Abstract
Human cytomegalovirus (HCMV) infection causes a rapid induction of c-Fos and c-Jun, the major subunits of activator protein 1 (AP-1), which in turn have been postulated to activate the viral immediate-early (IE) genes. Accordingly, the major IE promoter (MIEP) enhancer, a critical control region for initiating lytic HCMV infection and reactivation from the latent state, contains one well-characterized AP-1 site and a second candidate interaction site. In this study we explored the role of these AP-1 elements in the context of the infection. We first show that the distal candidate AP-1 motif binds c-Fos/c-Jun heterodimers (AP-1 complex) and confers c-Fos/c-Jun-mediated activity to a core promoter. Site-directed mutagenesis studies indicate that both AP-1 response elements are critical for 12-O-tetradecanoylphorbol-13-acetate (TPA)-enhanced MIEP activity in transient-transfection assays. In marked contrast to the results obtained with the isolated promoter, disruption of the AP-1 recognition sites of the MIEP in the context of the infectious HCMV genome has no significant influence on the expression of the MIE protein IE1 or viral replication in different cell types. Moreover, a chimeric murine CMV driven by the HCMV MIEP (hMCMV-ES) with the two AP-1 binding sites mutated is not compromised in virulence, is able to grow and disseminate to different organs of the newborn mice as efficiently as the parental virus, and is competent in reactivation. We show, however, that combined inactivation of the enhancer AP-1 and NF-κB recognition sites leads to an attenuation of the hMCMV-ES in the neonatal murine infection model, not observed when each single element is abolished. Altogether, these results underline the functional redundancy of the MIEP elements, highlighting the plasticity of this region, which probably evolved to ensure maximal transcriptional performance across many diverse environments.
Human cytomegalovirus (HCMV) replication begins with the expression of the major immediate-early (MIE) gene products IE1 and IE2, which are multifunctional proteins mainly involved in regulating both viral and cellular gene expression (reviewed in reference 51). The MIE proteins are essential for the progression of the replication cycle and crucial determinants of the transition from latency to reactivation (62, 63). Hence, the regulation of their expression is a key point in controlling the outcome of the HCMV infectious programs.
Transcription of the HCMV MIE genes is driven by a complex and potent promoter, the MIE promoter (MIEP), which comprises different functional units including a basal promoter, the enhancer region, and the modulator (23). The MIEP contains binding sites for a diverse set of signal-regulated stimulatory and inhibitory transcription factors, such as NF-κB, ATF/CREB, activator protein 1 (AP-1), YY1, Sp1/Sp3, and retinoic acid receptor (RAR)/retinoid X receptor (RXR), most of them densely packed in the enhancer region (48). In addition, viral tegument proteins and the MIE proteins themselves have also been shown to modulate MIEP activity. During latency, the MIEP is associated with markers of repressed heterochromatin, remaining silent (53, 59). Cellular differentiation and alterations in the levels of specific transcription factors by a variety of stimuli promote the activation of the MIEP and thereby the expression of downstream MIE genes. Consequently, MIEP activity is dependent on cell type, cellular differentiation stage, and the activity of specific signaling transduction pathways. Work with transgenic mice carrying a LacZ reporter under the control of the HCMV MIEP enhancer indicated that the expression of the MIEP is restricted to specific cell types in multiple organs, paralleling tissues normally infected by HCMV in the natural host (5, 6, 41). A number of studies in the last several years have addressed the relevance of different segments of the MIEP for MIE gene expression and viral replication (27, 33, 34, 47, 49). While the more distal component of the enhancer (spanning from −550 to −300 relative to the transcription start site [+1] of the MIEP) has been shown only to partially contribute to viral replication at a low multiplicity of infection (MOI) (47), progressive deletions starting from the distal end of the proximal segment of the enhancer (spanning from −300 to −39) resulted in recombinant viruses that replicated slower and with more-reduced efficiencies (33). Notably, HCMVs with the complete enhancer region eliminated fail to replicate in cultured fibroblasts (27), demonstrating a crucial genetic role for this regulatory region. The contribution of a number of binding sites for cellular transcription factors in the enhancer has been extensively assessed in cell reporter assays and shown to affect promoter function, and more recently in specific cases, the roles of particular sites have been examined in the context of infection (7, 16, 17, 27, 32, 37, 38, 43, 46, 55). However, due to the strict species specificity associated with HCMV, the impact of disrupting enhancer elements in a natural infection cannot be approached. Thus, we are still far from understanding the mechanisms of action of the MIEP in vivo.
In the related mouse cytomegalovirus (MCMV), we showed that enhancerless viruses are viable but exhibit an MOI-dependent growth phenotype in permissive fibroblasts. Most importantly, these enhancer-deficient viruses are drastically attenuated even in immunocompromised SCID mice (24). In fact, we have recently reported that enhancerless MCMVs are capable of establishing a low-level maintenance infection but fail to grow exponentially in tissues of the severely immunodeficient host (56). These results demonstrate a key role of the enhancer in viral multiplication and pathogenesis in its natural host. Although the MIEPs diverge across CMV species in complexity, number, and distribution of transcription factor binding sites and in the primary sequence, they are conserved at the functional level and share several common regulatory elements (15, 19, 63). On this basis, and to investigate the regulation of the HCMV MIEP in latency and pathogenesis, infectious model systems were developed based on chimeric MCMVs in which the native MIEP enhancer, with or without the core promoter, was replaced by the corresponding region from HCMV (4, 25, 26). These hybrid viruses are highly competent to grow in permissive fibroblasts but exhibit a partial defect in dissemination and replication in specific organs during acute infection. To get around this limitation, we have recently used the neonatal mouse model and established conditions of infection for the enhancer swap virus (hMCMV-ES) that mostly reproduce the in vivo growth characteristics and latency/reactivation properties of wild-type (wt) MCMV (26). Thus, this system allows us now to analyze the involvement of specific genetic elements of the HCMV enhancer in tissue tropism and pathogenesis and, for the first time, in the establishment of latency and subsequent reactivation.
The AP-1 family of transcription factors participates in the regulation of multiple genes involved in control of cell proliferation, differentiation, apoptosis, immune responses, and tumorigenesis in response to a wide array of stimuli, including stress, growth factors, and proinflammatory cytokines (20). AP-1 is a dimeric transcription factor that includes members of the Jun (c-Jun, JunB, and JunD) and Fos (c-Fos, FosB, Fra-1, and Fra-2) families (2), which form homo- and heterodimers that bind and activate transcription at 12-O-tetradecanoylphorbol-13-acetate (TPA) response elements (TRE) with the consensus sequence 5′-TGAC/GTCA-3′ (3; reviewed in reference 60). Some DNA viruses have developed different strategies to manipulate AP-1, most likely destined to enhance viral replication, and several lines of evidence indicate that this could be the case for HCMV. Early studies demonstrated that HCMV induces AP-1 in a biphasic mode. The initial interaction of viral particles with the cell leads to a rapid induction of both c-Fos and c-Jun (8-10), the major components of AP-1, and later in the infection a second increase of c-Fos RNA, in which the viral IE1 and IE2 proteins have been involved, can be observed (28, 52). Furthermore, in independent studies using transfection assays, IE1 has been reported to stimulate AP-1 activity, mainly through c-Jun phosphorylation by c-Jun N-terminal kinase (39, 65). In addition, the MIEP enhancer region displays a consensus AP-1 element between positions −174 and −168 (relative to the transcription start site [+1] of the MIEP), which has been previously shown to be important for promoter activity in transient-transfection assays (44) but recently reported not to substantially contribute to viral replication in vitro (17), and a potential nonconsensus AP-1 motif between positions −239 and −233, which remains unexplored. The virion pp71 protein has been reported to strongly stimulate transcription from promoters with ATF/CREB and AP-1 elements, including the MIEP (45). Moreover, besides the proposed positive role of AP-1 in HCMV replication, AP-1 has also been implicated in viral reactivation. Work by Hummel and Abecassis (30) in the mouse model provided evidence that allogeneic transplantation induces activation of AP-1, which in turn could stimulate MCMV IE1 gene expression, the first step required in viral reactivation. Notably, the MCMV enhancer harbors several scattered AP-1 sites (19). In addition, using the HCMV MIEP enhancer transgenic mouse model, signaling mediated by ischemia/reperfusion injury has been shown to contribute to the activation of the HCMV enhancer most likely through stimulation of AP-1 (40).
In the present study, we have sought to directly test the role of the AP-1 motifs in the HCMV enhancer during a CMV infection, both in tissue culture and in a mouse model. Through the generation and use of HCMV and MCMV enhancer swap recombinants containing mutations in the AP-1 response elements of the MIEP enhancer, we show that disruption of these sites has neutral effects on MIE gene expression or viral replication in culture cells and does not lead to reduced viral growth and dissemination among different organs or altered mortality in mice. Moreover, the mutated enhancer swap virus is not notably impaired in its reactivation capacity. We report, however, that simultaneous elimination of the enhancer AP-1 and NF-κB recognition sites results in reduced growth of the enhancer swap virus during acute infection. Thus, altogether our results demonstrating the dispensability of the enhancer AP-1 binding motifs in the infectious life cycle of CMV and its in vivo compensation through the NF-κB elements evidence the functional robustness of the MIEP regulation by host factors.
MATERIALS AND METHODS
Cells and viruses.
The human lung fibroblast cell line HEL299, the U373 MG cell line derived from glioblastoma, the endothelial cell line SVEC-4, the epithelial tumor cell line C127I, the macrophagic cell line RAW 264.7, and primary mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mM glutamine, 1 mM sodium pyruvate, 50 U of penicillin per ml, 50 μg of streptomycin per ml, and 10% fetal bovine serum (FBS). The myelomonocytic U937 cells were cultured in RPMI 1640 medium supplemented as stated above and 10% FBS. The retinal pigment epithelium (RPE) cells were grown in DMEM-Ham's F-12 (1:1) medium supplemented with 2 mM glutamine, 50 U of penicillin per ml, 50 μg of streptomycin per ml, 0.25% sodium bicarbonate, and 5% FBS. The murine liver-derived cell line MMH (1) was cultured on collagen-coated plates in RPMI 1640 medium supplemented with 2 mM glutamine, 50 U of penicillin per ml, 50 μg of streptomycin per ml, 100 ng/ml of epidermal growth factor, 16 ng/ml of insulin-like growth factor II, 10 μg/ml of insulin, and 10% FBS. The parental HCMV strain used in this study is the recombinant virus HCMV-GFP (green fluorescent protein) (11) originating from the bacterial artificial chromosome (BAC)-cloned HCMV AD169 genome (14). The parental hMCMV-ES, a chimeric MCMV in which the natural enhancer (sequences from nucleotides [nt] −48 to −1191) has been replaced by the HCMV enhancer (sequences from −52 to −667), was generated from a full-length MCMV Smith-BAC (pSM3fr) (64) as described in the work of Gustems et al. (26). The enhancerless MCMV, MCMVdE, and the enhancer swap MCMV containing the enhancer NF-κB sites disrupted, hMCMV-ES.NFkB, have been previously described (7, 24). Viral stocks were prepared by infecting cells at a low MOI and titrated by standard plaque assays on HEL299 cells (HCMV-derived stocks), MEFs, or NIH 3T3-Bam25 cells (24) (hMCMV-ES-derived stocks).
Plasmid construction.
The reporter plasmid pMIEP(−66/+7)CAT, which encompasses MIEP sequences between nucleotide positions −66 and +7 in front of the chloramphenicol acetyltransferase (CAT) reporter gene, has been described previously (22). This construct essentially contains the MIEP TATA and cis repression sequence (crs) element but lacks the upstream enhancer and downstream transcriptional control domains. The reporter plasmid pMIEP(−66/+7)2Ap1CAT was generated by inserting two copies of the double-stranded oligonucleotides 5′-GTA TTA GTC ATC GCT ATT ACC ATG-3′ and 5′-CAT GGT AAT AGC GAT GAC TAA TAC-3′, corresponding to the AP-1−239 recognition site and flanking natural sequences, at the HindIII site of the pMIEP(−66/+7)CAT. The reporter constructs pMIEP.Luc, pMIEPAp1−239.Luc, pMIEPAp1−239/−174.Luc, and pMIEPNFkB.Luc contain the luciferase gene under the control of wild-type (wt) MIEP sequences from −1145 to +112 or these same sequences containing mutations in the AP-1−239, both AP-1−239 and AP-1−174 (see Fig. 2A), or NF-κB (27) binding sites, respectively. Plasmids pMIEP(−1145/+112)CAT.Ap1 and pIE111H.Ap1 were generated by disrupting through site-directed mutagenesis the two AP-1 elements in the HCMV MIEP enhancer in pMIEP(−1145/+112)CAT (21) and pIE111H (4), respectively. To generate the shuttle plasmid pST76-AsacB.MIEP.Ap1, the 2.1-kbp MluI fragment from pIE111H.Ap1 was transferred into pST76-ASacB as described in the work of Benedict et al. (7). Plasmid pST-SNRΔNsi is a shuttle plasmid containing the HCMV MIEP enhancer (from nt −52 to nt −667) flanked by sequences from nt 178699 to nt 182940 and from nt 184086 to nt 187889 of the MCMV genome (58) and where an NsiI site (originally introduced during hMCMV-ES generation) (4) between the HCMV MIEP position −667 and the MCMV MIEP position −1192 (nt 184086 of the MCMV genome) has been disrupted by site-directed mutagenesis. The shuttle plasmid pST-ES.NFkB/Ap1 was constructed by replacing in pST-SNRΔNsi a 630-bp NotI/BlpI fragment, including HCMV MIEP sequences from nt −52 to nt −667, with the same enhancer sequences containing the two AP-1 and the four NF-κB binding sites mutated, previously obtained by site-directed mutagenesis. The identities of all recombinant plasmids were confirmed by DNA sequencing.
Transfection assays.
Transfections of U373 cells and MEFs were performed by the calcium phosphate coprecipitation method as described previously (22), while U937 cells were electroporated (960 μF, 200 V). Cellular extracts were prepared and assayed for β-galactosidase, luciferase, or CAT activity (22). Equal final concentrations of transfected DNA were achieved with pUC19 plasmid. pSV-c-jun expression clone and pBK28 (human c-Fos cDNA expression vector) were a gift from I. Verma (The Salk Institute, California). For the CAT assays, cell extracts containing the same amount of β-galactosidase were used. The CAT activity was quantified by liquid scintillation counting. Luciferase activity was measured with the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Firefly luciferase activities in lysates were normalized using the corresponding internal Renilla luciferase activities (provided by the tk-Renilla plasmid pRL-TK).
Generation of CMV mutants.
HCMV.Ap1 mutants were generated starting from the HCMV-GFP BAC (11), which contains the enhanced GFP (E-GFP) open reading frame under the control of the MCMV MIEP inserted within the US2-US11 region, by using two sequential ET mutagenesis steps (13, 54). First, enhancer sequences from −52 to −667 were replaced by the rpsL-neo cassette amplified from plasmid pRpsL-neo (Gene Bridges, Germany) with oligonucleotides Henh-rpsl.for (5′-CAC TAA ACG AGC TCT GCT TAT ATA GAC CTC CCA CCG TAC ACG CCT GGC CTG GTG ATG ATG GCG GGA TC-3′) and Henh-rpsl.rev (5′-CAT TGG TTA TAT AGC ATA AAT CAA TAT TGG CTA TTG GCC ATA TCA GAA GAA CTC GTC AAG AAG G-3′). In the second step, the enhancer sequences were reintroduced using the linear MluI/EagI fragment from plasmid pMIEP(−1145/+112)CAT.Ap1. Two independent BACs were obtained and called HCMV.Ap1a and HCMV.Ap1b. The hMCMV-ES.Ap1 BAC and hMCMV-ES.NFkB/Ap1 BAC were generated by the two-step mutagenesis procedure as described in the work of Benedict et al. (7) using MCMV pSM3fr BAC and shuttle plasmid pST76-AsacB.MIEP.Ap1 or pST-ES.NFkB/Ap1, respectively. To construct hMCMV-ES.Ap1-rev, first the enhancer sequences from −52 to −667 were deleted by ET mutagenesis using a linear fragment amplified from plasmid pGP704-kan (13) with oligonucleotides EnhBLPIdel.for (5′-GTA CCG ACG CTG GTC GCG CCT CTT ATA CCC ACG TAG AAC GCA GCT CAG CAG GAC GAC GAC GAC AAG TAA-3′) and EnhNDEIdel.rev (5′-GAC TTT TTA CCC AAT TTC CCA AGC GGA AAG CCC CCT AAT ACA CTC ATA TGA CAG GAA CAC TTA ACG GCT GA-3′). Then the enhancer sequences were reintroduced by the two-step mutagenesis procedure using the shuttle plasmid pST-SNRΔNsi. The resulting BAC was called hMCMV-ES.Ap1-rev and is identical to parental hMCMV-ES, except that it lacks an NsiI site adjacent to the HCMV MIEP enhancer. To generate hMCMV-ES.NFkB/Ap1-rev, first the enhancer sequences from −52 to −667 were deleted by ET mutagenesis using a linear fragment amplified from plasmid pOri6K-F5 (12) with oligonucleotides MMIEPK6.for (5′-GCG GAA AGC CCC CTA ATA CAC TCA TAA AAT GCA TAA AGC GGC CGC GAA AAG TGC CAC CTG CAG AT-3′) and MMIEPK6.rev (5′-CGA CGC TGG TCG CGC CTC TTA TAC CCA CGT AGA ACG CAG CTC AGC CAG GAA CAC TTA ACG GCT GA-3′). Then the enhancer sequences were reintroduced as indicated in the construction of hMCMV-ES.Ap1-rev, resulting in an hMCMV-ES.NFkB/Ap1-rev BAC identical to parental hMCMV-ES except that the NsiI site adjacent to the HCMV MIEP enhancer is absent. BACs were transfected into HEL299 cells (HCMVs) or MEFs (hMCMV-ESs) using the calcium phosphate transfection technique. Progeny virus obtained from the transfections were amplified, subjected to three rounds of plaque purification, and used for the preparation of viral stocks. The integrity of the viruses generated was confirmed by restriction enzyme analysis, and enhancer regions were sequenced (data not shown). Enhancer sequences were as expected for all the recombinants, except in hMCMV-ES.Ap1, where sequences between −607 and −602 in the MIEP enhancer were CGCCAT instead of GGTACG.
Mobility shift assays.
The mobility shift assay was performed essentially as described previously (22). Probe MIEP-239, containing the AP-1 element at position −239, consisted of double-stranded oligonucleotides 5′-GTA TTA GTC ATC GCT ATT ACC ATG-3′ and 5′-CAT GGA AAT AGC GAT GAC TAA TAC-3′. Probe AP-1cons, containing a consensus AP-1 motif, consisted of oligonucleotides 5′-AGC TTA GCT ATG ACT CAT CCG GAA GCT-3′ and 5′-AGC TTC CGG ATG AGT CAT AGC TAA GCT-3′. The probe bearing the AP1−174 site consisted of oligonucleotides 5′-GAT AGC GGT TTG ACT CAC GGG GAT-3′ and 5′-ATC CCC GAG AGT CAA ACC GCT ATC-3′. The probe containing the NF-κB motif consisted of oligonucleotides 5′-ATG TGA GGG GAC TTT CC C AGG C-3′ and 5′-GCC TGG GAA AGT CCC CTC AAC T-3′. For in vitro transcription of RNA for c-Jun and c-Fos, RNA was synthesized from linearized pGEM-cjun and pGEM-cfos plasmids using T7 RNA polymerase and a standard in vitro transcription protocol. In vitro-transcribed RNA (50 μl) for each transcript was mixed with 60 μl of rabbit reticulocyte lysate (Amersham). The reaction volume was adjusted to 150 μl (depending on single or cotranslation mixtures) and incubated at 30°C for 1 h. The c-Fos and c-Jun proteins were preincubated in binding buffer either independently, in combination, or as cotranslation products at room temperature for 15 min, and subsequently the 32P-labeled oligonucleotide probes were added to the reaction mixtures and further incubated at room temperature for 30 min. To get first-order kinetics of binding, labeled oligonucleotide probes in the reaction mixtures were added in excess. Reaction mixtures were analyzed on 4% polyacrylamide gels. After electrophoresis, the gel was dried and autoradiographed.
Viral nucleic acid isolation and analysis.
Preparation of total DNA from infected cells, restriction analysis, and gel electrophoresis were essentially done as described previously (4). BAC DNA was prepared by the alkaline lysis procedure (13). To analyze whether an appropriate excision of the BAC vector sequences from the recombinant hMCMV-ES genomes had occurred, PCRs were performed as described previously (64) using genomic DNA isolated either from cells infected with the recombinant viruses or from the full-length MCMV BAC (data not shown). To confirm the correct mutagenesis of the AP-1 binding sites of the MIEP enhancer in HCMV.Ap1 and hMCMV-ES.Ap1 recombinants, enhancer sequences were amplified with primers dN and dBlp (7) from viral stocks by PCR and the amplified fragments were subjected to endonuclease treatment with ApaI or BglII. Appropriate disruption of the MIEP NF-κB binding sites in hMCMV-ES.NFkB, or both NF-κB and AP-1 binding elements in hMCMV-ES.NFkB/Ap1, was verified by PCR amplification of enhancer sequences from viral stocks using primers M182 (5′-GAC GGT ACC GAC GCT GGT-3′) and AA3 (5′-CTG GCT CTC CGC CCA AGG GCC CCC GCC C-3′) and subsequent treatment with StuI and ApaI endonucleases.
Determination of CMV replication kinetics.
Multistep growth in vitro was analyzed by infecting different cell types in 24-well plates with the recombinant viruses at the indicated multiplicities of infection. MMH cells were treated for 10 days before infection with 2% dimethyl sulfoxide (DMSO). After a 2-h adsorption period, cells were washed with phosphate-buffered saline (PBS) and incubated in DMEM supplemented with 3% FBS. At specific time points postinfection (p.i.), the supernatants of the infected cells were harvested, cleared of cellular debris, and frozen at −70°C. Infectious virus was determined by standard plaque assay on MEFs (recombinant hMCMV-ESs) or HEL299 cells (recombinant HCMVs). In the case of U373 cells, intracellular viral titers were measured. For the growth kinetics assays performed under starvation conditions, cells were cultured in medium containing 0.5% FBS from 24 h before the infection and until the end of the experiment. 12-O-Tetradecanoylphorbol-13-acetate (TPA) at 20 ng/ml or the corresponding vehicle (DMSO) was added to the culture medium containing 0.5% FBS 2 h before infection and maintained until the end of the experiment.
Western blot analysis.
HEL299 cells were either mock infected or infected with parental or mutant HCMVs at the different MOIs indicated. In specific cases, cells were also treated with TPA as indicated above. At 6, 12, 24, 48, and 72 h p.i., samples were lysed in protein sample buffer and boiled for 5 min. Cell lysates were subjected to SDS-PAGE (7% polyacrylamide) and transferred to nitrocellulose filters. Equal quantities of total protein were analyzed per lane. Filters were incubated with anti-HCMV IE1 or anti-β-actin specific monoclonal antibodies, MAB810 (Chemicon) and A2066 (Sigma), respectively. As secondary antibodies, horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (W402B; Promega) or peroxidase-conjugated goat anti-rabbit immunoglobulin G (111-035-003; Jackson) was used. Blots were developed by using the Immuno-Star horseradish peroxidase (HRP) substrate (Bio-Rad) according to the manufacturer's protocol.
Mouse infections.
Three-day-old BALB/cOlaHsd mice were obtained from Harlan (Netherlands) and housed in the vivarium (University of Barcelona) under specific-pathogen-free conditions. To assess levels of virulence, 3-day-old BALB/c mice were inoculated intraperitoneally (i.p.) with increasing doses of tissue culture-derived hMCMV-ES recombinants (ranging from 1 × 103 to 2 × 106 PFU per mouse in a final volume of 40 μl) and their survival was monitored daily for at least 40 days after infection. Data points were fitted to a sigmoidal function available at Origin software, and the 50% lethal dose (LD50) values were estimated from these curves. To analyze the replication of the hMCMV-ES recombinants in vivo, mice were i.p. inoculated with 5 × 104 PFU of tissue culture-propagated virus. At designated times after infection, mice were sacrificed, and specific organs were removed and harvested as a 10% (wt/vol) tissue homogenate. Tissue homogenates were sonicated and centrifuged, and viral titers from the supernatants were determined by standard plaque assays, including centrifugal enhancement of infectivity (29) on MEFs. An ex vivo assay was utilized to detect reactivated virus from spleen and lung, as previously described (35, 57). Briefly, mice were i.p. inoculated with 5 × 104 PFU of tissue culture-derived hMCMV-ES recombinants and maintained for 4 months to establish latency. Animals were sacrificed, and their spleens, lungs, and salivary glands were harvested. Reactivation was examined from spleens and lungs, which were manually minced and placed with medium in wells of a six-well multiwell plate. Cultures were kept (with fresh medium addition when required) for 50 days. Every 5 days, part of the supernatant was transferred to MEF monolayers (including a centrifugal-enhancement-of-infectivity step) for detection of infectious virus. The identity of the reactivated viruses was confirmed by PCR analysis using two primers that bind to viral sequences flanking the MCMV enhancer and as templates viral DNAs from the indicator MEF cultures when cytopathic effect was reached (shown in Fig. S1 in the supplemental material). In addition, PCR products from a representative sample of each group and organ were cloned and sequenced. Salivary glands were analyzed to ensure the clearance of infectious virus at the time of explantation, by sonicating them as a 10% (wt/vol) tissue homogenate and subsequently transferring them to MEF monolayers.
RESULTS
Characterization of the AP-1−239 binding site in the HCMV MIEP.
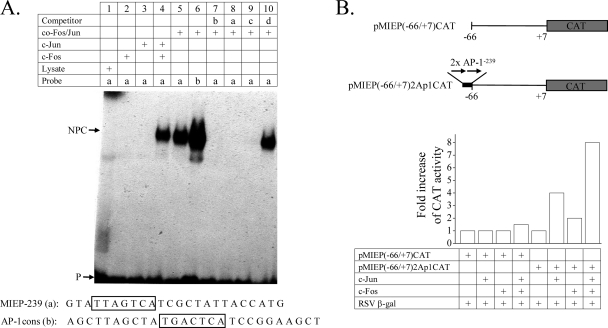
In addition to the already-described AP-1 recognition motif (44) (referred to here as AP-1−174) located from nt −174 to −168 (relative to the ie1/ie2 transcription start site), the MIEP enhancer harbors another putative binding site for AP-1 situated from nt −239 to −233 (referred to here as AP-1−239) that remains unexplored. While the AP-1−174 motif (TGACTCA) is a perfect match to the AP-1 consensus sequence (TGAC/GTCA), the AP-1−239 element (TTAGTCA) differs in 1 nucleotide from it. We sought to examine the ability of c-Jun and c-Fos to directly bind to this AP-1−239 motif by performing electrophoretic mobility shift assays (EMSAs) using a radiolabeled oligonucleotide probe containing the potential AP-1−239 recognition sequence (MIEP-239 probe). As a control, we carried out an EMSA using the consensus sequence for AP-1 binding (here named the AP-1cons probe). As depicted in Fig. 1 A, no specific DNA-protein complex was formed when the radiolabeled MIEP-239 oligonucleotide was incubated with either c-Fos or c-Jun (lanes 2 and 3). However, a retarded complex was observed when the MIEP-239 probe was incubated with both combined c-Fos and c-Jun (lane 4) or cotranslated AP-1 complex (lane 5). This complex had a mobility similar to that obtained when c-Fos and c-Jun were incubated with the AP-1cons oligonucleotide (lane 6). To verify the specificity of the AP-1 binding to the MIEP motif, we first performed competition assays using excess unlabeled AP-1cons probe, MIEP-239 probe, a probe containing the AP-1−174 sequence (probe c), or a nonspecific probe containing an NF-κB binding site (probe d). As expected, specific competition with the AP-1cons probe (lane 7), the MIEP-239 probe (lane 8), and the probe bearing the AP-1−174 motif (lane 9) abolished the formation of the DNA-protein complex, while the nonspecific probe did not alter the retarded complex (lane 10). In additional experiments we also performed supershift assays with anti-c-Jun, resulting in a decreased mobility of the retarded DNA-protein complex (data not shown). In conclusion, the results indicate that c-Jun and c-Fos proteins interact with the MIEP AP-1−239 element.
FIG. 1.
The AP-1−239 motif of the HCMV MIEP binds c-Jun/c-Fos heterodimers and confers c-Jun/c-Fos-mediated responsiveness. (A) Mobility shift assay demonstrating the interaction of c-Jun/c-Fos heterodimers with the AP-1−239 motif of the HCMV MIEP. All lanes contained the radiolabeled probe with the MIEP AP-1−239 site (MIEP-239; a) or with a consensus AP-1 motif (AP-1cons; b) and in vitro-translated c-Fos, c-Jun, or cotranslational AP-1 complex (co-Fos/Jun), except lane 1, which contained rabbit reticulocyte lysate as a control, as indicated. In addition, a 10-fold molar excess of unlabeled AP-1cons probe (b, lane 7), MIEP-239 probe (a, lane 8), a probe containing MIEP−174 (c, lane 9), or a nonspecific probe containing the NF-κB site (d, lane 10) was included. The positions of the major specific nucleoprotein complexes (NPC) corresponding to the c-Fos/c-Jun complex and of the free probe (P) are indicated. The nucleotide sequences of MIEP-239 and AP-1cons probes, with the corresponding AP-1 motifs in boxes, are shown at the bottom of the autoradiograph. (B) The AP-1−239 element of the HCMV MIEP confers c-Fos/c-Jun-mediated responsiveness. A scheme of the reporter constructs pMIEP(−66/+7)CAT and pMIEP(−66/+7)2Ap1CAT is shown, with the CAT gene represented by a shaded box, the location of the two copies of the AP-1−239 motif inserted at the −66 site of the MIEP marked by a black box, and their orientation indicated by arrows. Coordinates are given with respect to the HCMV ie1/ie2 transcriptional start site (+1). U373 cells were cotransfected with pMIEP(−66/+7)CAT or pMIEP(−66/+7)2Ap1CAT, along with a c-Jun, a c-Fos, or both c-Jun and c-Fos expression vectors, and 36 h later CAT activity present in the extracts was determined. Transfection efficiency was standardized by cotransfection of pRSV-β-Gal. The fold stimulation of CAT activity, calculated for each construct by taking as 1.00 the activity of the reporter gene in the absence of c-Fos and c-Jun, is shown. Data derived from a representative CAT assay (from three independent experiments) are presented.
To analyze whether this element could confer c-Jun and c-Fos inducibility on a basal promoter, two copies of the AP-1−239 oligonucleotide bearing the distal AP-1 site were inserted in a recombinant plasmid, pMIEP(−66/+7)CAT, containing the core MIEP (comprising sequences between nucleotide positions −66 and +7) linked to the CAT reporter gene. In this construct, named pMIEP(−66/+7)2Ap1CAT, which essentially contains the TATA and crs element but lacks the upstream enhancer and downstream transcriptional control domains, the two copies of the distal AP-1 site were cloned 5′ at position −66 of the promoter (Fig. 1B). U373 cells were transfected with pMIEP(−66/+7)2Ap1CAT or the control plasmid pMIEP(−66/+7)CAT in the presence or absence of cotransfected c-Fos, c-Jun, or both. In the absence of c-Fos- and c-Jun-expressing plasmids, low levels of transcriptional activity from pMIEP(−66/+7)2Ap1CAT could be detected, while barely no transcriptional activity was observed from pMIEP(−66/+7)CAT under the same conditions (Fig. 1B and data not shown). As seen in Fig. 1B, when each transcription factor alone was cotransfected, a significant increase (4-fold in the presence of c-Jun and 2-fold in the presence of c-Fos) in CAT activity from pMIEP(−66/+7)2Ap1CAT was obtained, but none from the control reporter construct pMIEP(−66/+7)CAT. The observed activations most likely resulted from the association of c-Fos and c-Jun transiently expressed with their corresponding endogenous heterodimeric partners, c-Jun and c-Fos. However, simultaneous overexpression of c-Fos and c-Jun resulted in substantial activation (8-fold) of pMIEP(−66/+7)2Ap1CAT that was not detected with pMIEP(−66/+7)CAT. Thus, these results illustrate the functional involvement of c-Fos and c-Jun in the activation of the distal MIEP AP-1 site.
Direct involvement of the AP-1 motifs in the activation of the HCMV MIEP.
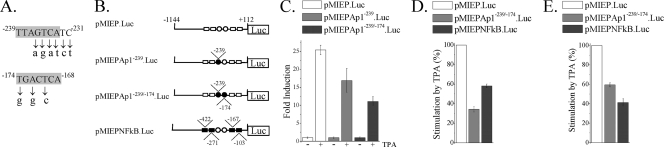
As a first step to test the functional involvement of the HCMV enhancer AP-1 sites in viral infection, point mutations (shown in Fig. 2 A) were introduced within the core of the two elements present in the MIEP and analyzed in transient-transfection assays. We also sought to examine the individual contribution of the AP-1−239 site to MIEP activity. For this purpose, U937 cells were transfected with plasmids containing sequences from −1144 to +112 of either the wild-type MIEP, the MIEP mutated in AP-1−239, or the MIEP mutated in both AP-1−174 and AP-1−239 fused with the luciferase reporter gene (illustrated in Fig. 2B) and treated with TPA, an activator of AP-1. As shown in Fig. 2C, disruption of the AP-1−239 site resulted in a significant inhibition of the TPA-induced levels of luciferase activity (from 25- to 17-fold). As expected, disruption of both AP-1−174 and AP-1−239 motifs of the MIEP led to a further reduction (from 25- to 11-fold) of the TPA-induced luciferase activity. In none of the cases, however, was the basal transcription of the MIEP mutated constructs altered compared to that of the wild-type MIEP reporter construct. The absence of a complete elimination of the TPA-induced MIEP activity could be due to the fact that other elements, such as the 4 NF-κB sites, which remained intact in the AP-1 mutated MIEP reporter construct, could be contributing to this stimulation. Accordingly, when U937 cells were transfected with an NF-κB mutated MIEP reporter plasmid, the TPA-induced promoter response was significantly decreased in comparison to that exhibited by the wild-type MIEP-driven reporter construct (Fig. 2B and D). To further explore whether these observations could be extended to murine cells, transfection assays were carried out in MEFs. As shown in Fig. 2E, mutation of either the AP-1 or NF-κB element of the MIEP resulted in decreased TPA-induced luciferase activity, indicating that the AP-1 elements within the HCMV enhancer are functionally competent in murine cells. These data are consistent with previous observations reporting a reduction in lipopolysaccharide (LPS)-induced MIEP activity after abrogation of the AP-1−174 element in RAW 264.7 cells, although in this case a slight reduction of basal MIEP activity was also observed (44). Thus, these results indicate that the two AP-1 elements of the MIEP are involved in transcriptional activation in response to TPA.
FIG. 2.
Effect of the elimination of the two enhancer AP-1 sites on MIEP activity. (A) The nucleotide sequences and locations of the two AP-1 sites lying within the MIEP enhancer are shown in shaded boxes. Below the wild-type sequence, the specific point mutations introduced in each AP-1 element are indicated in lowercase letters. Coordinates refer to the HCMV ie1/ie2 transcriptional start site. (B) Structures of the luciferase (Luc) reporter plasmids containing MIEP sequences from −1144 to +112 without (pMIEP.Luc) or with the AP-1−239 site (pMIEPAp1−239.Luc), the AP-1−239 and AP-1−174 sites (pMIEPAp1−239/−174.Luc), or the NF-κB sites (pMIEPNFkB.Luc) altered. AP-1 and NF-κB recognition sites are marked by circles and rectangles, respectively, and their positions within the MIEP are indicated. Open symbols, wild-type sequence; black symbols, mutated sequences. (C) U937 cells were electroporated with the indicated luciferase reporter constructs along with the internal control plasmid pRL-TK. Fifteen hours later, cells were serum starved for 24 h and then treated with TPA (20 ng/ml; +) or vehicle (DMSO; −) for 6 h. Cells were lysed, and luciferase activity was determined. Each value represents the average ± standard deviation of four determinations. The results are presented as the fold induction, taking as 1.00 the activity exhibited by the cells transfected with plasmid pMIEP.Luc in the absence of TPA. (D) Similar to the experiment shown in panel C, except that the results are presented as percentages of TPA stimulation of cells transfected with the corresponding reporter construct relative to the activity of TPA-induced cells transfected with pMIEP.Luc (100%). (E) MEFs were transfected with pMIEP.Luc, pMIEPAp1−239/−174.Luc, or pMIEPNFkB.Luc and processed as indicated for panel D. Fold induction of the pMIEP.Luc after TPA stimulation ranged from 4 to 9 in different experiments carried out in MEFs.
Construction of HCMVs with disruption of the AP-1 motifs of the enhancer.
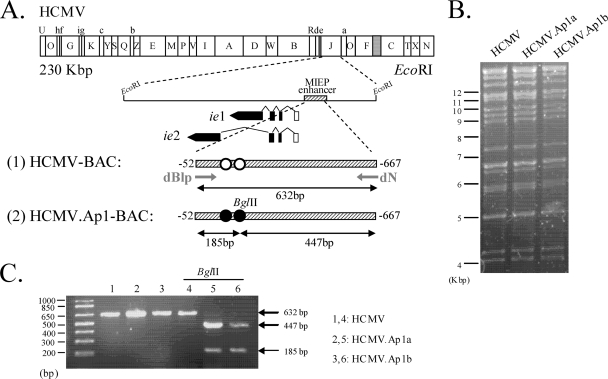
To directly address whether the AP-1 motifs within the MIEP play a role in HCMV replication, we generated two independent HCMVs bearing the AP-1 enhancer elements disrupted. A schematic representation of the parental and mutated BAC genomes is shown in Fig. 3 A. The recombinant HCMV BACs constructed were transfected into HEL299 cells, and the corresponding viruses (named HCMV.Ap1a and HCMV.Ap1b) were recovered. The integrity of the recombinant HCMV genomes generated was analyzed by restriction analysis (Fig. 3B and data not shown). As expected, the EcoRI restriction patterns of the parental and mutated viral genomes were identical. In addition, since a unique BglII restriction site was introduced in the MIEP when the AP-1−239 element was disrupted (shown in line 2, Fig. 3A), we used it to confirm the successful mutagenesis of the HCMV.Ap1 viruses. A 632-bp fragment corresponding to the HCMV enhancer regions of HCMV and HCMV.Ap1 was amplified by PCR and subsequently digested with BglII. As expected, the two predicted BglII fragments of 447 and 185 bp were detected after digestion of the PCR-amplified product from the HCMV.Ap1 genomes, while the PCR-amplified product from the parental virus remained unaltered after treatment with this restriction enzyme (Fig. 3C). Mutations were also corroborated by sequencing the complete enhancer and flanking regions of the recombinant viruses (data not shown). Altogether these results indicate that the designed modifications have been introduced within the MIEP region of the HCMV.Ap1 mutants and that no major unwanted rearrangements have occurred elsewhere in the viral genome.
FIG. 3.
Construction of HCMV mutants with the AP-1 sites in the MIEP enhancer disrupted. (A) Schematic representation of the HCMV-BAC genomes generated. The top line represents the EcoRI map of the parental HCMV-BAC genome, with the BAC region and the E-GFP gene indicated by a gray box. The EcoRI J fragment encompassing the HCMV MIE region is enlarged below, with the structures of the ie1 and ie2 transcripts indicated. Coding and noncoding exons are depicted as black and white boxes, respectively. The crosshatched bar in line 1 represents the MIEP enhancer (nucleotides −52 to −667 relative to the ie1/ie2 transcription start site) of the parental HCMV genome, with the AP-1 sites indicated in white. Line 2 represents the MIEP enhancer of the HCMV.Ap1-BAC genomes with the AP-1 sites disrupted indicated in black. The BglII restriction site introduced when mutating the MIEP AP-1 site at −239 is shown. Primers dN and dBlp, used to amplify the enhancer region, are marked. The sizes of the PCR-amplified products from the HCMV or the HCMV.Ap1 genomes after digestion with BglII are indicated below lines 1 and 2, respectively. The illustration is not drawn to scale. (B) Ethidium bromide-stained agarose gel of EcoRI-digested parental HCMV, HCMV.Ap1a, and HCMV.Ap1b genomes after separation on a 0.6% agarose gel. Size markers are shown at the left. (C) Verification of the appropriate mutagenesis of the AP-1 recognition sites within the HCMV MIEP. The enhancer sequences from the parental HCMV and mutant HCMV.Ap1 viruses were PCR amplified using the dN and dBlp primers. Shown are the amplified products separated on a 2% agarose gel before and after treatment with BglII. Size markers are shown at the left, and product sizes are shown at the right.
Disruption of the enhancer AP-1 recognition sites does not alter IE1 gene expression in HCMV-infected fibroblasts.
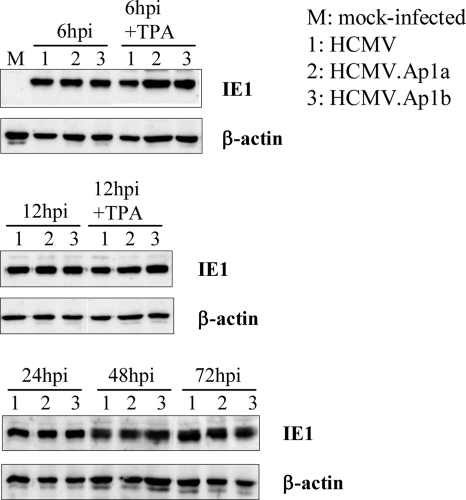
Having generated two independent HCMV.Ap1 recombinants, we then examined the consequences of disrupting the AP-1 enhancer motifs for MIE gene expression in the context of the HCMV infection in HEL299 fibroblasts. For this purpose, HEL299 cells were infected throughout a 72-h period with parental or mutant viruses at an MOI of 0.1, except for the 6- and 12-h time points, in which an MOI of 0.6 was used to allow detection of the IE1 protein, and cell lysates were subjected to Western blot assays using a monoclonal antibody specific for this MIE protein. As shown in Fig. 4, significant differences could not be detected in the expression of the IE1 protein between the HCMV and HCMV.Ap1 mutants at any of the time points analyzed. Furthermore, treatment of cells with the AP-1 inducer TPA (Fig. 4) did not result in differential expression of IE1 in cells infected with the mutant viruses compared to the ones infected with the parental HCMV. Therefore, in contrast to the results obtained in transient-transfection assays with the isolated MIEP, abrogation of the enhancer AP-1 motifs did not significantly influence IE1 expression in infected fibroblasts.
FIG. 4.
Analysis of IE1 expression in HCMV.Ap1-infected cells. HEL299 fibroblasts were mock infected or infected at an MOI of 0.6 (for the 6- and 12-h-p.i. time points) or 0.1 (for the 24-, 48-, and 72-h-p.i. time points) with HCMV, HCMV.Ap1a, and HCMV.Ap1b. When indicated, cells were treated with TPA. At designated times p.i., cells were lysed and subjected to Western blot analysis with an HCMV IE1-specific antibody. Anti-β-actin antibody was used as an internal control.
The AP-1 binding sites within the MIEP are not required during HCMV replication in cultured cells in vitro.
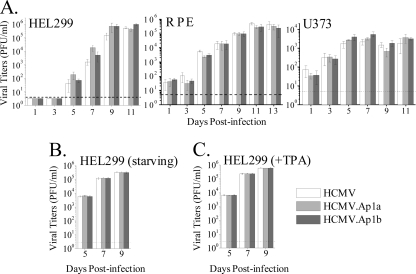
To determine the impact that disrupting the enhancer AP-1 consensus elements has on HCMV growth, kinetic studies were performed on different cell types. HEL299 fibroblasts, retinal pigment epithelium (RPE) cells, and U373 MG cells derived from glioblastoma were infected with the parental HCMV, HCMV.Ap1a, or HCMV.Ap1b. An MOI of 0.025 was used to infect the fully permissive fibroblasts, while RPE and U373 MG cells (which support viral replication to reduced levels only) were infected at an MOI of 1, resulting in around 10 to 25% GFP-positive cells in the culture at 48 h p.i. Subsequently, at different days p.i., extracellular (HEL299 and RPE) or cell-associated (U373) infectious virus was quantified from the cultures by standard plaque titration assays. As shown in Fig. 5 A, both mutant viruses grew in a fashion comparable to that of the parental HCMV in the three cell types tested. We next examined the phenotype exhibited by the HCMV.Ap1 mutants under starvation conditions, by exposing cultures from 24 h prior to infection until the end of the kinetic experiment to medium supplemented with 0.5% FBS. Again, under these conditions, parental and mutant viruses replicated equivalently in HEL299 cells (Fig. 5B). Finally, we analyzed the growth characteristics of the HCMV.Ap1 mutants after treatment with TPA. No significant differences in the replication capacity of HCMV.Ap1a, HCMV.Ap1b, and the parental virus were revealed in HEL cells under these conditions (Fig. 5C). Altogether these findings indicate that the AP-1 binding sites within the enhancer region do not independently contribute in a significant manner to HCMV growth in different cell types and conditions in culture.
FIG. 5.
Growth kinetics of HCMV.Ap1 mutants in different cell types. (A) Cells were infected at an MOI of 0.025 (HEL299) or 1 (U373 and RPE) with HCMV, HCMV.Ap1a, and HCMV.Ap1b. At the indicated days p.i. cultures were collected and the amounts of extracellular (HEL299 and RPE) or cell-associated (U373) infectious virus present were determined. Each data point represents the average and standard deviation of three separate cultures. Dashed lines represent the limits of detection. (B) Similar to the experiment shown in panel A, except that HEL299 cells were serum starved as indicated in Materials and Methods. (C) Similar to the experiment shown in panel B, except that HEL299 cells were treated with TPA.
Generation of a chimeric hMCMV-ES containing mutations in the AP-1 recognition sites of the HCMV enhancer.
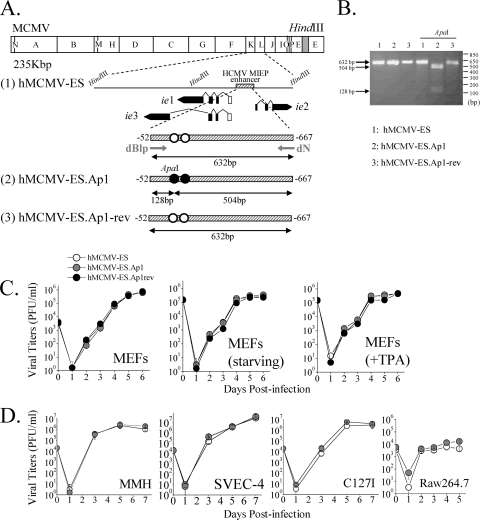
Although the AP-1 sites within the MIEP do not seem to play a role during HCMV replication in vitro, they could be required during in vivo infection. We have previously established a system for studying HCMV enhancer functions in the context of acute and latent infections, based on the hybrid enhancer swap hMCMV-ES virus in which the native MCMV MIEP enhancer (sequences from −48 to −1191) has been replaced by the HCMV enhancer (sequences from −52 to −667), and the neonatal mouse model (26). This system allows us to directly evaluate the role of the AP-1 binding sites present on the HCMV enhancer during viral infection in vivo through the generation of an hMCMV-ES with the two AP-1 binding sites abrogated (hMCMV-ES.Ap1). hMCMV-ES.Ap1 was constructed using the MCMV BAC system (13, 50), and a schematic representation of the parental and recombinant viral genomes is shown in Fig. 6 A (lines 1 and 2). As a control, we generated a revertant BAC genome by restoring the HCMV enhancer in hMCMV-ES.Ap1 (hMCMV-ES.Ap1-rev [Fig. 6A, line 3]). The constructed viral BACs were transfected into MEFs, and the corresponding viruses were recovered. To confirm that the mutated HCMV enhancer was present in the hMCMV-ES.Ap1 virus, and taking advantage of a unique ApaI restriction site (Fig. 6A) that was introduced in the HCMV enhancer when the AP-1−174 element was disrupted, a 632-bp fragment corresponding to the HCMV enhancer regions of hMCMV-ES, hMCMV-ES.Ap1, and hMCMV-ES.Ap1-rev was PCR amplified and subsequently digested with ApaI. As expected, and shown in Fig. 6B, after treatment of the PCR-amplified product obtained from hMCMV-ES.Ap1 with ApaI, two bands of 504 and 128 bp were detected, while the amplified products from hMCMV-ES and hMCMV-ES.Ap1-rev were not altered following ApaI treatment.
FIG. 6.
Construction and in vitro growth analysis of parental hMCMV-ES, hMCMV-ES.Ap1, and hMCMV-ES.Ap1-rev viruses. (A) Schematic illustrations of the parental hMCMV-ES, hMCMV-ES.Ap1, and hMCMV-ES.Ap1-rev genomes. The HindIII map of the parental MCMV genome is given at the top with the BAC sequences represented by a gray box. The enlarged map below (line 1) represents the MIE locus of the hMCMV-ES genome, carrying a 616-bp fragment (shaded rectangle) corresponding to the HCMV MIEP enhancer replacing the MCMV enhancer, with the structure of the MIE transcripts (ie1, ie2, and ie3). Coding and noncoding exons are depicted as black and white boxes, respectively. AP-1 sites are indicated in white. In hMCMV-ES.Ap1 (line 2), the AP-1 binding sites within the enhancer were mutated (black circles). A unique ApaI restriction site, introduced when the MIEP AP-1−174 recognition site was altered, is indicated. In hMCMV-ES.Ap1-rev (line 3), the native HCMV enhancer sequences were reintroduced in hMCMV-ES.Ap1 in replacement of the mutated HCMV enhancer. The sizes of the expected PCR-amplified fragments with primers dN and dBlp (marked by arrows) flanking the MIEP enhancer before and after ApaI treatment are indicated. Coordinates are given relative to the HCMV ie1/ie2 transcription start site. The illustration is not drawn to scale. (B) To verify the appropriate mutagenesis of the AP-1 sites, enhancer sequences were amplified from hMCMV-ES (lanes 1), hMCMV-ES.Ap1 (lanes 2), and hMCMV-ES.Ap1-rev (lanes 3) viruses by PCR using dN and dBlp primers. The amplified products were digested with restriction enzyme ApaI. Shown are the amplified products separated on a 2% agarose gel before and after ApaI treatment. Size markers are shown at the right, and product sizes are shown at the left. (C) Growth kinetics of hMCMV-ES.Ap1 and control viruses in MEFs under different conditions. MEFs were infected at an MOI of 0.025 PFU/cell with hMCMV-ES, hMCMV-ES.Ap1, and hMCMV-ES.Ap1-rev viruses in DMEM-3% FBS, under starvation conditions (DMEM-0.5% FBS), or in the presence of TPA, as indicated. At the designated days p.i., cell supernatants were collected and titrated on MEFs. (D) Replication of hMCMV-ES.Ap1 in different cell types. MMH cells, SVEC-4 cells, and C127I cells were infected at an MOI of 0.025, and RAW 264.7 cells were infected at an MOI of 0.1, with hMCMV-ES and hMCMV-ES.Ap1. The level of infectious virus present in culture supernatants at the designated days p.i. was determined. Each data point represents the average and standard deviation of three separate cultures.
We next sought to analyze whether removal of the AP-1 motifs within the HCMV MIEP enhancer altered the growth phenotype of the hMCMV-ES. To this end, we infected mouse embryo fibroblasts (MEFs) with parental hMCMV-ES, hMCMV-ES.Ap1, or hMCMV-ES.Ap1-rev at a low MOI. Figure 6C shows no delay in replication kinetics or reduction of viral yields associated with hMCMV-ES.Ap1 in comparison to the parental or revertant viruses. Furthermore, hMCMV-ES.Ap1 and control viruses replicated in similar manners under low-serum conditions and in the presence of the AP-1 inducer TPA in MEFs (Fig. 6C). To examine the growth behavior of hMCMV-ES.Ap1 in other murine cell types (different than MEFs), we infected the liver-derived cell line MMH, the endothelial cell line SVEC-4, the epithelial cell line C127I, and the macrophagic cell line RAW 264.7 with hMCMV-ES.Ap1 and the parental virus under low-MOI conditions. As shown in Fig. 6D, no significant differences could be detected in the replication abilities of these two viruses in any of the different cell types tested. Thus, elimination of the AP-1 motifs within the HCMV MIEP enhancer does not result in impaired replication kinetics in different cultured cells of the mutant hMCMV-ES.Ap1 virus.
Growth of hMCMV-ES.Ap1 in neonatal mice.
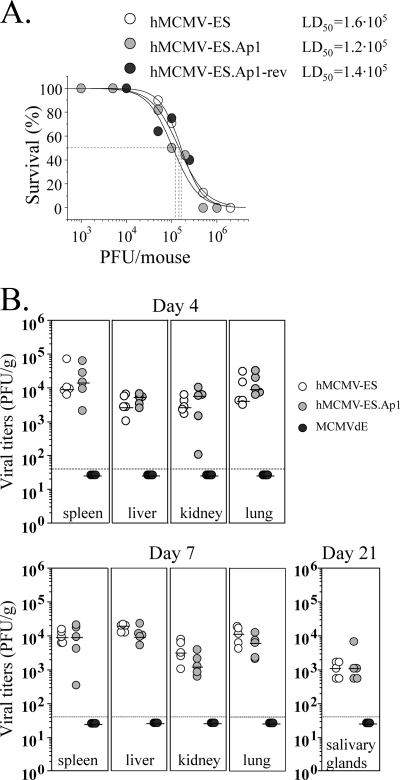
We proceeded to examine the behavior of hMCMV-ES.Ap1 in the neonatal BALB/c mouse. We have previously shown that hMCMV-ES is able to replicate in a variety of relevant target organs for MCMV in this animal model, exhibiting an LD50 around 1 × 105 PFU (26). We first assessed the virulence level of hMCMV-ES.Ap1 in comparison to the corresponding parental and revertant viruses. Taking into account the LD50 value reported for hMCMV-ES, we i.p. inoculated groups of 3-day-old mice with doses ranging from 1 × 103 to 2 × 106 PFU per mouse of hMCMV-ES.Ap1 or the parental or the revertant viruses and monitored them for their survival daily for at least a 6-week period. As shown in Fig. 7 A, the estimated LD50s were similar for the three viruses analyzed (1.2 × 105 PFU for hMCMV-ES.Ap1, 1.6 × 105 PFU for hMCMV-ES, and 1.4 × 105 PFU for hMCMV-ES.Ap1-rev), highlighting the absence of a virulence alteration associated with the abolition of the enhancer AP-1 sites in hMCMV-ES.
FIG. 7.
Infection of neonatal BALB/c mice with hMCMV-ES.Ap1, parental, and revertant viruses. (A) Virulence of hMCMV-ES.Ap1 in newborn mice. Groups of 4 to 11 3-day-old BALB/c mice were i.p. inoculated with increasing doses of parental hMCMV-ES, hMCMV-ES.Ap1, and hMCMV-ES.Ap1-rev. Animals were monitored daily for survival, and the corresponding LD50 was calculated as indicated in Materials and Methods. (B) Growth kinetics of hMCMV-ES.Ap1 in the newborn mice. Three-day-old BALB/c mice were i.p. inoculated with 5 × 104 PFU of hMCMV-ES, hMCMV-ES.Ap1, or MCMVdE. Animals were sacrificed at the indicated times after infection, and viral titers from selected organs were determined. Horizontal bars indicate the median values. The dashed lines indicate the limit of detection of the assay. The observed differences did not reach statistical difference (P > 0.05) as determined by the Mann-Whitney test (two-tailed).
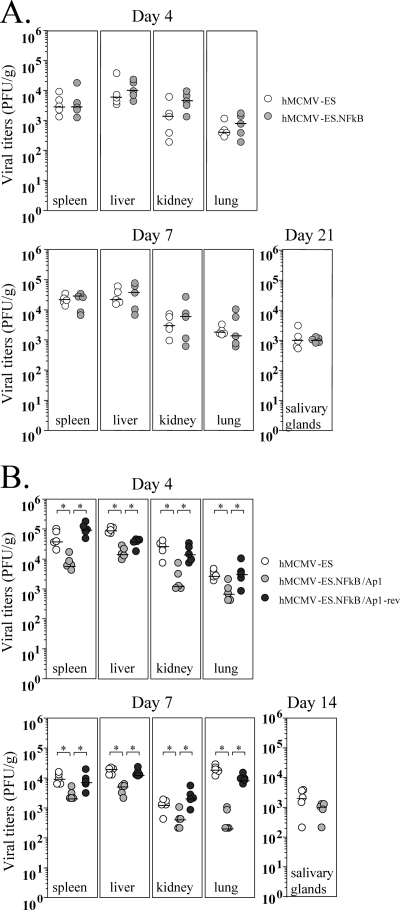
Next, we evaluated the growth of hMCMV-ES.Ap1 in key target organs of MCMV. For this purpose, 3-day-old mice were infected with 5 × 104 PFU of hMCMV-ES.Ap1 or hMCMV-ES, and at days 4, 7, and 21 after infection, their spleens, livers, kidneys, lungs, and salivary glands were extracted and examined for the presence of infectious virus. Figure 7B shows that hMCMV-ES.Ap1 grew to levels comparable and followed a course similar to those of the parental virus. In this experiment, as a control, an MCMV bearing a deletion of the entire enhancer region (MCMVdE [24]) was included. Under the conditions of the experiment, infectious MCMVdE could not be detected in any of the organs analyzed, proving that the hMCMV-ES model has the potential to unravel contributions of specific MIEP elements to viral replication during acute infection in neonates (Fig. 7B). These data demonstrate that the AP-1 sites within the HCMV enhancer region do not substantially contribute to hMCMV-ES growth and virulence in the neonatal mouse.
Reactivation properties of hMCMV-ES.Ap1 in spleen and lung explant cultures.
We have also reported that hMCMV-ES is capable of establishing latency and of reactivating in the neonatal murine model. Therefore, we sought to examine the reactivation properties of hMCMV-ES.Ap1 in comparison to hMCMV-ES. Groups of 10 to 12 3-day-old mice were infected with 5 × 104 PFU of hMCMV-ES.Ap1 or parental hMCMV-ES and maintained for a minimum of 4 months. At this time, no persistent virus could be detected in salivary gland homogenates and viral reactivation was analyzed in spleen and lung explants. As shown in Table 1, reactivation in spleens was detected in 4 out of 11 (36%) animals infected with the parental virus and in 4 out of 10 (40%) animals infected with hMCMV-ES.Ap1, while the frequency of reactivation in the lungs was 30% and 50% for the mutant and the parental virus, respectively. Thus, these results support the dispensability of the AP-1 elements in the establishment of latency and subsequent reactivation of the hMCMV-ES.
TABLE 1.
Reactivation of hMCMV-ES.Ap1 virusa
| Virus | % reactivation in explant: |
|
|---|---|---|
| Spleen | Lung | |
| hMCMV-ES | 36 (4/11) | 50 (6/12) |
| hMCMV-ES.Ap1 | 40 (4/10) | 30 (3/10) |
Percentages of viral reactivation from spleen and lung explants accumulated at day 50 postexplantation. Numbers of positive mice versus the numbers of total animals per group are shown in parentheses.
Compensation between the AP-1 and NF-κB recognition sites of the HCMV MIEP enhancer during acute infection.
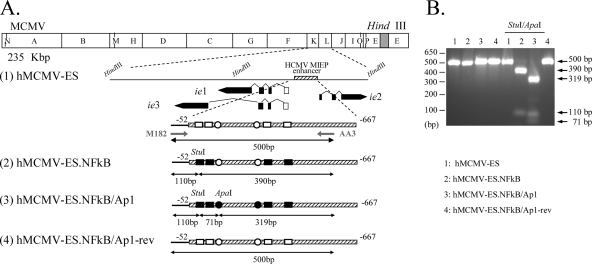
A possibility accounting for the apparently silent role of the AP-1 sites in MIEP activation during viral infection could be related, at least in part, to compensatory actions between transcription factors operating in this region. In fact, convergence of the NF-κB and AP-1 pathways has been observed in different biological systems (36). Thus, we sought to determine whether this event was taking place by analyzing the effect of inactivating the enhancer NF-κB binding sites in conjunction with the AP-1 elements on viral growth in the in vivo model. For that purpose, hMCMV-ES.NFkB/Ap1, having the four NF-κB and two AP-1 binding sites within the enhancer disrupted, was constructed using the MCMV BAC system (Fig. 8 A, line 4). As a control, a revertant BAC genome was constructed by restoring the HCMV enhancer in hMCMV-ES.NFkB/Ap1 (named hMCMV-ES.NFkB/Ap1-rev) (Fig. 8A, line 3). The constructed viral BACs were transfected into MEFs, and the corresponding viruses were recovered. For the analysis, we also included an hMCMV-ES containing the enhancer NF-κB sites mutated (hMCMV-ES.NFkB). The original hMCMV-ES.NFkB BAC genome is shown in Fig. 8A (line 2). To confirm that the appropriate HCMV enhancer version was present in the recombinant viruses generated, we took advantage of the unique ApaI and StuI restriction sites (Fig. 8B) that were introduced in the HCMV enhancer when the AP-1−174 element was disrupted and the NF-κB site at position −101, respectively. A 500-bp fragment corresponding to the HCMV enhancer regions of hMCMV-ES, hMCMV-ES.NFkB, hMCMV-ES.NFkB/Ap1, and hMCMV-ES.NFkB/Ap1-rev was PCR amplified and subsequently digested with both enzymes. As expected, and shown in Fig. 8B, after digestion of the PCR-amplified products obtained from hMCMV-ES.NFkB and hMCMV-ES.NFkB/Ap1 with ApaI and StuI, two bands of 390 and 110 bp in the case of hMCMV-ES.NFkB and three bands of 319, 110, and 71 bp in the case of hMCMV-ES.NFkB/Ap1 were detected. On the other hand, the amplified products from hMCMV-ES and hMCMV-ES.NFkB/Ap1-rev were not altered following ApaI and StuI treatment.
FIG. 8.
Construction of hMCMV-ES.NFkB/Ap1 and hMCMV-ES.NFkB/Ap1-rev viruses. (A) Schematic illustrations of the parental hMCMV-ES, hMCMV-ES.NFkB, hMCMV-ES.NFkB/Ap1, and hMCMV-ES.NFkB/Ap1-rev genomes. The HindIII map of the parental MCMV genome is given at the top with the BAC sequences represented by a gray box. The enlarged map below (line 1) represents the MIE locus of the hMCMV-ES genome, carrying a 616-bp fragment (shaded rectangle) corresponding to the HCMV MIEP enhancer replacing the MCMV enhancer, with the structure of the MIE transcripts (ie1, ie2, and ie3). Coding and noncoding exons are depicted as black and white boxes, respectively. The AP-1 recognition sites within the enhancer are marked by circles, and the NF-κB ones are shown as rectangles. In hMCMV-ES.NFkB (line 2), the NF-κB binding sites within the enhancer were mutated (black squares), and in hMCMV-ES.NFkB/Ap1 (line 3), both the NF-κB and AP-1 sites were disrupted (black squares and circles, respectively). Unique StuI and ApaI restriction sites, introduced when the MIEP AP-1−174 and NF-κB (at position −101) elements were altered, are indicated. In hMCMV-ES.NFkB/Ap1-rev (line 4), the native HCMV enhancer sequences were reintroduced in hMCMV-ES.NFkB/Ap1 in replacement of the mutated HCMV enhancer. The sizes of the expected PCR-amplified fragments with primers M182 and AA3 (marked by arrows) after StuI and ApaI treatment are indicated. Coordinates are given relative to the HCMV ie1/ie2 transcription start site. The illustration is not drawn to scale. (B) To verify the appropriate mutagenesis of the NF-κB and AP-1 sites, enhancer sequences were amplified from hMCMV-ES (lanes 1), hMCMV-ES.NFkB (lanes 2), hMCMV-ES.NFkB/Ap1 (lanes 3), and hMCMV-ES.NFkB/Ap1-rev (lanes 4) viruses by PCR using M182 and AA3 primers. The amplified products were digested with restriction enzymes StuI and ApaI. Shown are the amplified products separated on a 2% agarose gel before and after StuI and ApaI treatment. Size markers are shown at the left, and product sizes are shown at the right.
We first analyzed whether elimination of the NF-κB motifs within the HCMV MIEP enhancer altered the growth phenotype of the hMCMV-ES.NFkB in neonatal mice. For this purpose, 3-day-old mice were inoculated with 5 × 104 PFU of hMCMV-ES.NFkB or hMCMV-ES, and at days 4, 7, and 21 after infection, their spleens, livers, kidneys, lungs, and salivary glands were removed to determine the presence of infectious virus. Figure 9 A shows no significant differences in hMCMV-ES.NFkB replication levels and kinetics in the different mouse organs analyzed compared with those exhibited by the parental virus. These results indicate that, in a manner similar to that shown for the AP-1 sites, inactivation of the NF-κB recognition sequences within the HCMV enhancer region does not result in a perceptible impact on hMCMV-ES growth in the neonatal mouse. We then examined the growth properties of hMCMV-ES.NFkB/Ap1. Three-day-old mice were infected with 5 × 104 PFU of hMCMV-ES.NFkB/Ap1 or control viruses hMCMV-ES and hMCMV-ES.NFkB/Ap1-rev, and viral yields in target organs were determined at different days after infection. Figure 9B shows an attenuated phenotype of the double mutant hMCMV-ES.NFkB/Ap1 compared to both the hMCMV-ES and the revertant virus. At day 4 after infection, hMCMV-ES.NFkB/Ap1 titers in spleen, liver, kidney, and lung were 6-, 6-, 22-, and 4-fold reduced, respectively, in comparison to hMCMV-ES. At day 7 postinfection, a 5-, 4-, 3-, and 90-fold decrease in spleen, liver, kidney, and lung, respectively, could also be observed. Finally, viral titers in the salivary gland at day 14 after infection were slightly lower (2-fold) than those of the hMCMV-ES, although this difference was not statistically significant. As expected, restoration of the natural HCMV enhancer in the revertant hMCMV-ES.NFkB/Ap1 virus reverted the hMCMV-ES.NFkB/Ap1 attenuated phenotype. Taking all data together, we can conclude that the enhancer AP-1 and NF-κB binding motifs are, at least to some extent, functionally redundant.
FIG. 9.
Infection of neonatal BALB/c mice with hMCMV-ES, hMCMV-ES.NFkB, hMCMV-ES.NFkB/Ap1, and hMCMV-ES.NFkB/Ap1-rev. (A) Growth kinetics of hMCMV-ES.NFkB in the newborn mice. Three-day-old BALB/c mice were i.p. inoculated with 5 × 104 PFU of hMCMV-ES or hMCMV-ES.NFkB. Animals were sacrificed at the indicated times after infection, and viral titers from selected organs were determined. All values were above the limits of detection of the assay (4 × 101 PFU/g). Horizontal bars indicate the median values. The observed differences did not reach statistical difference (P > 0.05) as determined by the Mann-Whitney test (two-tailed). (B) Growth kinetics of hMCMV-ES.NFkB/Ap1 in the newborn mice. The experiment was similar to that in panel A, except that mice were inoculated with 5 × 104 PFU of hMCMV-ES, hMCMV-ES.NFkB/Ap1, or hMCMV-ES.NFkB/Ap1-rev. Observed differences between hMCMV-ES.NFkB/Ap1 and control viruses hMCMV-ES and hMCMV-ES.NFkB/Ap1-rev that reached statistical differences (P > 0.05) as determined by the Mann-Whitney test (two-tailed) are indicated by an asterisk.
DISCUSSION
Unraveling the molecular mechanisms underlying transcriptional control of the MIE region is a central issue in understanding CMV pathogenesis and latency. Thus, in an effort to determine the functional requirements of specific signal-regulated control elements of the CMV MIEP, in the present study we have investigated the relevance of the HCMV enhancer AP-1 recognition motifs during infection.
The transcription factor AP-1 binds and upregulates genes containing the consensus sequence 5′-TGAC/GTCA-3′ (3). Earlier reports using transient-transfection assays documented the presence of a functional canonical AP-1 binding site (TGACTCA) located between positions −174 and −168 (AP-1−174) of the MIEP (44). Importantly, in this study we provide several lines of evidence to show that an additional candidate AP-1 binding site, representing an imperfect AP-1 element (TTAGTCA) spanning from position −239 to −233 (AP-1−239), also functionally binds AP-1 and activates the MIEP. First, mobility shift assays indicate that c-Fos and c-Jun heterodimers form a specific DNA-bound complex with this MIEP site at position −239. Second, we find that two copies of this AP-1 motif are able to confer activity to a core promoter, activity that is further increased following overexpression of c-Fos and c-Jun. Third, we show that disruption of the enhancer AP-1−239 motif significantly lowers (around 32%) the overall TPA-inducible activity of the MIEP in transfection experiments. Thus, on the basis of these results we conclude that the nonconsensus AP-1−239 element is able to bind c-Fos/c-Jun heterodimers and is involved in the regulation of the MIEP. Furthermore, elimination of the two AP-1 elements resulted in a more drastic reduction (approximately 56%) of the MIEP response to TPA. This is consistent with the results previously reported in RAW 264.7 cells (44) when the MIEP consensus site AP-1−174 was individually disrupted, although in this case an effect on the constitutive activation of the promoter was observed, which might correlate with the different pools of transcription factors associated with the distinct cell types used in the two studies. Altogether, our results and previous published studies illustrate the direct involvement of the two AP-1 sites in activation of the MIEP in transient-transfection assays.
To evaluate the functional requirements of the two AP-1 sites in the context of an infection, we engineered HCMV recombinants in which the AP-1 sites have been eliminated. An analysis of these recombinants in different cell systems shows that in marked contrast with the results obtained with the isolated promoter, loss of the two enhancer AP-1 binding sites in HCMV does not reduce the rate of MIE gene expression or viral replication in a variety of cell types. Thus, under the conditions tested, the enhancer AP-1 motifs are dispensable in the initiation and subsequent steps of the viral replication cycle. This is in line with recent observations made by Caposio and coworkers, showing the lack of contribution of the enhancer AP-1−174 motif to HCMV replication in cultured fibroblasts (17). It is possible, however, that under specific circumstances, such as a shortage of specific cellular transcription factors or strong activation of a particular signal transduction pathway, defined elements within the MIEP could play a more prominent role in viral replication. Indeed, Keller et al. (38) showed that while abrogation of the enhancer ATF/CREB elements in the HCMV MIEP failed to alter viral replication in proliferating cells, a minimal negative effect was observed after stimulation of the cyclic AMP (cAMP)/protein kinase A (PKA) signaling pathway in quiescently infected cells. Also, the NF-κB binding sites have been shown to play a neutral role during HCMV infection of different cell types in vitro, but a replication defect could be detected in recombinant HCMVs derived from a clinical isolate containing the enhancer NF-κB motifs eliminated when analyzed in the absence of serum growth factors (7, 16, 27). In this line, in a recent study, desilencing of the HCMV MIE genes during HCMV quiescence in NT-2 cells has been reported to be impaired by abrogation of the NF-κB and CREB sites (46). Our results, however, indicate that HCMV growth is not influenced by the absence of the enhancer AP-1 elements under starvation conditions or strong activation of the corresponding signaling pathway by TPA. We note that the experiments performed in the present study have been carried out with a culture-adapted HCMV strain, preventing the analysis of the derived mutants in a number of cell types of biological significance for the virus, such as endothelial and dendritic cells and monocytes/macrophages. Moreover, the extreme species specificity associated with CMV restricts the analysis of HCMV mutants in an in vivo setting. In this respect, murine CMV has been extensively used as an animal model system to examine different aspects of CMV biology, pathogenesis, and latency. However, the highly repetitive nature of the MCMV MIEP complicates its manipulation (19). In addition, and as stated in the introduction, although the MCMV and HCMV enhancers share common regulatory pathways, their primary sequences and architectures are dissimilar (15, 19). In an attempt to overcome these limitations, and taking into consideration the importance of the MCMV enhancer region during acute infection (24), recombinant MCMVs in which the HCMV enhancer replaced the native one were generated some years ago (4, 25, 26). Although these enhancer swap viruses are partially attenuated in the mouse, we have recently defined conditions of infection in neonatal mice that result in comparable active replication of the chimeric virus in target organs, the establishment of latency, and its subsequent reactivation (26). Taking advantage of this system, we assessed the relevance of the HCMV MIEP AP-1 motifs in the context of the infection in neonatal mice by constructing and analyzing enhancer swap viruses having the enhancer AP-1 sites eliminated (hMCMV-ES.Ap1). We present data that underline that inactivation of the HCMV MIEP AP-1 sites does not result in reduced virulence or any measurable attenuation of the hMCMV-ES in the newborn mice. The mutant virus exhibits a replication phenotype in different organs and dissemination properties comparable to those of the parental virus.
The molecular mechanisms by which reactivation from latency, a key hallmark of the herpesviruses, occurs remain largely unknown. CMV reactivation is a multistep process in which induction of MIE gene expression is a crucial checkpoint (42). Therefore, the cellular network of transcription factors operating through the MIEP is central in determining the switching on of this regulatory region. In this regard, treatment of latently infected mice with inducers of diverse signal transduction pathways, such as LPS, tumor necrosis factor alpha (TNF-α), or interleukin 1β (IL-1β), triggers IE expression and MCMV reactivation in immunocompetent mice (18, 61). Moreover, it has been postulated that as a result of the allogeneic stimulation and ischemia reperfusion injury associated with organ transplantation, specific transcription factors would get activated, including NF-κB and AP-1, which in turn would induce MIE gene expression through binding to the corresponding response elements in the MIEP (30, 31). According to this model, one could predict a relevant role of the AP-1 signaling pathway and corresponding binding sites within the MIEP in stimulating CMV reactivation. Alternatively, AP-1 could negatively regulate MIE gene expression during latency. We have been unable, however, to detect in our experiments a substantial contribution of the enhancer AP-1 motifs to viral reactivation. Thus, after inactivation of the HCMV enhancer AP-1 elements, hMCMV-ES.Ap1 is not significantly altered in its ability to reactivate from latency, when assessed in spleen and lung explant cultures.
Altogether, despite the substantial involvement of the AP-1 sites in enhancing MIEP transcription in transient-transfection assays, we have not been able to unveil a dominant function of these elements in the viral life cycle. We cannot discard the possibility that HCMV could use AP-1 signaling through the enhancer region to augment its growth in vivo or increment the efficiency of reactivation under circumstances not analyzed here. In addition, one has to take into account that besides components of the host transcriptional machinery, MIEP activity is modulated by viral proteins, which might be species specific. However, we considered the hypothesis that the enhancer's insensitivity to mutations is due to its inherent robustness. The absence of an overtly measurable role of the enhancer AP-1 motifs in the context of the infection could be masked in part by compensation of additional cellular factors in the enhancer that perform redundant functions. For example, both NF-κB and ATF/CREB binding activities could compensate for the lack of AP-1 binding. In fact, some compensatory actions in the MIEP enhancer have been recently reported during HCMV infection in vitro (17). Here, we investigated the potential redundancy among MIEP enhancer transcription factor binding sites, by generating an hMCMV-ES.NFkB/Ap1 containing two classes of elements in this region eliminated, the four NF-κB and two AP-1 recognition sites, and analyzing it in the context of the neonatal mouse model. In contrast to recombinant viruses containing each single class of element mutated, which resulted mostly in a silent phenotype, hMCMV-ES bearing the combined mutations showed a replication deficiency in the majority of the organs of the infected animal examined. Thus, compensatory mechanisms may help to explain the absence of a significant phenotype associated with the individual disruption of either the NF-κB or the AP-1 element under the experimental conditions tested. Due to the fact that the pools of transcription factors differ among distinct tissues and fluctuate with the various physiological conditions encountered, HCMV may have evolved a generalistic redundant enhancer that supplies transcriptional activity across many diverse environments. The nature of this promoter also provides an explanation for the fact that despite the variability in the content and distribution of transcription binding sites among enhancers from different CMV species, they all activate transcription at equivalent high levels (63).
There is an urgent need of developing experimental in vivo systems to study the role of the MIEP in CMV pathogenesis. To our knowledge, this is the first time that specific genetic elements within the HCMV MIEP enhancer have been directly assessed in the context of an in vivo infection. The uniqueness of the enhancer swap MCMV-neonatal mouse system has allowed us to unveil the dispensability of the HCMV enhancer AP-1 recognition motifs during acute infection and in the process of reactivation, and more importantly, unveil the presence of compensatory mechanisms through the NF-κB sites operating in this region. These results indicate that one may need to simultaneously disrupt more than one host transcription factor binding site in order to ultimately define the combination of extracellular signal inputs, host factors, and target genetic elements that dominate in the control of the MIEP.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministerio de Educación y Ciencia (SAF2008-00382 to A.A.), the Wellcome Trust (66784/Z/02/Z), and the German Research Council (DFG; collaborative research grant 587, individual project A13 to M.M.).
We thank Ray Jupp for help with the EMSAs, Emi Guilletti for CAT assays, and Susana Kalko for assistance with statistics.
Footnotes
Published ahead of print on 24 November 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Amicone, L., et al. 1997. Transgenic expression in the liver of truncated Met blocks apoptosis and permits immortalization of hepatocytes. EMBO J. 16:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angel, P., and M. Karin. 1991. The role of Jun, Fos, and the AP-1 complex in cell proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 3.Angel, P., et al. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 4.Angulo, A., M. Messerle, U. H. Koszinowski, and P. Ghazal. 1998. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J. Virol. 72:8502-8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baskar, J. F., et al. 1996. Developmental analysis of the cytomegalovirus enhancer in transgenic animals. J. Virol. 70:3215-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baskar, J. F., et al. 1996. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J. Virol. 70:3207-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benedict, C. A., et al. 2004. Neutrality of the canonical NF-κB-dependent pathway for human and murine cytomegalovirus transcription and replication in vitro. J. Virol. 78:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boldogh, L., S. AbuBakar, and T. Albrecht. 1990. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science 247:561-564. [DOI] [PubMed] [Google Scholar]
- 9.Boldogh, L., S. AbuBakar, C. Z. Deng, and T. Albrecht. 1991. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J. Virol. 65:1568-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boldogh, L., M. P. Fons, and T. Albrecht. 1993. Increased levels of sequence-specific DNA-binding proteins in human cytomegalovirus-infected cells. Biochem. Biophys. Res. Commun. 197:1505-1510. [DOI] [PubMed] [Google Scholar]
- 11.Borst, E. M., and M. Messerle. 2000. Development of a cytomegalovirus vector for somatic gene therapy. Bone Marrow Transplant. 25:80-82. [DOI] [PubMed] [Google Scholar]
- 12.Borst, E. M., and M. Messerle. 2005. Analysis of human cytomegalovirus oriLyt sequence requirements in the context of the viral genome. J. Virol. 79:3615-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borst, E. M., C. Benkartek, and M. Messerle. 2007. Use of bacterial artificial chromosomes in generating targeted mutations in human and mouse cytomegaloviruses. Curr. Protoc. Immunol. 77:10.32.1-10.32.30. [DOI] [PubMed] [Google Scholar]
- 14.Borst, E., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boshart, M., et al. 1985. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41:521-530. [DOI] [PubMed] [Google Scholar]
- 16.Caposio, P., A. Luganini, G. Hahn, S. Landolfo, and G. Gribaudo. 2007. Activation of the virus-induced IKK/NF-kB signalling axis is critical for the replication of human cytomegalovirus in quiescent cells. Cell. Microbiol. 9:2040-2054. [DOI] [PubMed] [Google Scholar]
- 17.Caposio, P., A. Luganini, M. Bronzini, S. Landolfo, and G. Gribaudo. 2010. The Elk-1 and serum response factor binding sites in the major immediate-early promoter of human cytomegalovirus are required for efficient viral replication in quiescent cells and compensate for inactivation of the NF-kappaB sites in proliferating cells. J. Virol. 84:4481-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook, C. H., J. Trgovcich, P. D. Zimmerman, Y. Zhang, and D. D. Sedmak. 2006. Lipopolysaccharide, tumor necrosis alpha, or interleukin-1 beta triggers reactivation of latent cytomegalovirus in immunocompetent mice. J. Virol. 80:9151-9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorsch-Hasler, K., et al. 1985. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 82:8325-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eferl, R., and E. F. Wagner. 2003. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3:859-868. [DOI] [PubMed] [Google Scholar]
- 21.Ghazal, P., et al. 1992. Retinoic acid receptors initiate induction of the cytomegalovirus enhancer in embryonal cells. Proc. Natl. Acad. Sci. U. S. A. 89:7630-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghazal, P., and J. A. Nelson. 1991. Enhancement of RNA polymerase II initiation complex by a novel DNA control domain downstream from the cap site of the cytomegalovirus major immediate-early promoter. J. Virol. 65:2299-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghazal, P., and J. A. Nelson. 1993. Transcription factors and viral regulatory proteins as potential mediators of human cytomegalovirus pathogenesis, p. 360-383. In Y. Becker, G. Darai, and E.-S. Huang (ed.), Molecular aspects of human cytomegalovirus diseases. Springer-Verlag Publishers, Heidelberg, Germany.
- 24.Ghazal, A., M. Messerle, K. Osborn, and A. Angulo. 2003. An essential role of the enhancer for murine cytomegalovirus in vivo growth and pathogenesis. J. Virol. 77:3217-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grzimek, N. K., et al. 1999. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J. Virol. 73:5043-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gustems, M., A. Busche, M. Messerle, P. Ghazal, and A. Angulo. 2008. In vivo competence of murine cytomegalovirus under the control of the human cytomegalovirus major immediate-early enhancer in the establishment of latency and reactivation. J. Virol. 82:10302-10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gustems, M., et al. 2006. Regulation of the transcription and replication cycle of human cytomegalovirus is insensitive to genetic elimination of the cognate NF-κB binding sites in the enhancer. J. Virol. 80:9899-9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagemeier, C., S. M. Walker, P. J. G. Sissons, and J. H. Sinclair. 1992. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J. Gen. Virol. 73:2385-2393. [DOI] [PubMed] [Google Scholar]
- 29.Hudson, J. B. 1988. Further studies on the mechanism of centrifugal enhancement of cytomegalovirus infectivity. J. Virol. Methods 19:97-108. [DOI] [PubMed] [Google Scholar]
- 30.Hummel, M., and M. M. Abecassis. 2002. A model for reactivation of CMV from latency. J. Clin. Virol. 25:S123-S136. [DOI] [PubMed] [Google Scholar]
- 31.Hummel, M., et al. 2001. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J. Virol. 75:4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isomura, H., et al. 2005. Two sp1/sp3 binding sites in the major immediate-early proximal enhancer of human cytomegalovirus have a significant role in viral replication. J. Virol. 79:9597-9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isomura, H., T. Tsurumi, and M. F. Stinski. 2004. Role of the proximal enhancer of the major immediate-early promoter in human cytomegalovirus replication. J. Virol. 78:12788-12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isomura, H., and M. F. Stinski. 2003. The human cytomegalovirus major immediate-early enhancer determines the efficiency of immediate-early gene transcription and viral replication in permissive cells at low multiplicity of infection. J. Virol. 77:3602-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan, M. C., and V. L. Mar. 1982. Spontaneous activation of latent cytomegalovirus from murine spleen explants: role of lymphocytes and macrophages in release and replication of virus. J. Clin. Invest. 70:762-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karin, M., and M. Delhase. 1998. JNK or IKK, AP-1 or NF-kappaB, which are the targets for MEK kinase 1 action? Proc. Natl. Acad. Sci. U. S. A. 95:9067-9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keller, M. J., D. G. Wheeler, E. Cooper, and J. L. Meier. 2003. Role of the human cytomegalovirus immediate-early promoter's 19-base-pair-repeat cyclic AMP-response element in acutely infected cells. J. Virol. 77:6666-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keller, M. J., et al. 2007. Reversal of human cytomegalovirus major immediate-early enhancer/promoter silencing in quiescently infected cells via the cyclic AMP signalling pathway. J. Virol. 81:6669-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, S., et al. 1999. Human cytomegalovirus IE1 protein activates AP-1 through a cellular kinase(s). J. Gen. Virol. 80:961-969. [DOI] [PubMed] [Google Scholar]
- 40.Kim, S. J., et al. 2005. Renal ischemia/reperfusion injury activates the enhancer domain of the human cytomegalovirus major immediate early promoter. Am. J. Transplant. 5:1606-1613. [DOI] [PubMed] [Google Scholar]
- 41.Koedood, M., A. Fichtel, P. Meier, and P. J. Mitchell. 1995. Human cytomegalovirus (HCMV) immediate-early enhancer/promoter specificity during embryogenesis defines target tissues of congenital HCMV infection. J. Virol. 69:2194-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurz, S. K., and M. J. Reddehase. 1999. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J. Virol. 73:8612-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lashmit, P., S. Wang, H. Li, H. Isomura, and M. F. Stinski. 2009. The CREB site in the proximal enhancer is critical for cooperative interaction with the other transcription factor binding sites to enhance transcription of the major intermediate-early genes in human cytomegalovirus-infected cells. J. Virol. 83:8893-8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, Y., W.-J. Sohn, D.-S. Kim, and H.-J. Kwon. 2004. NF-kB- and c-Jun-dependent regulation of human cytomegalovirus immediate-early gene enhancer/promoter in response to lipopolysaccharide and bacterial CpG-oligodeoxynucleotides in macrophage cell line RAW 264.7. Eur. J. Biochem. 271:1094-1105. [DOI] [PubMed] [Google Scholar]
- 45.Liu, B., and M. F. Sinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 66:4434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, X., et al. 2010. Phorbol ester-induced human cytomegalovirus MIE enhancer activation through PKC-delta, CREB, and NF-κB de-silences MIE gene expression in quiescently infected human pluripotent NTera2 cells. J. Virol. 84:8495-8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meier, J. L., and J. A. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J. Virol. 74:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meier, J. L., and M. F. Stinski. 2006. Major immediate-early enhancer and its gene products, p. 151-166. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Wymondham, Norwich, United Kingdom.
- 49.Meier, J. L., M. K. Keller, and J. J. McCoy. 2002. Requirement of multiple cis-acting elements in the human cytomegalovirus major immediate-early distal enhancer for viral gene expression and replication. J. Virol. 76:313-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U. S. A. 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 52.Monick, M. M., L. J. Geist, M. F. Stinski, and G. W. Hunninghake. 1992. The immediate early genes of human cytomegalovirus upregulate expression of the cellular genes myc and fos. Am. J. Respir. Cell Mol. Biol. 7:251-256. [DOI] [PubMed] [Google Scholar]
- 53.Murphy, J., W. Fischle, E. Verdin, and J. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muyrers, J. P., et al. 2000. Point mutation of bacterial artificial chromosomes by ET recombination. EMBO Rep. 3:239-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Netterwald, J., et al. 2005. Two gamma interferon-activated site-like elements in the human cytomegalovirus major immediate-early promoter/enhancer are important for viral replication. J. Virol. 79:5035-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Podlech, J., et al. 2010. Enhancerless cytomegalovirus is capable of establishing a low-level maintenance infection in severely immunodeficient host tissues but fails in exponential growth. J. Virol. 84:6254-6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Presti, R. M., J. L. Pollock, A. J. Dal Canto, A. K. O'Guin, and H. W. Virgin. 1998. Interferon-g regulation of acute, chronic, and latent murine cytomegalovirus infection and disease of the great vessels. J. Exp. Med. 178:577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reeves, M. B., P. A. MacAri, P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. U. S. A. 102:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 61.Simon, C. S., C. K. Seckert, D. Dreis, M. J. Reddehase, and N. K. A. Grzimek. 2005. Role of tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs. J. Virol. 79:326-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinclair, J., and P. Sissons. 2006. Latency and reactivation of HCMV. J. Gen. Virol. 87:1763-1779. [DOI] [PubMed] [Google Scholar]
- 63.Stinski, M. F., and H. Isomura. 2008. Role of the cytomegalovirus major immediate early enhancer in acute infection and reactivation from latency. Med. Microbiol. Immunol. 197:223-231. [DOI] [PubMed] [Google Scholar]
- 64.Wagner, M. S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, X., and G. E. Sonenshein. 2005. Induction of the RelB NF-κB subunit by the cytomegalovirus IE1 protein is mediated via Jun kinase and c-Jun/Fra-2 AP-1 complexes. J. Virol. 79:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.