NS, rgCkYM-PA, rgCkYM-PB1, rgCkYM-PB2, rgCkYM-NP, or rgDkYK10). All the chickens were observed every 12 h until they died, and the kinetics of intranasal pathogenicity index (INPI) scores are represented by the average ± standard deviation for eight chickens per group as follows: 0, no clinical signs; 1, ruffled feathers and/or falling down; 2, other clinical signs or lesions; 3, death. (B) Each of eight chickens were inoculated intranasally with 106 EID50 of each reassortant virus based on the DkYK10 background (rgCkYM7, rgDkYK-PB2Ck/NPCk, rgDkYK-NPCk, rgDkYK-PB2Ck, and rgDkYK10). The INPI score kinetics for each reassortant group are as described for panel A. The average fever (C) and MDT (D) of eight inoculated chickens in each reassortant group shown in panel B were compared among the groups. Fever was based on the increase above the basal body temperature of each chicken, and the time to death of each chicken was defined as the time when the body temperature fell below 30°C after inoculation. The fever and MDT shown are the mean ± standard deviation for each group. *, significant difference (P < 0.05) compared to rgDkYK10.
The MDTs for chickens infected with the two parent viruses and eight CkYM7-based reassortant viruses are shown in Table 1. The MDTs for chickens infected with rgCkYM-PB2Dk and rgCkYM-NPDk were 56.9 ± 4.3 h and 61.3 ± 9.6 h, respectively, whereas MDTs of chickens infected with rgCkYM-HADk, rgCkYM-NSDk, and rgCkYM-PB1Dk were 50.5, 48.1, and 46.8 h, respectively. Other three reassortant viruses and rgCkYM7 had MDTs of ≤43 h.
TABLE 1.
Comparison of the MDT for, fever produced by, and infectivity of rgCkYM7, rgDkYK10, and reassortant viruses based on CkYM7a
| Virus | MDTb (hpi) | Feverc (°C) | Virus titerd (EID50/g) |
||
|---|---|---|---|---|---|
| Lung | Spleen | Brain | |||
| CkYM7 | 41.1 ± 1.7 | 1.1 ± 0.6 | 8.0 ± 0.5 | 8.2 ± 0.6 | 6.5 ± 0.0 |
| rgCkYM7 | 37.6 ± 2.4 | 1.3 ± 0.4 | 7.8 ± 0.3 | 8.0 ± 1.0 | 6.7 ± 0.3 |
| rgCkYM-HADk | 50.5 ± 2.8 | 1.9 ± 0.3 | 7.3 ± 0.8 | 6.7 ± 0.8 | 6.7 ± 0.3 |
| rgCkYM-NADk | 43.1 ± 5.7 | 1.4 ± 0.5 | 7.8 ± 0.8 | 8.8 ± 0.8 | 6.8 ± 0.3 |
| rgCkYM-MDk | 36.9 ± 3.1 | 1.6 ± 0.3 | 7.8 ± 0.6 | 8.5 ± 0.0 | 7.2 ± 0.8 |
| rgCkYM-NSDk | 48.1 ± 4.1 | 1.7 ± 0.3 | 7.8 ± 0.6 | 8.7 ± 0.3 | 7.2 ± 0.8 |
| rgCkYM-PADk | 46.8 ± 2.7 | 1.4 ± 0.3 | 7.7 ± 0.3 | 7.3 ± 0.3 | 6.0 ± 1.3 |
| rgCkYM-PB1Dk | 40.3 ± 3.2 | 1.5 ± 0.4 | 8.0 ± 0.5 | 8.2 ± 0.3 | 6.5 ± 0.0 |
| rgCkYM-PB2Dk | 56.9 ± 4.3 | 1.7 ± 0.8 | 7.0 ± 0.5 | 6.7 ± 0.3 | 6.7 ± 1.3 |
| rgCkYM-NPDk | 61.3 ± 9.6 | 1.5 ± 0.4 | 7.3 ± 0.4 | 7.2 ± 0.3 | 6.7 ± 0.3 |
| DkYK10 | 99.0 ± 16.3 | 3.0 ± 0.3 | 6.7 ± 0.6 | 5.5 ± 1.0 | 7.5 ± 0.0 |
| rgDkYK10 | 90.5 ± 12.4 | 3.1 ± 0.5 | 6.8 ± 0.6 | 6.3 ± 0.3 | 7.3 ± 0.3 |
Each of eight chickens was inoculated with 106 EID50 of each reassortant.
Mean death time (MDT) values are the means ± SD from eight chickens.
Values are the means ± SD for each organ from eight chickens. Fever was estimated as the temperature above the basal body temperature of each chicken, which was estimated as the mean body temperature for 24 h before inoculation.
Tissues were harvested soon after death, and the virus infectivity of each sample was determined with embryonated chicken eggs. The virus titers are the means ± SD for each organ from three chickens.
The final virus titers for lungs, spleens, and brains were similar among chickens inoculated with one of several reassortant viruses, and there was no significant difference in regard to fever (Table 1).
Determination of the role of PB2 and NP in the pathogenicity of CkYM7.
Next, PB2, NP, or NP plus PB2 from CkYM7 was combined with the remaining seven or six genes of DkYK10 to generate three reassortant viruses: rgDkYK-PB2Ck, -NPCk, and -PB2Ck/NPCk, respectively. The INPI kinetics of these three viruses were compared to those of the parental viruses. The INPI kinetics of rgCkYM7 increased quickly, but the kinetics of DkYK10 slowly reached a score of 3. The kinetics of rgDkYK-PB2Ck and rgDkYK-NPCk were between those of the two parents, and the kinetics of rgDkYK-PB2Ck/NPCk were similar to those of rgCkYM7 (Fig. 1B).
The temperatures of chickens infected with rgCkYM7, rgCkYM-PB2Ck/NPCk, rgCkYM-NPCk, rgDkYK-PB2Ck, or rgDkYK10 were 1.3 ± 0.4°C, 1.5 ± 0.5°C, 2.6 ± 0.4°C, 3.1 ± 0.5°C, and 3.1 ± 0.7°C, respectively, above basal body temperature (i.e., fever) (Fig. 1C). The virus strain that caused a longer MDT in chickens had a tendency to induce a higher fever. The MDTs for chickens infected with rgCkYM7, rgDkYK-PB2Ck/NPCk, rgDkYK-NPCk, rgDkYK-PB2Ck, or rgDkYK10 were 37.6 ± 2.4 h, 46.0 ± 3.9 h, 65.6 ± 5.2 h, 76.0 ± 5.0 h, and 90.5 ± 12.4 h, respectively (Fig. 1D).
Replication kinetics of reassortant viruses in chickens.
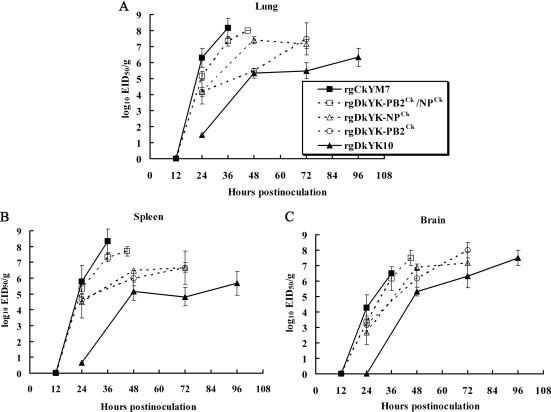
To compare the viral growth kinetics of the reassortant viruses in chickens, the lungs, spleens, and brains were collected sequentially after virus inoculation, and the virus titers of the tissue homogenates were determined with embryonated chicken eggs.
In the lungs (Fig. 2 A), rgCkYM7 replicated very rapidly and efficiently, and rgDkYK-PB2Ck/NPCk was the second most rapid, followed by rgDkYK-NPCk and rgDkYK-PB2Ck. The replication of rgDkYK10 was the slowest among the five reassortant viruses. Some association was found between the rapidity of virus replication in the lungs and the final virus titer. The final virus titers for rgDkYK-PB2Ck/NPCk, rgDkYK-NPCk, and rgDkYK-PB2Ck in the lungs were higher than those of rgDkYK10. The virus titers of rgCkYM7 and rgDkYK-PB2Ck/NPCk at 36 hpi were 108.2 EID50/g and 107.3 EID50/g, respectively. At 48 hpi, the titer of rgDkYK-NPCk was roughly 100 times higher than that of rgDkYK-PB2Ck and reached 107.4 EID50/g at 48 hpi, whereas those of rgDkYK-NPCk and rgDkYK-PB2Ck were similar at 72 hpi (∼107.5 EID50/g). rgDkYK10 grew slowly in the lungs, and the final virus titer was 106.3 EID50/g. The replication kinetics of these five reassortant viruses in the spleen were similar to the kinetics in the lungs (Fig. 2B), except there was no remarkable difference between the titers of rgDkYK-NPCk and rgDkYK-PB2Ck.
FIG. 2.
Comparison of the replication kinetics in chickens among DkYK10-based reassortant viruses. Four-week-old chickens were divided into five groups (n = 9 or 12 chickens per group) and inoculated intranasally with 106 EID50 of rgCkYM7, rgDkYK-PB2Ck/NPCk, rgDkYK-NPCk, rgDkYK-PB2Ck, or rgDkYK10. Three chickens per group at each sampling point shown were euthanized, and lungs, kidneys, and brains were harvested. Ten percent tissue homogenates were prepared, and the supernatants were used for infectivity titration with embryonated eggs. Average virus titers ± standard deviations for lungs (A), spleen (B), and brain (C) of three chickens at each time point are shown (log10 EID50/g).
In contrast, viral growth in the brain was not much different among the five reassortant viruses, and the final virus titers were roughly 107.0 EID50/g for all five virus strains (Fig. 2C). The only clear difference among the five reassortant viruses was rapidity. rgCkYM7 and rgDkYK-PB2Ck/NPCk replicated rapidly in the brain, the virus titers of rgDkYK-PB2Ck and rgDkYK-NPCk reached 107.0 EID50/g at 72 hpi, and rgDkYK10 replicated slowly and its final titer was attained at 96 hpi.
Detection of virus antigen in tissues.
To compare the distribution of viral antigens in tissues of chickens infected with the reassortant viruses, the lungs, spleen, liver, heart, and brain were collected sequentially and stained immunohistochemically with anti-M antibody (Table 2) (Fig. 3).
TABLE 2.
Distribution of viral antigen in tissues from 4-week-old SPF chickens inoculated intranasally with rgCkYM7, rgDkYK10, or reassortant viruses based on DkYK10
| Virus | Tissuea | Detection of AI virus matrix antigenb |
|||||
|---|---|---|---|---|---|---|---|
| 12 hpi | 24 hpi | 36 hpi | 45 or 48 hpic | 72 hpi | 96 hpi | ||
| rgCkYM7 | Lung | −, −, − | −, −, + | +++, +++, +++ | |||
| Spleen | −, −, − | +, −, ++ | +++, +++, +++ | ||||
| Liver | −, −, − | +, −, + | ++, +++, + | ||||
| Heart | −, −, − | −, −, − | ++, +++, ++ | ||||
| Brain | −, −, − | −, −, − | +, ++, + | ||||
| rgDkYK-PB2Ck/NPCk | Lung | −, −, − | −, −, − | +++, ++, ++ | +++, +++, +++ | ||
| Spleen | −, −, − | −, −, − | +++, ++, ++ | +++, +++, +++ | |||
| Liver | −, −, − | −, −, − | +, +, + | ++, ++, + | |||
| Heart | −, −, − | −, −, − | +, +, + | +++, ++, +++ | |||
| Brain | −, −, − | −, −, − | ±, +, + | +++, ++, + | |||
| rgDkYK-NPCk | Lung | −, −, − | +, +, + | +, ++, + | |||
| Spleen | −, −, − | −, +, − | −, −, − | ||||
| Liver | −, −, − | −, −, − | −, −, − | ||||
| Heart | −, −, − | ±, ±, ± | +, +, + | ||||
| Brain | −, −, − | ±, ±, ± | ++, ++, ++ | ||||
| rgDkYK-PB2Ck | Lung | −, −, − | +, +, + | +, +, ++ | |||
| Spleen | −, −, − | −, −, − | +, −, + | ||||
| Liver | −, −, − | −, −, − | +, −, − | ||||
| Heart | −, −, − | ±, ±, ± | ++, +, + | ||||
| Brain | −, −, − | +, ++, ++ | ++, +++, +++ | ||||
| rgDkYK10 | Lung | −, −, − | −, −, − | −, −, + | +, ++, ++ | ||
| Spleen | −, −, − | −, −, − | −, −, − | −, +, ± | |||
| Liver | −, −, − | −, −, − | −, −, − | −, ±, + | |||
| Heart | −, −, − | ±, ±, − | +, ±, ± | +, +++, +++ | |||
| Brain | −, −, − | −, −, − | ++, −, +++ | +++, +++, +++ | |||
Tissues were obtained from the chickens used for the virus growth curve in Fig. 2.
Three chickens were sacrificed at each time point, and their tissues were examined for AI virus matrix antigen. Grading was by the level of matrix protein detected: −, none; ±, slight; +, mild; ++, moderate; +++, severe.
Tissues from chickens infected with rgDkYK-PB2Ck/NPCk were harvested at 45 hpi, whereas those infected with the other viruses were harvested at 48 hpi.
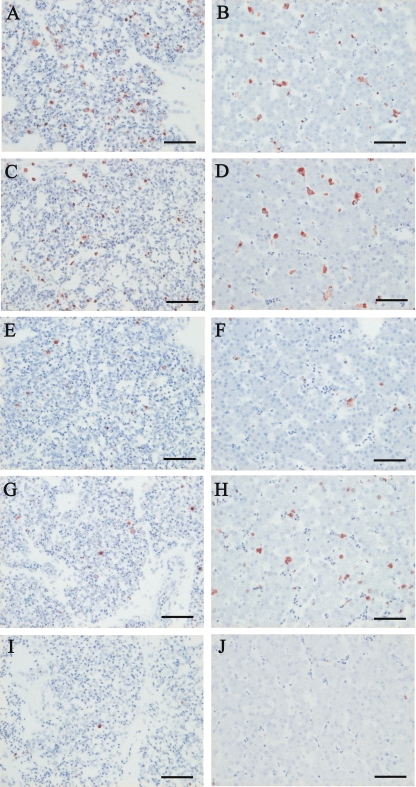
FIG. 3.
Immunohistochemical staining of the AI virus matrix protein in lung and brain tissues from chickens inoculated with each reassortant virus. Each of three chickens was inoculated intranasally with 106 EID50 of rgCkYK7, rgDkYK-PB2Ck/NPCk, and rgDkYK-NPCk, rgDkYK-PB2Ck, or rgDkYK10, and the lungs (A, C, E, G, and I) and brains (B, D, F, H, and J) were harvested for immunohistochemical staining at 36 hpi from chickens inoculated with rgCkYK7 (A and B) or rgDkYK-PB2Ck/NPCk (C and D) and at 48 hpi from chickens inoculated with rgDkYK-NPCk (E and F), rgDkYK-PB2Ck (G and H), or rgDkYK10 (I and J). Scale bars, 50 μm.
M antigen was first detected in the lungs, spleen, and liver 24 hpi in chickens infected with rgCkYM7 and was abundant in all of the tissues tested at 36 hpi except the brain tissue (Table 2). The main M antigen-positive cells in these tissues were vascular endothelial cells and macrophages (Fig. 3A). However, M antigen was slightly scattered in vascular endothelial cells and neuronal cells in the brains at 36 hpi (Fig. 3B).
M antigen was detected in the lungs and spleens of chickens infected with rgDkYK-PB2Ck/NPCk at 36 hpi and rapidly spread to other tissues at 45 hpi (Table 2). The M antigen-positive cells were macrophages in the lungs (Fig. 3C) and spleens. M antigen was detected more frequently in the brain at 45 hpi than at 36 hpi, and the positive cells were neuronal cells (Fig. 3D).
A small focal aggregation of M antigen-positive cells was first detected at 48 hpi in the lungs and brains from the rgDkYK-NPCk group (Table 2) (Fig. 3E and F). M antigen spread moderately to the lungs and broadly throughout the brains by 72 hpi (Table 2). M antigen was detected frequently in the lungs and brain of the rgDkYK-PB2Ck group at 48 hpi (Fig. 3G and H) and was detected even more frequently at 72 hpi (Table 2).
The antigen was hardly detected at 48 hpi in the tissues from chickens infected with rgDkYK10 (Table 2) (Fig. 3I and J) and substantially detected in the brains at 72 hpi; this infection spread to the lungs and heart by 96 hpi (Table 2). The M antigen was detected prominently in the brains, hearts, and lungs of chickens infected with rgDkYK10 (Table 2).
Detection of apoptotic cells in tissues.
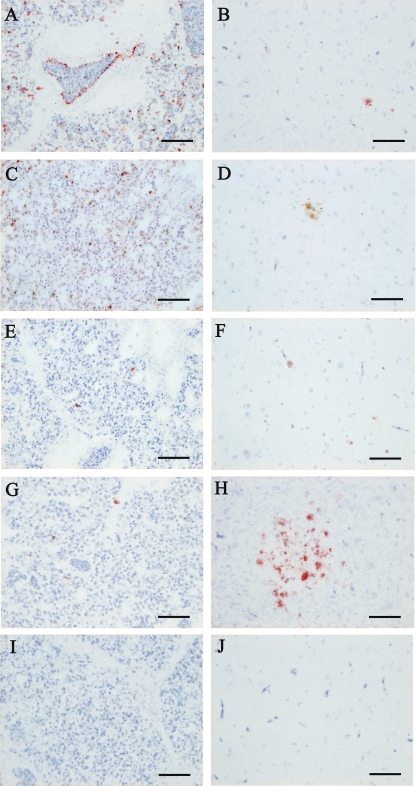
Apoptotic cells in the lungs and livers of chickens infected with the reassortant viruses were determined at each sampling point by using the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) method (Table 3) (Fig. 4). In both tissues, apoptotic cell numbers increased quickly in chickens infected with rgCkYM7 or rgDkYK-PB2Ck/NPCk, whereas those in chickens infected with rgDkYK-NPCk or rgDkYK-PB2Ck increased moderately, and the rate of apoptotic cell numbers increased slowly in chickens infected with rgDkYK10. Apoptotic cells were detected frequently in the lungs and livers collected at 36 hpi from chickens infected with rgCkYM7 (Fig. 4A and B) or rgDkYK-PB2Ck/NPCk (Fig. 4C and D) and at 48 hpi from chickens infected with rgDkYK-PB2Ck (Fig. 4G and H). Photographs of the lungs and livers collected at 36 hpi from chickens infected with rgCkYM7 (Fig. 4A and B) or rgDkYK-PB2Ck/NPCk (Fig. 4C and D) and at 48 hpi from chickens infected with rgDkYK-NPCk (Fig. 4E and F), rgDkYK-PB2Ck (Fig. 4G and H), or rgDkYK10 (Fig. 4I and J) are shown. Based on histopathological observations, most of the apoptotic cells might be macrophages in the lungs and Kupffer cells in livers.
TABLE 3.
Detection of apoptotic cells in tissues from 4-week-old SPF chickens inoculated intranasally with rgCkYM7, rgDkYK10, or reassortant viruses based on DkYK10
| Virus | Tissuea | Detection of apoptotic cellsb |
|||||
|---|---|---|---|---|---|---|---|
| 12 hpi | 24 hpi | 36 hpi | 45 or 48 hpic | 72 hpi | 96 hpi | ||
| rgCkYM7 | Lung | +, +, + | +, +, + | ++, +++, +++ | |||
| Liver | +, ±, + | +++, +++, ++ | |||||
| rgDkYK-PB2Ck/NPCk | Lung | +, +, + | +, +, ± | ++, +++, ++ | +++, +++, +++ | ||
| Liver | −, −, − | −, −, − | ++, ++, +++ | +++, +++, +++ | |||
| rgDkYK-NPCk | Lung | ±, −, ± | +, ++, ++ | ++, +++, +++ | |||
| Liver | ±, ±, ± | ±, +, + | +, ++, ++ | ||||
| rgDkYK-PB2Ck | Lung | +, ±, ± | +, ++, ++ | +++, ++, ++ | |||
| Liver | +, +, ± | +, ++, ++ | ++, +++, ++ | ||||
| rgDkYK10 | Lung | +, +, − | ±, ±, + | ++, +, ++ | ++, ++, ++ | ||
| Liver | −, −, − | −, −, − | −, −, − | ++, ++, ++ | |||
Tissues were obtained from the chickens used for the virus growth curve in Fig. 2.
Three chickens were sacrificed at each time point, and their tissues were examined for the distribution of apoptotic cells. Grading was as follows: −, none; ±, slight; +, mild; ++, moderate; +++, severe.
Tissues from chickens infected with rgDkYK-PB2Ck/NPCk were harvested at 45 hpi, whereas those infected with the other viruses were harvested at 48 hpi.
FIG. 4.
The detection of apoptotic cells in lungs and livers of chickens inoculated with each reassortant virus shown in Fig. 2. Three chickens were inoculated intranasally with 106 EID50 of rgCkYK7, rgDkYK-PB2Ck/NPCk, rgDkYK-NPCk, rgDkYK-PB2Ck, or rgDkYK10, and the lungs (A, C, E, G, and I) and livers (B, D, F, H, and J) were harvested for TUNEL staining at 36 hpi from chickens inoculated with rgCkYK7 (A and B) or rgDkYK-PB2Ck/NPCk (C and D) and at 48 hpi from chickens inoculated with rgDkYK-NPCk (E and F), rgDkYK-PB2Ck (G and H), or rgDkYK10 (I and J). Scale bars, 50 μm.
Comparison of virus growth kinetics in CEFs and DEFs.
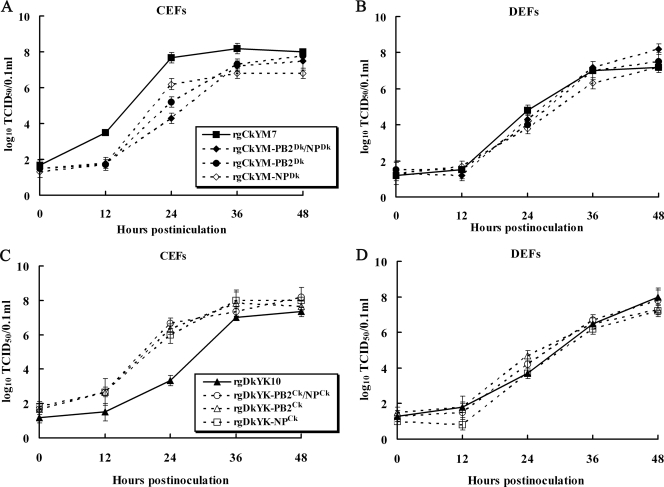
To determine the roles of PB2Ck and NPCk genes in replication of HPAI virus in cultured cells, the viral growth kinetics of the three CkYM7-based reassortants (rgCkYM-PB2Dk, -NPDk, -PB2Dk/NPDk) in CEFs were compared with those of parental rgCkYM7. Although the final virus titers of these reassortants at 48 hpi were similar, three CkYM7-based reassortants replicated more slowly in CEFs than the parental rgCkYM7 (Fig. 5 A). However, replication kinetics of these reassortants were similar to those of rgCkYM7 in DEFs (Fig. 5B). Inversely, another three reassortants possessing PB2Ck, NPCk, or both genes in the DkYK10 background replicated quickly in CEFs compared with replication of the parental rgDkYK10 (Fig. 5C); however, these reassortants replicated as quickly as the rgDkYK10 in DEFs (Fig. 5D). These results showed that both PB2Ck and NPCk contributed an increased replication of CkYM7 in CEFs but not in DEFs.
FIG. 5.
Comparison of the growth kinetics of reassortant viruses in CEFs and DEFs and polymerase activity in 293T cells. CEFs (A) or DEFs (B) were inoculated with rgCkYM7, rgCkYM-PB2Dk/NPDk, rgCkYM-NPDk, or rgCkYM-PB2Dk at an MOI of 0.001, and the average virus titers of the culture supernatants from three wells were determined at each time point using embryonated chicken eggs. CEFs (C) or DEFs (D) were inoculated with rgDkYK10, rgDkYK-PB2Ck/NPCk, rgDkYK-NPCk, or rgDkYK-PB2Ck at an MOI of 0.001, and the average virus titers were determined as described above.
Identification of critical amino acids of NP for high pathogenicity.
To identify the critical amino acids of NP responsible for the different pathogenicities in chickens, five DkYK10 mutants with single amino acid substitutions of NP, termed rgDkYK-NPG34S, rgDkYK-NPI67V, rgDkYK-NPK77R, rgDkYK-NPM105V, and rgDkYK-NPS482N, were first compared for their kinetics of INPI and MDT. As shown in Fig. 7 A, a mutant virus, rgDkYK-NPM105V, was more virulent than other four mutants or parental rgDkYK10. Similarly, another two single amino acid mutants, termed rgDkYK-NPT373A and rgDkYK-NPS377N, were shown to be of a pathogenicity similar to that of the parental rgDkYK10 (Fig. 7B). MDTs of chickens inoculated with rgDkYK-NPCk, rgDkYK-NPM105V, and rgDkYK10 were 66.3 ± 8 h, 77.1 ± 8 h, and 99.3 ± 9 h, respectively, while those of other six mutants were similar to that of rgDkYK10 (Fig. 7C). Replication of the rgDkYK-NPM105V in chickens was compared with that of rgDkYK-NPCk and rgDkYK10 by titration of virus infectivity in lungs at 48 and 60 hpi. The assay showed that rgDkYK-NPM105V replicated quickly in chickens, and the virus titers at 48 and 60 hpi were similar to those of the rgDkYK-NPCk reassortant (Fig. 7D). Quick replication of rgDkYK-NPM105V in CEFs was also observed compared with that for the parental rgDkYK10 (Fig. 7E). These results showed that the valine at position 105 in NPCk contributed to the pathogenicity of CkYM7 in chickens.
FIG. 6.
Polymerase activity of several viral gene combinations in 293T and DF-1 cells. (A) Polymerase activity in 293T cells was assayed 24 h after cotransfection with the pHH21-lucR reporter plasmid and four pHW2000 plasmids containing the PB2, PB1, PA, or NP gene from CkYM7 or DkYK10. The luciferase activity was determined as the mean count per second (CPS) ± standard deviation from triplicate wells, and the relative polymerase activities of each combination versus parental DkYK10 minireplicon are shown. (B and C) The polymerase activity of each combination in chicken DF-1 cells was determined as described above with pCk-polI-lucR plus four pHW2000 plasmids containing the PB2, PB1, PA, and NP genes from CkYM7 or DkYK10 (B) or plus eight plasmids (C).
FIG. 7.
Identification of critical amino acids in NP responsible for high pathogenicity in chickens by comparison of pathogenicity of seven DkYK10 mutants with a single amino acid substitution in NP. (A and B) Each of eight chickens were inoculated with each of the NP mutant viruses (106 EID50/chicken), and their INPI kinetics were compared with those of rgDkYK-NPCk and rgDkYK10 as described in the legend to Fig. 1. (C) The MDTs of chickens after inoculation with each of seven NP mutant viruses (106 EID50/chicken) are indicated. The values are the mean ± standard deviation for the eight chickens in each group. *, significant difference (P < 0.05) compared to rgDkYK10. (D) Each of three chickens was inoculated intranasally with rgDkYK-NPCk, rgDkYK-NPM105V, or rgDkYK-10 (106 EID50/chicken). Virus infectivity titers in lungs harvested at 48 and 60 hpi were determined with embryonated chicken eggs and compared among the virus groups. (E) The growth kinetics of rgDkYK-NP105V in CEFs was compared with those of rgDkYK-NPCk and rgDkYK10. CEFs were inoculated with each recombinant virus at an MOI of 0.001, and virus titers of the culture supernatants were determined at each time point using CEFs and expressed as an average result from three wells per sampling point. (F) The amino acid residue at position 105 on the body domain of the NP crystal structure is shown in red, while that at position 184 in the RNA binding domain of NP that relates to pathogenicity in chickens is plotted in blue (52).
Influence of PB2Ck and NPCk on polymerase activity of DkYK10.
In order to analyze the influence of PB2Ck and NPCk on polymerase activity in 293T cells, we performed a luciferase assay by using reporter plasmid pHH21-lucR-expressed luciferase under the control of the human RNA polymerase I promoter with several combinations of PB1, PB2, PA, and NP genes of rgCkYM7 or rgDkYK10 (a human minireplicon system). As shown in Fig. 6 A, the polymerase activity of DkYK10 in 293T cells was enhanced significantly by PB2Ck but only slightly by NPCk.
Next, we further analyzed the influence of NPCk on the polymerase activity of DkYK10 in DF-1 cells with a chicken minireplicon system (PA, PB1, PB2, NP, and pCk-polI-lucR) (Fig. 6B). However, the activity was not enhanced by NPCk or NP105V but was enhanced by PB2Ck. Since the activity should be enhanced in chicken cells infected with rgDkYK-NPCk, we compared the luciferase activity in the presence of all eight gene segments of DkYK10 in DF-1 cells. Finally, it was demonstrated that the polymerase activity of DkYK10 in chicken cells was increased by NPCk or NP105V as well as by PB2Ck (Fig. 6C). We confirmed that transfection of all eight gene segments of DkYK10 in DF-1 cells did not result in the production of infectious viruses because no viral RNA (vRNA) template of all eight gene segments was expressed in DF-1 cells by a human RNA polymerase I promoter in pHW2000. These results demonstrated that PB2Ck directly enhanced the polymerase activity of DkYK10 in both 293T cells and DF-1 cells, while NPCk or NP105V increased the activity indirectly in chicken cells, and the presence of HA, NA, M and NS genes was essential for the increased polymerase activity.
Molecular epidemiological analysis.
Involvement of NP105V identified in this study and NP184K detected previously (52) was determined for adaptation of duck AI viruses to chickens using amino acid sequences of 1,025 NP genes of viruses isolated in Asia, which were downloaded from Influenza Virus Resource. As shown in Table 4, most chicken viruses and most Asian H5N1 HPAI viruses possessed valine at position 105 of NP, in contrast to the methionine seen for most wild duck viruses. In contrast, almost all of the viruses from both ducks and chickens had lysine at position 184 of NP, and only a minor population of avian viruses contained alanine. These results strongly suggest that the NP105V mutation may be involved in the adaptation of duck AI viruses to chickens.
TABLE 4.
Database search for the amino acid at position 105 and position 184 in NP of avian influenza viruses isolated from ducks and chickens in Asia
| Subtype | Host | No. of genes with the indicated amino acid at position: |
|||||
|---|---|---|---|---|---|---|---|
| 105 |
184 |
||||||
| Met | Val | Other aa | Lys | Ala | Other aa | ||
| All subtypes | Duck | 225 | 31 | 8 | 263 | 0 | 1 |
| except for H5N1 | Chicken | 13 | 210 | 8 | 231 | 0 | 0 |
| H5N1 | Duck | 8 | 215 | 6 | 229 | 0 | 0 |
| Chicken | 9 | 288 | 4 | 300 | 0 | 1 | |
DISCUSSION
Knowledge of the molecular mechanisms of H5N1 HPAI viruses of waterfowl origin for further acquiring virulence in chickens is important for an understanding of the molecular pathogenesis of HPAI viruses in chickens. Although host specificity is affected by multiple genes, analyzing two HPAI viruses with limited numbers of mutations enabled us to determine the critical genes contributing to the increased pathogenicity of the H5N1 HPAI viruses in chickens or the interspecies barrier between ducks and chickens. Our study revealed that PB2 and NP genes contributed dramatically to high levels of virus replication in chickens, although HA, NS, and PB1 might be involved in pathogenicity to some extent. Our study also showed that the enhanced replication of rgDkYK-PB2CK and rgDkYK-NPCk in chickens was associated with the quick replication in CEFs, although PB2CK and NPCk did not enhance the replication of DkYK10 in DEFs. The polymerase activity of DkYK10 in 293T cells was enhanced dramatically by PB2Ck and slightly by NPCk. These results suggest that both PB2Ck and NPCk genes may contribute to the high pathogenicity of CkYM7 in chickens or adaptation to chickens. Previously, the amino acids at position 515 in PA and position 436 in PB1 were shown to be associated with lethality in ducks (16), and the lysine at position 184 of NP is critical for the high pathogenicity in chickens (51, 52). Our study demonstrated that valine at position 105 in NP was the critical amino acid for the high pathogenicity. These results enable a deepened understanding of the molecular pathogenicity and host range restriction of HPAI viruses between ducks and chickens.
The quick replication of AI viruses in chickens and cultured cells may be the most important parameter for assessing the pathogenicity of AI viruses in chickens. Also, severe apoptosis of macrophage-like cells and polymerase assay in host cells may be important parameters to identify the genes.
Influenza virus RNA polymerase is a trimeric complex comprised of three different subunits (PA, PB1, and PB2) and plays a key role in transcription/replication in cells. For viral RNA transcription and replication, PA is involved in the assembly of functional viral RNA polymerase complexes (18) and endonuclease activity (10). PB1 contains the RNA polymerase motif and catalyzes the sequential addition of nucleotides during RNA chain elongation (3). PB2 binds to the cap-1 structures of host pre-mRNAs and is involved in transcription initiation by the cap-stealing mechanism (2, 50). The primary function of NP is to encapsidate the viral genome and binds PB1 and PB2, but not PA (1), to form viral ribonucleoprotein (vRNP) complexes with three polymerase subunits for the purpose of RNA transcription, replication, and packaging (37). Also, NP plays a key role in the switch from capped RNA-primed transcription to unprimed viral RNA replication (1, 32). These viral RNA polymerase subunits and NP also play a central role in the regulation of host range or adaptation to new hosts (1, 5, 7, 19, 38).
The NP has a crescent-like structure consisting of three domains (54), the head (positions 150 to 272, 438 to 452), body (positions 21 to 149, 273 to 396, 453 to 489), and tail (positions 402 to 428) (54). Although the NP184K at the RNA binding groove of NP (52) may be essential for the replication and pathogenicity of AI viruses in both ducks and chickens, the amino acid may not be involved in adaptation from ducks to chickens, as shown in Table 4. In contrast, the NP105V is associated with quick replication and high pathogenicity of H5N1 virus in chickens and may be the first residue of NP involved in the adaptation of AI viruses from ducks to chickens. The NP105V is placed at the surface of the NP body domain. The NP binds to PB1 and PB2, and the NP body domain is a major binding site at least for PB2 (37). The interaction of NP with PB2 may regulate the polymerase activity or viral RNA synthesis of influenza viruses in mammalian hosts (1, 19). Recently, it was reported that cellular factors, such as RNA polymerase II and importin α1, may play a key role in the regulation of the NP-viral polymerase interaction and host range restriction (7, 37, 38). However, molecular interaction of NP with viral polymerase subunits or cellular proteins may not play a role in the increased pathogenicity of DkYK10 to chickens. Polymerase activity of DkYK10 was not enhanced by NPCk or NP105V in the presence of PB2 or cellular proteins in the chicken minigenome system, suggesting that enhancement of polymerase activity by NPCk or NP105V in chicken cells may not be caused by the direct binding to viral polymerase subunits or cellular proteins. The NP-derived enhancement of polymerase activity of DkYK10 in chicken cells required other viral proteins or at least M and NS gene products (data not shown), which may imply that there are more complicated mechanisms of pathogenicity or adaptation to new hosts. It is known that NS1 acts to modulate viral RNA replication (9).
PB2 plays a critical role in the initiation of transcription by the cap-stealing mechanism after binding the cap on host pre-mRNA molecules (2, 50). Although the crystal structure of PB2 has not been reported, four functional domains have been identified. PB2 consists of the PB1 binding domain (amino acid [aa] residues 1 to 259 and 580 to 759) (34, 36), RNA cap binding domain (aa residues 242 to 252, 318 to 483, and 533 to 577) (4, 13, 20), RAF-1/Hsp90 binding domain (aa residues 258 to 401) (29, 31), and nuclear localization signals (aa residues 448 to 496 and 736 to 739) (30). Amino acid residues at positions 504, 591, 627, and 701 of PB2 are known to regulate the replication of avian viruses in mammalian hosts (5-7, 11, 40, 53). Recently, lysine at position 627 in PB2 has been reported to play a key role in the interaction with NP and adaptation of influenza A viruses from avian to mammals (19, 38). However, the roles of PB2 in host range restriction and pathogenicity between ducks and chickens are largely unknown. Only one report has described the involvement of PB2 in the pathogenicity of AI viruses from ducks to chickens, but the critical domain or amino acids have not been identified (51). Our study showed that the enhanced polymerase activity of PB2 of CkYM7 in 293T and DF-1 cells may contribute to its quick replication in both CEFs and chickens. Comparing the amino acids of PB2 between CkYM7 and DkYK10 showed that 10 different amino acids, at positions of 9, 64, 66, 67, 108, 339, 340, 453, 615, and 731 in PB2, are scattered on four functional domains. The first five amino acids (9, 64, 65, 67, 108) are located on the binding site to NP and PB2, and next three amino acids (339, 340, 453) are present on the homo-oligomerization site, while the amino acids at positions 615 and 731 are on the biding site to NP/PB1 and PB1/importin α1, respectively. It is possible that the critical amino acid in PB2 for enhancing the polymerase activity may play a role in direct binding to these viral or cellular proteins. To understand the molecular mechanisms of H5N1 HPAI viruses that underlie the increased pathogenicity in chickens or the adaptation from ducks to chickens, the critical amino acids of PB2 need to be identified.
Acknowledgments
We gratefully acknowledge R. G. Webster (St. Jude Children's Research Hospital, Memphis, TN) and Yoshihiro Kawaoka (University of Wisconsin, Madison, WI) for kindly providing us with plasmids pHW2000 and pHH21, respectively.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1.Biswas, S. K., P. L. Boutz, and D. P. Nayak. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 72:5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaas, D., E. Patzelt, and E. Kuechler. 1982. Identification of the cap binding protein of influenza virus. Nucleic Acids Res. 10:4803-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braam, J., I. Ulmanen, and R. M. Krug. 1983. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell 34:609-618. [DOI] [PubMed] [Google Scholar]
- 4.Fechter, P., et al. 2003. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 278:20381-20388. [DOI] [PubMed] [Google Scholar]
- 5.Gabriel, G., et al. 2007. Differential polymerase activity in avian and mammalian cells determines host range of influenza virus. J. Virol. 81:9601-9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabriel, G., et al. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. U. S. A. 102:18590-18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabriel, G., A. Herwig, and H. D. Klenk. 2008. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 4:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Govorkova, E. A., et al. 2005. Lethality to ferrets of H5N1 influenza viruses isolated from humans and poultry in 2004. J. Virol. 79:2191-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hale, B. G., R. E. Randall, J. Ortin, and D. Jackson. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359-2376. [DOI] [PubMed] [Google Scholar]
- 10.Hara, K., F. I. Schmidt, M. Crow, and G. G. Brownlee. 2006. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 80:7789-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 13.Honda, A., K. Mizumoto, and A. Ishihama. 1999. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells 4:475-485. [DOI] [PubMed] [Google Scholar]
- 14.Horimoto, T., and Y. Kawaoka. 1997. Biologic effects of introducing additional basic amino acid residues into the hemagglutinin cleavage site of a virulent avian influenza virus. Virus Res. 50:35-40. [DOI] [PubMed] [Google Scholar]
- 15.Horimoto, T., and Y. Kawaoka. 1994. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J. Virol. 68:3120-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hulse-Post, D. J., et al. 2007. Molecular changes in the polymerase genes (PA and PB1) associated with high pathogenicity of H5N1 influenza virus in mallard ducks. J. Virol. 81:8515-8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hulse, D. J., R. G. Webster, R. J. Russell, and D. R. Perez. 2004. Molecular determinants within the surface proteins involved in the pathogenicity of H5N1 influenza viruses in chickens. J. Virol. 78:9954-9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi, A., T. Naito, and K. Nagata. 2005. Involvement of influenza virus PA subunit in assembly of functional RNA polymerase complexes. J. Virol. 79:732-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labadie, K., E. Dos Santos Afonso, M. A. Rameix-Welti, S. van der Werf, and N. Naffakh. 2007. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 362:271-282. [DOI] [PubMed] [Google Scholar]
- 20.Li, M. L., P. Rao, and R. M. Krug. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Z., et al. 2005. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 79:12058-12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Z., et al. 2006. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J. Virol. 80:11115-11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, C., M. C. Eichelberger, R. W. Compans, and G. M. Air. 1995. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J. Virol. 69:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mase, M., et al. 2005. Isolation of a genotypically unique H5N1 influenza virus from duck meat imported into Japan from China. Virology 339:101-109. [DOI] [PubMed] [Google Scholar]
- 25.Mase, M., et al. 2005. Characterization of H5N1 influenza A viruses isolated during the 2003-2004 influenza outbreaks in Japan. Virology 332:167-176. [DOI] [PubMed] [Google Scholar]
- 26.Massin, P., P. Rodrigues, M. Marasescu, S. van der Werf, and N. Naffakh. 2005. Cloning of the chicken RNA polymerase I promoter and use for reverse genetics of influenza A viruses in avian cells. J. Virol. 79:13811-13816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitnaul, L. J., M. R. Castrucci, K. G. Murti, and Y. Kawaoka. 1996. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J. Virol. 70:873-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitnaul, L. J., et al. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 74:6015-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Momose, F., et al. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277:45306-45314. [DOI] [PubMed] [Google Scholar]
- 30.Mukaigawa, J., and D. P. Nayak. 1991. Two signals mediate nuclear localization of influenza virus (A/WSN/33) polymerase basic protein 2. J. Virol. 65:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naito, T., F. Momose, A. Kawaguchi, and K. Nagata. 2007. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 81:1339-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newcomb, L. L., et al. 2009. Interaction of the influenza a virus nucleocapsid protein with the viral RNA polymerase potentiates unprimed viral RNA replication. J. Virol. 83:29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palese, P., K. Tobita, M. Ueda, and R. W. Compans. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61:397-410. [DOI] [PubMed] [Google Scholar]
- 34.Perales, B., S. de la Luna, I. Palacios, and J. Ortin. 1996. Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J. Virol. 70:1678-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perdue, M. L., M. Garcia, D. Senne, and M. Fraire. 1997. Virulence-associated sequence duplication at the hemagglutinin cleavage site of avian influenza viruses. Virus Res. 49:173-186. [DOI] [PubMed] [Google Scholar]
- 36.Poole, E., D. Elton, L. Medcalf, and P. Digard. 2004. Functional domains of the influenza A virus PB2 protein: identification of NP- and PB1-binding sites. Virology 321:120-133. [DOI] [PubMed] [Google Scholar]
- 37.Portela, A., and P. Digard. 2002. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 83:723-734. [DOI] [PubMed] [Google Scholar]
- 38.Rameix-Welti, M. A., A. Tomoiu, E. Dos Santos Afonso, S. van der Werf, and N. Naffakh. 2009. Avian influenza A virus polymerase association with nucleoprotein, but not polymerase assembly, is impaired in human cells during the course of infection. J. Virol. 83:1320-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed, L., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 40.Rolling, T., et al. 2009. Adaptive mutations resulting in enhanced polymerase activity contribute to high virulence of influenza A virus in mice. J. Virol. 83:6673-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salomon, R., et al. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203:689-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senne, D. A., et al. 1996. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 40:425-437. [PubMed] [Google Scholar]
- 43.Seo, S. H., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950-954. [DOI] [PubMed] [Google Scholar]
- 44.Solorzano, A., et al. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song, M. S., et al. 2009. The polymerase acidic protein gene of influenza a virus contributes to pathogenicity in a mouse model. J. Virol. 83:12325-12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steel, J., A. C. Lowen, S. Mubareka, and P. Palese. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 5:e1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki, K., et al. 2009. Association of increased pathogenicity of Asian H5N1 highly pathogenic avian influenza viruses in chickens with highly efficient viral replication accompanied by early destruction of innate immune responses. J. Virol. 83:7475-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swayne, D. E. 2007. Understanding the complex pathobiology of high pathogenicity avian influenza viruses in birds. Avian Dis. 51:242-249. [DOI] [PubMed] [Google Scholar]
- 49.Tanimura, N., et al. 2006. Pathology of fatal highly pathogenic H5N1 avian influenza virus infection in large-billed crows (Corvus macrorhynchos) during the 2004 outbreak in Japan. Vet. Pathol. 43:500-509. [DOI] [PubMed] [Google Scholar]
- 50.Ulmanen, I., B. A. Broni, and R. M. Krug. 1981. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc. Natl. Acad. Sci. U. S. A. 78:7355-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wasilenko, J. L., et al. 2008. NP, PB1, and PB2 viral genes contribute to altered replication of H5N1 avian influenza viruses in chickens. J. Virol. 82:4544-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wasilenko, J. L., L. Sarmento, and M. J. Pantin-Jackwood. 2009. A single substitution in amino acid 184 of the NP protein alters the replication and pathogenicity of H5N1 avian influenza viruses in chickens. Arch. Virol. 154:969-979. [DOI] [PubMed] [Google Scholar]
- 53.Yamada, S., et al. 2010. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS pathogens 6:e1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye, Q., R. M. Krug, and Y. J. Tao. 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444:1078-1082. [DOI] [PubMed] [Google Scholar]