Abstract
Vaccinia virus (VV) mutants lacking the double-stranded RNA (dsRNA)-binding E3L protein (ΔE3L mutant VV) show restricted replication in most cell types, as dsRNA produced by VV activates protein kinase R (PKR), leading to eIF2α phosphorylation and impaired translation initiation. Here we show that cells infected with ΔE3L mutant VV assemble cytoplasmic granular structures which surround the VV replication factories at an early stage of the nonproductive infection. These structures contain the stress granule-associated proteins G3BP, TIA-1, and USP10, as well as poly(A)-containing RNA. These structures lack large ribosomal subunit proteins, suggesting that they are translationally inactive. Formation of these punctate structures correlates with restricted replication, as they occur in >80% of cells infected with ΔE3L mutant VV but in only 10% of cells infected with wild-type VV. We therefore refer to these structures as antiviral granules (AVGs). Formation of AVGs requires PKR and phosphorylated eIF2α, as mouse embryonic fibroblasts (MEFs) lacking PKR displayed reduced granule formation and MEFs lacking phosphorylatable eIF2α showed no granule formation. In both cases, these decreased levels of AVG formation correlated with increased ΔE3L mutant VV replication. Surprisingly, MEFs lacking the AVG component protein TIA-1 supported increased replication of ΔE3L mutant VV, despite increased eIF2α phosphorylation and the assembly of AVGs that lacked TIA-1. These data indicate that the effective PKR-mediated restriction of ΔE3L mutant VV replication requires AVG formation subsequent to eIF2α phosphorylation. This is a novel finding that supports the hypothesis that the formation of subcellular protein aggregates is an important component of the successful cellular antiviral response.
Eukaryotic cells have evolved stress-responsive pathways to cope with various environmental challenges. A central feature of the cellular stress response is the reprogramming of mRNA translation (21). One of several signaling pathways that control translation during stress is the eIF2α pathway, which regulates the recruitment of the initiator methionine by the translation initiation factor eIF2. A family of protein kinases phosphorylate a common site in eIF2α (serine 51), the regulatory subunit of eIF2. This phosphorylation inhibits eIF2 function, limiting preinitiation complex formation and reducing translation initiation (48). Each of the eIF2α kinases, which include protein kinase R (PKR), heme-regulated eIF2α kinase, general control nonderepressible 2, and PKR-like endoplasmic reticulum (ER) kinase, is activated in response to different environmental stresses such as oxidative and ER stress, heat shock, amino acid deprivation, and hemin deficiency. Some are also activated by immune signaling cascades and/or assault by pathogens, including viruses (26, 31, 37). A key consequence of eIF2α phosphorylation is the formation of cytoplasmic stress granules (SGs) (4), foci in which stalled mRNP (messenger ribonucleoprotein) complexes accumulate following translational arrest. SGs are sites of mRNA triage where mRNPs are assigned specific fates through their interactions with an array of RNA-binding proteins and their interactors. These include factors involved in RNA editing and processing, translational silencing, and microRNA processing.
The formation of SGs can be triggered by translational stalling caused by eIF2α phosphorylation, during which ribosomes run off the mRNA transcript, leaving an abundance of mRNA molecules released from polysomes (2). These nonpolysomal mRNA molecules interact with specific RNA-binding proteins, which direct their assembly into SGs or other types of RNA granules (e.g., processing bodies). The protein composition of the mRNP complex largely determines the type of RNA granule into which it is assembled, and protein-protein interactions between RNA-binding proteins appear to drive this process. The cellular proteins TIA-1 and G3BP play critical roles in SG assembly, forming aggregates via inter- and intramolecular interactions (5, 16, 46). These proteins interact with various other factors, including, in the case of G3BP, USP10 (a deubiquitinating enzyme) and caprin-1 (a cell cycle-associated RNA-binding protein with several putative functions, including vaccinia virus [VV] intermediate gene transcription [20, 43]). Other factors found in SGs include translation initiation factors such as eIF3 and small ribosomal subunits. Large ribosomal subunits are notably absent from SGs, because translational stalling occurs at a stage before the large subunit can join the translation complex (5).
To date, both pro- and antiviral activities have been ascribed to SG formation or inhibition, and this is often indicative of viral subversion of cellular stress responses (9). Several viruses alter SG formation and composition in order to promote their replicative cycle (9, 35, 41). For example, orthoreovirus particles induce and localize to SGs at early times in infection, and this step may promote the viral life cycle (36). The poliovirus proteases actively cleave G3BP, PABP, and eIF4G during infection (49), but nevertheless, stable SGs containing TIA-1 and positive-sense mRNAs form; these altered SGs apparently have no antiviral effect and may instead be proviral, promoting host cell shutoff (35).
Conversely, several viruses appear to inhibit SG formation, either throughout the cytoplasm like Sendai virus and rotavirus (18, 33) or in a localized area of the cell like Semliki Forest virus (32). West Nile virus (WNV) recruits TIA-1 and TIA-R to the 3′ stem-loop of its negative-sense RNA, and TIA-R appears to promote viral replication, as TIA-R knockout (KO) mouse embryonic fibroblasts (MEFs) display reduced viral replication (14). However, TIA-1 KO MEFs exhibit normal WNV replication, and no SG formation during infection is observed. TIA-1 KO MEFs showed increased viral production using vesicular stomatitis virus, Sindbis virus, and herpes simplex virus type 1 (HSV-1) (28).
VV is a large DNA virus with a double-stranded DNA genome and is the prototypic member of the Orthopoxvirus family, which also includes variola virus (the causative agent of smallpox) and monkeypox virus (an emerging zoonotic pathogen, [15]). Poxviruses have evolved elaborate ways to evade both the adaptive and passive immune responses and to prevent host cell antiviral responses from interrupting their replication (42). VV has not previously been thought to induce canonical SG formation, although the SG marker protein G3BP and its interacting partner caprin-1 have previously been shown to localize inside the viral replication factories during wild-type (WT) VV infection (19, 47). In addition to the report of G3BP and caprin-1 localization inside factories, the translation factors eIF4E, eIF4G, and PABP have been reported to accumulate in factories during a productive infection. Little is known about the behavior of these proteins during a nonproductive infection.
Infection of cells with VV lacking the E3L gene is a well-established model of nonproductive virus infection. The E3L protein is a double-stranded RNA (dsRNA)-binding protein that is thought to prevent host antiviral responses by sequestering dsRNA during infection. Cells infected with viruses lacking the E3L gene (ΔE3L mutant viruses) have previously been shown to activate the dsRNA-binding kinase PKR (11, 27). Replication of the ΔE3L mutant virus in vitro typically shows a 1- to 2-log10 defect compared to WT viruses (51). This defect is largely attributed to reduced viral protein translation following PKR activation, which is believed to block VV replication at the intermediate gene expression step (30, 51). Viral gene expression can be recovered by expressing proteins that sequester dsRNA (8). This has established the importance of host factor recognition of dsRNA as an initiating factor in the suppression of VV replication.
We were interested in investigating the events downstream of dsRNA recognition and PKR activation that contribute to the inhibition of VV replication. Specifically, we were interested in whether eIF2α phosphorylation is the final important step required for restricting VV replication or whether there is evidence that other functional proteins downstream of PKR activation and eIF2α phosphorylation contribute to the antiviral response. Specifically, we investigated whether the mechanism of translation inhibition during ΔE3L mutant virus infection might involve SG formation or antiviral functions of SG-associated cellular proteins.
MATERIALS AND METHODS
Cell lines.
U2OS cells were the kind gift of David Sabatini (Whitehead Institute for Biomedical Research). HeLa, BHK-21, and Vero E6 cells were obtained from ATCC. MEF cell lines lacking PKR, or expressing mutant eIF2αS51A, with their corresponding WT partners, were the kind gift of R. Kaufman (University of Michigan, Ann Arbor) (39, 40). TIA-1 KO MEFs and control lines were generated in our lab. All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with glutamine and 10% fetal calf serum (FCS, D10). MEF cell lines were immortalized via transfection with simian virus 40 large T antigen and passaged <20 times during the course of the studies described here to avoid any changes in their genetic material. Yellow fluorescent protein (YFP)-tagged ribosomal large-subunit protein 7a (YFP-RPL7a) cells were generated in our lab as previously described (23). Briefly, a construct of RPL7a incorporating the YFP coding sequence at its C terminus was generated using PCR and a template obtained from Open Biosystems. Stable transfectants were generated in U2OS cells selected by drug resistance (kanamycin) and subcloning by limiting dilution. The function of the YFP-tagged protein was confirmed by its inclusion in polysomal fractions (data not shown).
Viruses, viral infections, and plaque assays.
The Western Reserve (WR) strain of VV was obtained from B. Moss (NIAID, NIH) and grown in HeLa S3 cells. Virus stock titers were determined in triplicate on Vero cells, and virus was stored in aliquots at −80°C.
ΔE3L mutant virus was generated through homologous recombination to replace the entire E3L open reading frame (ORF) with the ORF of mCherry (Clontech 632523). Overlap PCR was used to generate a PCR product with the mCherry ORF flanked on either side by 500 bp of the sequences upstream and downstream of the E3L coding sequence. This PCR product was transfected into BHK-21 cells following infection with WT VV by using standard methods. Plaques containing recombinant viruses were identified by fluorescence microscopy following expression of mCherry and plaque purified three times on BHK-21 cells. Viral stocks were produced by inoculation of BHK-21 cells at a multiplicity of infection (MOI) of 0.01 and harvested by following standard protocols (13). Insertion sites and sequences were confirmed by directly sequencing a PCR product generated from infected cell lysates using primers flanking the insertion cassette. The ΔE3L mutant virus was replication competent in BHK-21 cells but not in HeLa or Vero cells, consistent with previous reports.
The ΔE3L-F17R-Venus virus was generated through homologous recombination by using the parental ΔE3L mutant virus described above. The F17R promoter was selected because this ORF has previously been shown to exhibit “late” gene expression kinetics (34). Briefly, overlap PCR was used to generate a PCR product with the F17R promoter sequence followed by the Venus ORF (Addgene 11931) immediately followed by a stop codon. This cassette was flanked on either side by 500 bp of the sequences upstream and downstream of the insertion site in the intergenic region between J4 and J5. This site has been previously shown to tolerate insertions well. Recombinant virus was generated, cloned, and sequenced as described for the ΔE3L mutant virus above.
For multiple-step growth curve experiments, cells were plated in six-well dishes at 90% confluence. Virus was added at an MOI of 0.01, and cells were incubated with frequent rocking for 1 h at 37°C. Cells were then washed five times with phosphate-buffered saline (PBS), and 1 ml DMEM-2% FCS (D2) was added. Infection times (hours postinfection [hpi]) were measured from this time point. At different time points, cells were harvested by scraping into the culture medium and stored at −80°C until plaque assays were performed. Single-cycle growth curve experiments were performed exactly as described for multiple-cycle experiments, except that virus was added at an MOI of 5. Cells were washed 10 times with PBS 1 h later to remove input virus.
Plaque assays were performed with confluent Vero (WT virus) or BHK-21 (ΔE3L mutant virus) cells in 12-well plates (for single-cycle assays, the titers of both viruses were determined on BHK cells). Samples were subjected to three cycles of rapid freeze-thawing using liquid nitrogen, with thawing at 37°C. Cell debris was removed by spinning at 1,000 rpm for 3 min, and samples were serially diluted 1:10 for plaque assay. An inoculum of 250 μl was used, with frequent rocking for 1 h at 37°C. Cells were then overlaid with D2 and incubated at 37°C for 36 h. Following incubation, cells were either stained using a 0.2% crystal violet solution (WT virus plaque assays) or fixed in 2% paraformaldehyde (PFA) in PBS (ΔE3L mutant virus assays). Plaques were counted by eye (Vero cells) or using mCherry expression as a marker for ΔE3L mutant virus replication. Note that under these fixation conditions (without permeabilization or prolonged washing), the mCherry protein was detectable by conventional fluorescence microscopy. ΔE3L mutant virus plaques were visualized as patches of mCherry-expressing cells with staining distributed throughout the cytoplasm.
IF and drug treatment.
For immunofluorescence (IF) experiments, virus was used to infect U2OS or HeLa cells at an MOI of 5. Cells were incubated at 37°C for 1 h, with frequent rocking. After 1 h, the inoculum was removed, cells were washed three times with PBS, and D2 was added. Infection times (hpi) were measured from this time point.
For experiments using sodium arsenite (NA; Sigma) and cycloheximide (CHX; Sigma), HeLa cells were infected with virus at an MOI of 5 as described above. At 5 hpi, NA was added as a dilution in PBS at a concentration of 0.1 mM. One hour later, medium was replaced with D2 containing 100 μg/ml CHX dissolved in dimethyl sulfoxide (DMSO). An equal volume of DMSO was added to the control medium. At 7 hpi, cells were fixed and processed for IF staining as described below.
At the indicated time points, cells were washed gently with PBS and fixed in 4% PFA (Electron Microscopy Sciences) for 20 min at room temperature. They were then washed in PBS and permeabilized using methanol at −20°C for 2 min with rocking. The methanol was removed, and the cells were incubated in blocking buffer (PBS containing 4% horse serum) overnight at 4°C. The next day, the cells were stained using antibodies diluted in blocking buffer for 1 h, followed by extensive washing. Secondary antibodies were diluted 1:1,000 in blocking buffer and applied for 30 min. It should be noted that the mCherry protein produced by the ΔE3L mutant virus was only weakly detectable by conventional fluorescence microscopy with the fixation and staining protocol used (data not shown). This allowed us to stain ΔE3L mutant virus-infected cells using antibodies coupled to red fluorophores, which were much brighter than the dim mCherry signal. Samples were washed and mounted using ProLong Antifade Gold (Invitrogen). Samples were viewed using a Zeiss Axioplan2 microscope with a 63× objective, an Apotome device, and Axiovision software or using a Perkin-Elmer Ultraview spinning-disc confocal microscope with a 100× objective. Montages were assembled using Adobe Photoshop.
Antibody to USP10 was obtained from Bethyl Laboratories (A300-900A). Antibodies to G3BP, eIF3b, and TIA-1 were obtained from Santa Cruz Biotechnology Inc. (catalog numbers sc70283, sc137215, and sc166246). Caprin antibody was obtained from the Protein Tech Group, the anti-FMRP antibody used was Santa Cruz sc-101048, the anti-FXR2 antibody used was Santa Cruz sc-32266, and the goat anti-FXR1 antibody used was sc-10544. DAPI (4′,6-diamidino-2-phenylindole) and secondary antibodies raised in donkeys and conjugated to Alexa 488 or 568 were obtained from Invitrogen.
Fluorescence in situ hybridization (FISH).
Briefly, cells were infected or mock infected as described above before fixation in 4% PFA for 20 min at room temperature. Cells were permeabilized in ice-cold methanol for 5 min, washed in PBS, and permeabilized further at 4°C in 70% RNase-free ethanol overnight. Coverslips were rehydrated in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer (Sigma S6639) before prehybridization with 2 ng probe [5′-fluorescein-oligo(dT) 50; Genelink] per coverslip in hybridization buffer (25% formamide [Ambion AM9342], 0.05 M EDTA, 10% dextran sulfate sodium salt [Sigma], 1× Denhardt's solution [Sigma D2532], 0.5 mg/ml tRNA [Sigma R9001], 1 mM ribonucleoside vanadyl complex [Fluka 94742]) for 1 h at 60°C. Hybridization was carried out in hybridization buffer plus 2 ng probe per coverslip for 5 min at 65°C and 4 h at 40°C in the dark. Cells were washed in 2× SSC, counterstained with DAPI to highlight DNA and viral replication factories, and mounted on microscope slides as described above. Samples were viewed on a Zeiss Axioplan 2 microscope with a 63× objective, an Apotome device, and Axiovision software.
SDS-PAGE and Western blotting.
Cells for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were plated in 12-well plates at 90% confluence and infected with WT or ΔE3L mutant virus at an MOI of 5. Following 1 h of incubation and three washes with PBS, the cells were overlaid with D2 and incubated at 37°C for 6 h, with the time starting from the last wash. To harvest samples, medium was aspirated and cells were lysed on ice using radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitors (Roche Complete Mini protease inhibitor cocktail) and phosphatase inhibitors (Sigma P2850) according to the manufacturer's instructions. SDS-PAGE running buffer was added, and samples were loaded onto 4 to 15% gradient Tris-HCl gels (Bio-Rad) after being heated at 95°C for 10 min. Rainbow Full Range protein gel markers were loaded onto each gel (GE Healthcare). Western blotting was carried out using the Bio-Rad minielectrophoresis system according to the manufacturer's instructions. Membranes were blocked overnight in PBS-0.2% Tween 20-4% nonfat dry milk before incubation with primary antibody. Antibody to eIF2αP was obtained from Cell Signaling Technology (catalog no. 9721). Anti-rabbit antibody conjugated to horseradish peroxidase was obtained from Santa Cruz Biotechnology Inc., and the signal was detected by incubation with ECL reagents (Invitrogen) and exposure to X-ray film (Kodak BioMax).
RESULTS
ΔE3L mutant VV replicates poorly in U2OS cells and displays altered viral factory (VF) morphology.
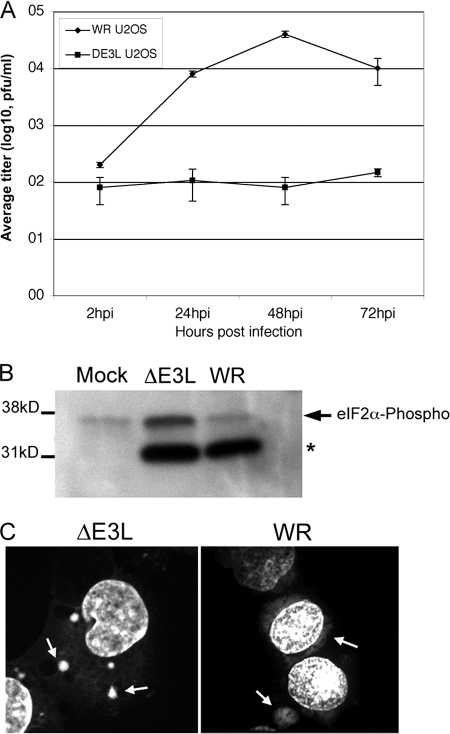
We constructed a mutant VV lacking the E3L gene (ΔE3L mutant VV) as described in Materials and Methods. Consistent with previous work, we found that this virus replicated poorly in U2OS and Vero cells but replicated to high titers in BHK cells (data not shown). We further characterized the replication of this virus in U2OS cells, as previous studies on SG structure and function have been performed with this cell line (Fig. 1 A). In a multiple-step growth curve, ΔE3L mutant VV replicated poorly in U2OS cells compared to the WT virus (WR strain). Titers of the ΔE3L mutant virus did not increase significantly across a 72-h period, whereas the WT virus grew to a titer of 5 × 106 PFU. Because the defect in ΔE3L mutant VV replication has previously been linked to its inability to prevent PKR activation (10-12), we collected samples for SDS-PAGE and probed Western blots for phosphorylated eIF2α (eIF2αP). Compared to those in mock-infected cells, the levels of eIF2α phosphorylation were not elevated in cells infected with the WT virus at 6 hpi (Fig. 1B). However, consistent with previous studies, the ΔE3L mutant virus caused a significant increase in levels of phosphorylated eIF2α (10-12). A cross-reactive polypeptide was detected in infected samples (*) but was easily distinguished from eIF2α by size.
FIG. 1.
Replication of WT and ΔE3L mutant VVs in U2OS cells. (A) ΔE3L mutant or WT virus was used to infect U2OS cells at an MOI of 0.01 in triplicate. At the time points indicated, cells were harvested and viral titers were measured by plaque assay on Vero E6 (WR = WT) or BHK-21 (ΔE3L mutant virus) cells. Error bars show 1 standard deviation. (B) Western blotting was performed using equal samples of U2OS cells infected with ΔE3L mutant or WT virus at an MOI of 5 for 6 h. Blots were probed with antibody to phosphorylated eIF2α. A cross-reactive smaller polypeptide bound the antibody in infected cells (*). (C) Replication factory formation was assessed in U2OS cells infected with ΔE3L mutant or WT virus at an MOI of 5. Cells were fixed at 6 hpi and stained using DAPI to detect DNA. Arrows indicate cytoplasmic viral replication factories. Z series were collected using a confocal microscope; a maximum-intensity projection algorithm was used to generate the images shown.
U2OS cells infected with ΔE3L mutant or WT VV were clearly identifiable by the presence of cytoplasmic viral replication factories (Fig. 1C, arrows). In ΔE3L mutant virus-infected cells, these replication factories were different in appearance from those routinely observed during WT VV replication. Factories in ΔE3L mutant virus-infected cells were more abundant, at an average frequency of 5.3 per cell (n = 25), while factories in WT-infected cells had an average frequency of 1.7 per cell (n = 25). In addition, factories in ΔE3L mutant virus-infected cells were generally smaller and of a more uniform round shape than those observed in WT-infected cells at the same time point.
G3BP forms cytoplasmic granules during infection with ΔE3L mutant VV infection.
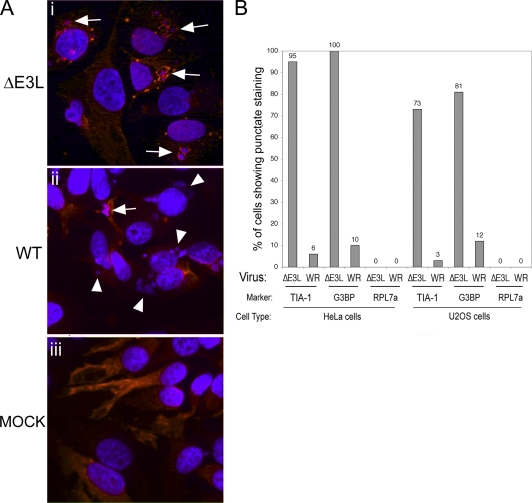
We were interested in how the SG-associated protein G3BP localized during the replication cycle of a ΔE3L mutant virus. To investigate this, both HeLa cells and U2OS cells were infected with ΔE3L mutant or WT VV or mock infected. At 7 hpi, a time when both WT and ΔE3L mutant virus infections should be well established, cells were fixed and stained for G3BP. In addition, we stained other infected cells for TIA-1 or the large ribosomal subunit protein RPL7a.
We found that during infection of HeLa or U2OS cells with WT VV, G3BP was generally found distributed throughout the cytoplasm in a majority of the cells (Fig. 2 A), similar to the staining seen in mock-infected cells, except that it formed distinct cytoplasmic granules in a small percentage of the cells (Fig. 2A, parts iii and ii). When this was quantified, there was punctate G3BP staining around approximately 10 to 12% of the DNA-stained VFs (Fig. 2B). In WT VV-infected cells, TIA-1 also occasionally formed cytoplasmic granules, though TIA-1 granules were slightly less frequent (3 to 6%). In contrast, during infection with the ΔE3L mutant virus, 81 to 100% of the factories found in cells contained cytoplasmic granules that stained with G3BP (Fig. 2A, part i, and B). A similar frequency of TIA-1 accumulation was observed during ΔE3L mutant VV infection (73 to 95% of factories). No factories containing RPL7a were observed during infection with WT or ΔE3L mutant VV in either cell line, suggesting that these accumulations do not represent sites of active translation. Owing to their higher frequency in cells infected with the replication-deficient ΔE3L mutant virus, we posit that they are part of the host antiviral response, and for simplicity's sake, we refer to these granules as antiviral granules (AVGs).
FIG. 2.
SG markers accumulate around VFs with different frequencies during ΔE3L mutant and WT VV infections. (A) HeLa cells were infected with ΔE3L mutant (i) or WT (ii) VV at an MOI of 5 or mock infected (iii). Cells were fixed at 7 hpi and stained with antibodies to G3BP (red) and with DAPI (blue). Fields of view collected at ×40 magnification are shown. Arrows show accumulations of G3BP in and around VFs. Arrowheads show VFs lacking obvious G3BP accumulations. (B) HeLa or U2OS cells were infected with ΔE3L mutant or WT VV at an MOI of 5 at 7 hpi, and cells were fixed and stained for the SG marker protein TIA-1 or G3BP or for the large ribosomal subunit protein RPL7a. DAPI was used to stain viral replication factories. DAPI-stained replication factories associated with each protein were counted by microscopy and expressed as a percentage of the total number of factories. Factories from 10 fields of view were counted for each sample (n > 50 in all cases).
AVGs contain SG-associated proteins and localize to viral replication factories in ΔE3L mutant virus-infected cells.
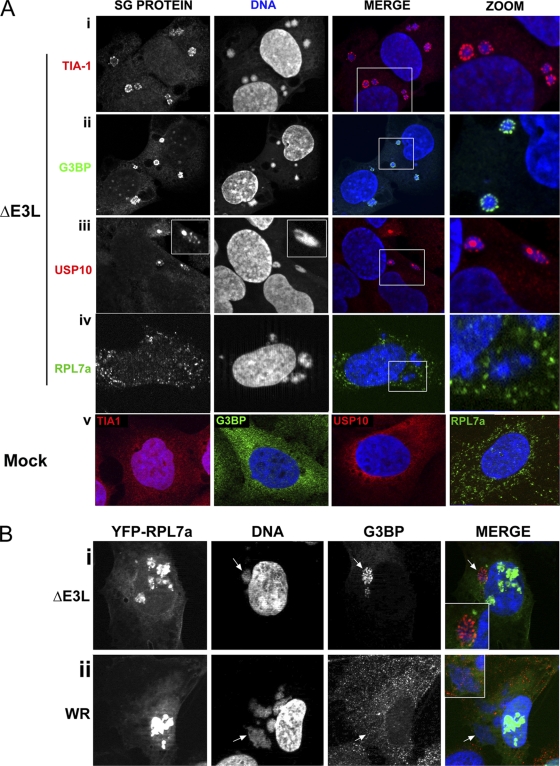
As elevated eIF2α phosphorylation was induced by ΔE3L mutant virus infection and proteins such as TIA-1 and G3BP are known to form cytoplasmic SGs in response to eIF2α phosphorylation, we hypothesized that the AVGs shown in Fig. 2 could represent small SGs. Therefore, we further characterized these AVGs by staining for additional protein markers of SGs (USP10) and for other SG components that are also G3BP binding partners, caprin-1 and USP10 (44). Cells were costained using DAPI as a marker for DNA to locate cytoplasmic viral replication factories.
In mock-infected cells, the SG marker TIA-1 was distributed in both the cytoplasm and the nuclei of mock-infected cells (Fig. 3 A, part v, red) (22, 24). G3BP was distributed diffusely throughout the cytoplasm (Fig. 3A, part v, green). In cells infected with the ΔE3L mutant virus, the pattern of TIA-1 and G3BP staining was dramatically altered. Both TIA-1 and G3BP were concentrated in distinct AVGs which were closely associated with the periphery of viral replication factories (Fig. 3A, parts i and ii). Serial Z sections confirmed that the AVGs were surrounding and in some cases embedded in replication factories; Z sectioning also showed that G3BP staining was observed in gaps in the DAPI staining that marks VFs, as described previously (19; see Fig. S1 in the supplemental material). This granule was not a cell type-specific response to VV infection, as similar granules containing TIA-1 and G3BP were observed when these experiments were performed with HeLa cells (data not shown and Fig. 2).
FIG. 3.
Characterization of granules during infection with ΔE3L mutant VV. (A) U2OS cells were infected with ΔE3L mutant virus at an MOI of 5 (i to iv) or mock infected (v). Cells were fixed at 7 hpi and stained with antibodies to the SG components TIA-1 (i and v, red), G3BP (i and v, green), and USP10 (i and v, red) or with antibody to the large ribosomal subunit protein RPL7a (iv, green). Cells were imaged at a magnification of ×63. (B) U2OS cells stably expressing a YFP-RPL7a protein (green) were infected with ΔE3L mutant (i) or WT VV (ii) at an MOI of 5. Cells were fixed at 7 hpi and stained with antibodies to G3BP (red). DAPI was used to stain DNA. Arrows identify a VF in ΔE3L mutant virus-infected cells. Boxed areas show zoomed regions.
USP10, a known interacting partner of G3BP (17, 44) also appeared to form AVGs that surround the ΔE3L mutant VF. In mock-infected cells, USP10 showed a diffuse, punctate cytoplasmic distribution (Fig. 3A, part v, red). During infection with the ΔE3L mutant virus, it displayed an altered distribution similar to that of G3BP; it accumulated in AVGs surrounding and embedded in viral replication factories (Fig. 3A, part iii). Caprin-1, an alternate binding partner of G3BP, also localized to punctate structures surrounding VFs in ΔE3L mutant virus-infected cells (data not shown) and the RNA-binding proteins fragile X (FMRP) and fragile X related (FXR1, FXR2) also localized to punctate structures (see Fig. S2 in the supplemental material).
To determine whether AVGs could represent areas of ongoing or stalled translation, we stained ΔE3L mutant virus-infected cells with antibodies to RPL7a, a large ribosomal subunit protein. In mock-infected cells, RPL7a distribution was diffusely cytoplasmic (Fig. 3A, part v, green). In ΔE3L mutant virus-infected cells, no change in this localization was observed (Fig. 3A, part iv, green). RPL7a staining remained cytoplasmic and did not accumulate in VFs or granules. In order to verify these results, we infected U2OS cells stably expressing YFP-tagged RPL7a with ΔE3L mutant or WT VV at an MOI of 5. No RPL7a-YFP protein was found to localize to WT VFs or to G3BP-labeled granules in ΔE3L mutant VV-infected cells (Fig. 3B). Instead, RPL7A-YFP was nucleolar (consistent with the synthesis of new ribosomes) and distributed throughout the cytoplasm. These results suggested that AVGs are not centers of translation.
ΔE3L mutant VV-induced AVGs are multiprotein structures.
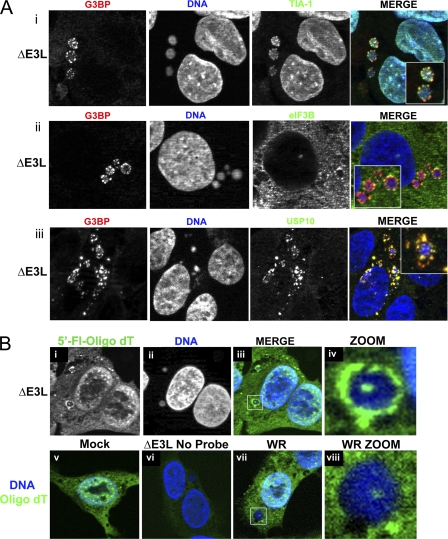
To determine whether all of the proteins we saw in granules were in separate granules, or if these proteins colocalized in the same granules during ΔE3L mutant VV infection, infected U2OS cells were double stained for G3BP and TIA-1 or for G3BP and eIF3b, a protein that can be found in SGs. Infected HeLa cells were double stained for G3BP and USP10. TIA-1 (green) and G3BP (red) showed partial colocalization in AVGs surrounding and embedded in viral replication factories (Fig. 4 A, part i). All AVGs appeared to contain both TIA-1 and G3BP, although the relative levels of the two proteins varied, resulting in a varied appearance of colocalization. Similar results were obtained when cells were double stained for G3BP and eIF3b (Fig. 4A, part ii). Here, the eIF3b (green) only partially coincided with G3BP (red). G3BP appeared to be the predominant component of AVGs, whereas eIF3b appeared only at their periphery. Moreover, AVGs containing G3BP frequently contained little or no detectable eIF3b (data not shown), suggesting that eIF3 is a minor component of AVGs. In HeLa cells, G3BP and USP10 showed only partial colocalization in granules, suggesting that these proteins may not physically interact at these structures (Fig. 4A, part iii, and data not shown). Similar colocalization studies were performed with some of the rare AVGs observed during WT VV infection to determine whether the structures formed were analogous; similar levels and similar patterns of colocalization were observed (see Fig. S3 in the supplemental material) (data not shown).
FIG. 4.
Multiple markers of SGs partially colocalize in granules, and granules contain poly(A) RNA. (A) U2OS (i and ii) or HeLa (iii) cells were infected with ΔE3L mutant virus at an MOI of 5. Cells were fixed at 7 hpi and costained with antibodies to the SG/granule components TIA-1 (i, green) and G3BP (i, red) or eIF3b (ii, green) and G3BP (ii, red) and with DAPI to stain DNA. (B) U2OS cells were infected with ΔE3L mutant (i to iv and vi) or WT (vii and viii) VV at an MOI of 5 or mock infected (v). At 7 hpi, cells were fixed and processed for FISH using a fluorescein-tagged oligo(dT) probe to localize mRNA. As a control, a sample infected with ΔE3L mutant virus was incubated without probe (vi). Maximum-intensity projections are shown. Boxed areas show zoomed regions.
Because both G3BP and TIA-1 are known to interact with mRNA to drive SG formation, we stained infected U2OS cells using a fluorescently tagged oligo(dT) RNA probe (Fig. 4B) to determine whether AVGs contain mRNA. Poly(A)-containing RNA accumulated in a pattern resembling that of AVGs in ΔE3L mutant virus-infected cells, showing a granular distribution around the periphery of the factory and occasionally within its center. Oligo(dT) probe colocalized with G3BP-positive AVGs in ΔE3L mutant virus-infected cells (data not shown). In WT VV-infected cells, oligo(dT) staining was detected in the center of VFs, in a distribution similar to that shown previously (19). However, probe was not detected surrounding the factories (Fig. 4B, part ii). Therefore, infection of U2OS and HeLa cells with the ΔE3L mutant VV induces the formation of cytoplasmic, translation-inactive AVGs. These AVGs contain poly(A) RNA, SG-associated cellular proteins and their known binding partners, and they form around (and occasionally inside) viral replication factories.
ΔE3L mutant VV-induced AVGs are not stress granules.
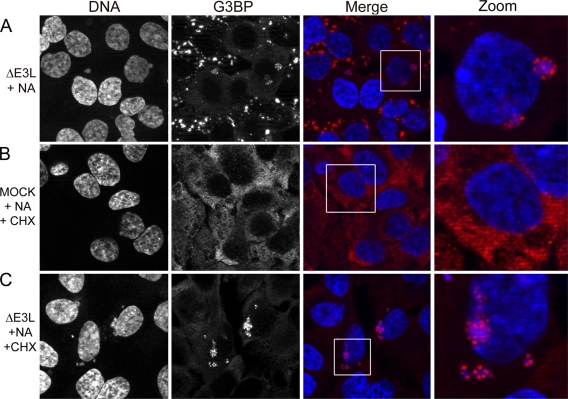
Because AVGs regularly stained positive for several SG-associated proteins (TIA-1, G3BP) but seemed to have less of other SG-associated proteins (e.g., eIF3), we asked whether granules are a uniquely localized type of SG or whether they represent a distinct structure uniquely triggered during ΔE3L mutant VV infection. To do this, HeLa or U2OS cells were infected with ΔE3L mutant VV at an MOI of 1 and treated for 2 h starting at 5 hpi with NA (a chemical known to stall translation and trigger SG formation via oxidative stress) to determine whether AVGs were redistributed throughout the cytoplasm. In ΔE3L mutant VV-infected cells, G3BP was found in AVGs surrounding VFs and not elsewhere in the cytoplasm following NA treatment (Fig. 5 A). In uninfected cells found in the same field of view, SG-like structures (scattered throughout the cytoplasm) were obvious (i.e., those lacking factories). Thus, AVGs were distinct from SGs in their localization, remaining closely associated with VFs even during NA treatment.
FIG. 5.
Granule formation appears to inhibit further SG formation, and granules are stable following treatment with CHX. U2OS cells were infected with ΔE3L mutant virus at an MOI of 1 (A, C) or mock infected (B). Some samples were treated with 0.1 mM NA at 5 hpi and fixed at 7 hpi (A). Further samples were treated with 0.1 mM NA at 5 hpi for 1 h and then treated with 100 μg/ml CHX for 1 h and fixed at 7 hpi (B, C). Postfixation, cells were stained for G3BP (red) and with DAPI (blue) to stain DNA. Maximum-intensity projections are shown.
This result suggested to us that AVGs might be highly stable structures. It is known that SGs are dismantled by treatment with CHX (22), which stalls translation by “freezing” ribosomes on mRNA molecules, preventing translation elongation and ribosomal runoff. This leads to the dismantling of cellular SGs because SGs are dynamic structures; without a source of new translation initiation complexes, SGs disperse (3). Mock-infected cells treated with CHX displayed complete dissolution of arsenite-induced SGs (Fig. 5B). In cells infected with ΔE3L mutant virus and treated with CHX at 6 hpi for 1 h, AVGs did not decrease in frequency and remained associated with viral replication factories in ΔE3L mutant VV-infected cells (Fig. 5C). The persistence of AVGs following a treatment that dissolves SGs indicates that AVGs make up a distinct cellular structure which is more stable than a canonical SG.
Granule formation requires the action of the PKR/eIF2α pathway.
Infection with ΔE3L mutant VV leads to PKR activation and eIF2α phosphorylation (11, 27, 50-52). As AVGs appeared only under conditions where eIF2α phosphorylation increased, we hypothesized that activation of the PKR/eIF2α pathway served as the trigger for AVG formation. If this hypothesis were correct, defects in the PKR signaling pathway should prevent both eIF2α phosphorylation and AVG formation, resulting in higher viral titers. Therefore, we asked whether ΔE3L mutant virus replication is rescued in MEFs lacking the PKR gene (PKR KO) or in MEFs expressing a mutant eIF2α protein where the regulatory phosphorylation site at serine 51 was mutated to the unphosphorylatable alanine (eIF2αS51A, AA cells) and whether this rescue correlated with a change in AVG formation.
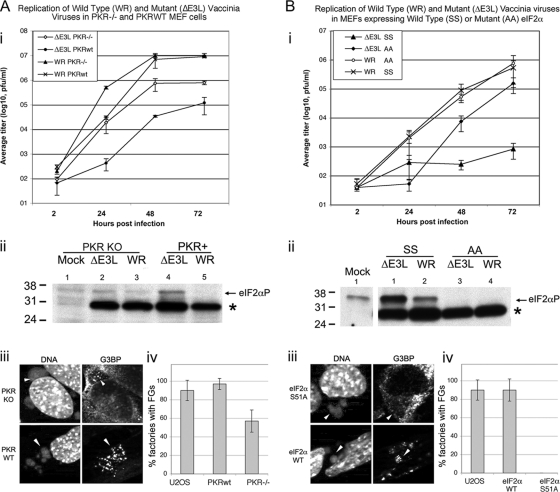
WT MEFs or PKR KO MEFs were infected with WT or ΔE3L mutant VV at an MOI of 0.01 in triplicate, and viral replication was measured at 2, 24, 48, and 72 hpi. WT VV replicated equally well in both MEF lines (Fig. 6 A, part i). ΔE3L mutant VV replicated poorly in WT MEFs. In PKR KO MEFs, replication of the ΔE3L mutant virus was increased by a factor of 6.6 relative to growth in WT MEFs at 72 hpi. This level of rescue is consistent with that previously shown in cells lacking PKR activity (50). In PKR KO cells, there was a decreased accumulation of phosphorylated eIF2α (Fig. 6A, part ii), but some phosphorylation was still observable.
FIG. 6.
Replication of WT and ΔE3L mutant VVs in (A) PKR KO and (B) eIF2αS51A MEFs and matched WT controls (A and B, parts i). Cells were infected with WT or ΔE3L mutant viruses at an MOI of 0.01 in triplicate. Samples were harvested at 2, 24, 48, and 72 hpi, and viral titers were measured by plaque assay on BHK (ΔE3L mutant virus) or Vero (WR) cells. Error bars show 1 standard deviation. (A and B, parts ii) Cells were infected with ΔE3L mutant or WR virus at an MOI of 5 or mock infected. At 6 hpi, cells were harvested for SDS-PAGE and Western blotting. Blots were probed with antibodies to phosphorylated eIF2α. A cross-reactive polypeptide was bound by the antibody in infected samples but was easily distinguishable from eIF2α (*). The values to the left are molecular sizes in kilodaltons. (A and B, parts iii) Cells were infected with ΔE3L mutant virus at an MOI of 5 and fixed at 6 hpi for staining with antibodies to G3BP and with DAPI to stain DNA. (A and B, parts iv) Cells were infected and stained as described above. Factories containing granules were counted and expressed as a percentage of the total number of factories. Error bars show 1 standard deviation.
We stained for G3BP in WT and KO PKR MEFs and calculated the frequency of AVG formation. Data from parallel experiments using U2OS cells were included for comparison (Fig. 6A and B, parts iv). In PKR WT MEFs, ΔE3L mutant virus infection triggered robust AVG formation, as determined using G3BP as a marker, with AVGs present around 97% of the VFs (Fig. 6A, parts iii [bottom] and iv). However, in PKR KO MEFs, AVG formation was reduced in frequency by ∼40% and qualitatively appeared smaller and less distinct (Fig. 6A, parts iii and iv).
We next asked whether eIF2α phosphorylation is required for AVG formation. WT (SS) and eIF2αS51A (AA) MEFs were infected with WT or ΔE3L mutant virus. At 72 hpi, production of ΔE3L mutant virions was increased by a factor of more than 180-fold in AA cells relative to that in matched WT MEFs (Fig. 6B, part i). The presence of unphosphorylatable eIF2α had no effect on WT VV replication. As expected, Western blot analysis showed that infection with the ΔE3L mutant virus stimulated eIF2α phosphorylation in WT MEFs but not in the AA knock-in cells (Fig. 6B, part ii). In MEFs containing WT eIF2α, granule formation was normal, but in MEFs lacking a phosphorylatable form of eIF2α, AVG formation was completely abolished. Thus, phosphorylation of eIF2α is critical for reducing the replication of VV lacking the E3L gene and for the formation of AVGs. Interestingly, the PKR KO cells do not rescue the virus as well as the AA cells, suggesting that PKR is not the only eIF2α kinase activated during VV infection.
Replication of ΔE3L mutant virus is increased in MEFs lacking the TIA-1 gene, despite elevated levels of eIF2α phosphorylation and AVG formation.
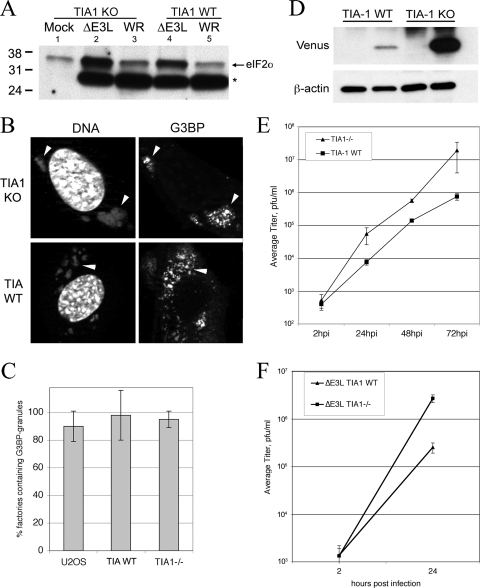
Based on the correlation we found between eIF2α phosphorylation levels and AVG frequency, we next tested whether AVGs actively inhibit viral replication downstream of eIF2α phosphorylation. We therefore asked whether TIA-1 (a component of AVGs) might be important for the antiviral activity of AVGs during ΔE3L mutant virus replication. We first determined whether MEFs lacking TIA-1 show altered eIF2α phosphorylation following ΔE3L mutant VV infection (Fig. 7 A). Western blot assays showed that ΔE3L mutant virus infection triggered a comparable increase in eIF2αP in both TIA-1 −/− and +/+ MEFs, relative to the level in the WT virus (Fig. 7A, ΔE3L mutant virus in lanes 2 and 4 and WT in lanes 3 and 5).
FIG. 7.
Replication of WT and ΔE3L mutant VVs in TIA-1 KO and WT MEF cells. (A) Cells were infected with ΔE3L mutant or WT VV at an MOI of 5 and harvested at 6 hpi for SDS-PAGE and Western blotting. Blots were probed with antibodies to phosphorylated eIF2α. A cross-reactive polypeptide was bound by the antibody in infected samples (*). The values to the left are molecular sizes in kilodaltons. (B) Cells were infected with ΔE3L mutant virus at an MOI of 5 and fixed at 6 hpi for staining with antibodies to G3BP and with DAPI. Arrowheads indicate virus factories. (C) Factories containing granules were counted and expressed as a percentage of the total number of factories. (D) Cells were infected with ΔE3L mutant virus expressing Venus under the control of the late F17R promoter at an MOI of 5. At 7 hpi, samples were harvested for SDS-PAGE and Western blotting and probed with antibodies to GFP (to detect Venus, upper panels) and beta-actin as a loading control (lower panels). (E) Cells were infected with ΔE3L mutant virus at an MOI of 0.01 in triplicate. Samples were harvested at 2, 24, 48, and 72 hpi, and viral titers were measured by plaque assay on BHK (ΔE3L mutant virus) or Vero (WR virus) cells. Error bars show 1 standard deviation. (F) Cells were infected with ΔE3L mutant virus at an MOI of 5 in triplicate. Samples were harvested at 2 and 24 hpi, and viral titers were measured by plaque assay on BHK cells. Error bars show 1 standard deviation.
Next, we quantified AVG formation in TIA-1−/− and TIA-1 +/+ MEFs infected with the ΔE3L mutant or WT virus, using G3BP to detect AVGs and DAPI to stain viral replication factories. No qualitative or quantitative difference in AVGs was detected between TIA-1−/− and TIA-1+/+ MEFs (Fig. 7B), consistent with the high levels of eIF2α phosphorylation seen in both cell types leading to AVG formation. However, it was notable that ΔE3L mutant VF morphology in the TIA-1−/− cells was more similar to the factories formed in WT virus-infected cells, suggesting that virus growth was being restored.
We investigated the possibility that TIA-1 KO MEFs were more permissive to ΔE3L mutant virus replication. Initially, we used a ΔE3L mutant virus that expressed the Venus fluorescent protein late during VV infection (using the F17R promoter). In WT MEFs, Western blotting for Venus expression showed that very little of the protein accumulated (Fig. 7D, lane 2) but in TIA-1 −/− MEFs, expression was significantly increased (lane 4), consistent with removal of this protein leading to restoration of DNA replication and late gene expression. In a multiple-cycle growth curve (Fig. 7E), the titer of the ΔE3L mutant virus was increased 25-fold in TIA-1 −/− MEFs compared to that in TIA-1 WT MEFs at 72 hpi. At 24 and 48 hpi, the titers of the ΔE3L mutant virus were increased 7-fold and 4-fold, respectively, compared to those seen during replication in TIA-1 +/+ MEFs. To confirm this phenotype, we also conducted single-cycle growth curve experiments with these cells to determine whether the improved virus growth was seen over a shorter time course of infection (Fig. 7F). These assays showed a similar increase in the replication of ΔE3L mutant VV. This confirmed that in the absence of TIA-1, the ΔE3L mutant virus evades the normal block of virus replication associated with the phosphorylation of PKR activation and eIF2α phosphorylation.
DISCUSSION
Here we show that cells respond to nonproductive VV infection by assembling cytoplasmic granules that surround viral replication factories. These granules require eIF2α phosphorylation and appear to be important for suppressing virus replication. These AVGs are similar to SGs in their protein content but are resistant to CHX-enforced disassembly, unlike SGs (1, 23). AVGs therefore appear more static and less dynamic than SGs. This suggests that AVGs are not responsible for holding mRNAs in a “paused” state for later release (like SGs) but that their function is solely to block VV replication.
AVGs appear to be distinct from other accumulations of host proteins that form at or in VV replication factories. Localized translation of viral mRNA has been proposed to occur inside VFs, as was elegantly demonstrated in cells that were infected with viruses expressing cyan fluorescent protein (CFP)- or YFP-tagged viral proteins (19). Similar reports describe localized recruitment of host translation initiation machinery (eIF4E, eIF4G, but not PABP) to specific subregions inside VFs (19, 47). This recruitment of host cell translation factors into VFs has been attributed to the successful viral hijacking of the host translational machinery. The AVGs that we define in these studies exhibit clear differences from the VF-internal structures described previously. (i) AVGs form primarily outside the VF (though there were some cases of these granules forming inside VFs). (ii) AVGs contain TIA-1, which was excluded from the VF-internal foci (47). (iii) AVGs appear earlier than the previously described foci. (iv) AVG assembly absolutely requires eIF2α phosphorylation, which is largely suppressed in WT VV-infected cells. (v) Most importantly, AVGs appear to be antiviral in nature, while the factory-internal granules are suggested to have a proviral function. AVGs are more abundant during ΔE3L mutant virus infection than during WT infection (AVGs surround factories in 73 to 100% of the cells infected with ΔE3L mutant VV, while only 3 to 12% of the WT-infected cells show similar structures). If active transcription or translation were associated with AVGs, we would expect to see more AVGs during a successful WT VV infection than during replication-limited ΔE3L mutant virus infection. Moreover, the requirement of increased eIF2α phosphorylation for the formation of AVGs argues strongly against their being a normal part of the VV replication cycle.
Requirements for AVG formation.
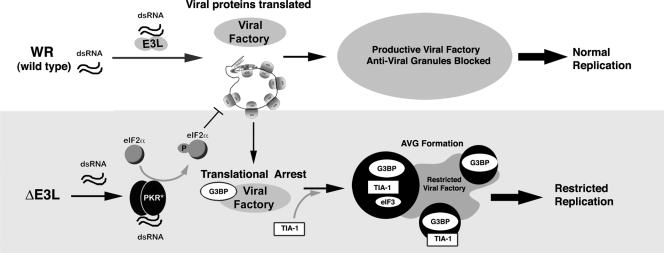
The requirement of eIF2α phosphorylation to trigger the formation of AVGs suggests a model for their assembly. We propose that E3L blockade of dsRNA sensing in WT VV infection allows the formation of large VFs that are not limited by AVGs in most cells (Fig. 8, top panel). In infections with VV lacking the E3L protein (bottom panel), dsRNA is available to activate PKR, which increases eIF2α phosphorylation and leads to AVG formation. A prediction of this model is that other VV mutants that induce eIF2α phosphorylation should also induce AVG formation (6, 29). It is not clear that eIF2α is actually a component of the AVGs, as IF staining did not conclusively show accumulation of this protein around ΔE3L mutant VFs, possibly owing to antibody cross-reactivity with an unknown viral protein. Rather, we speculate that phosphorylation of eIF2α may stall viral translation sufficiently to allow the proteins found in AVGs (RNA-binding proteins such as TIA-1, G3BP, FMRP, and FXR1) to form a nucleating structure on viral RNA, leading to AVG formation. Although speculative, this hypothesis is consistent with the known translational silencing properties of TIA-1, FXR1, and FMRP, the cage-like assemblies of AVGS that form around the VF, and their correlation with the inhibition of VV replication.
FIG. 8.
Model of the role of AVGs in the antiviral response to VV. Infection with WT virus (top) produces dsRNA, which is bound by E3L, preventing PKR activation and leading to a productive infection as eIF2α phosphorylation and AVG formation are suppressed by E3L. Infection with ΔE3L mutant virus (bottom) results in PKR activation, leading to eIF2α phosphorylation and delayed translation initiation. Factory-associated viral mRNAs are bound by the translational silencer TIA-1, which restricts viral replication. Abundant AVGs containing mRNA, G3BP, TIA-1, and associated proteins form.
There are two possible mechanisms of AVG function. One possibility is that AVGs sequester host factors required for replication away from the virus replication center. Our finding that G3BP and caprin-1 are concentrated in AVGs is of particular interest in this context, as G3BP and caprin-1 were proposed to facilitate the transcription of VV intermediate genes and to be required for intermediate viral transcription in vitro (20). G3BP and caprin-1 were detected inside WT VFs at an intermediate stage of VV infection (19). This recruitment was viewed as part of a successful infection. Though we observed little to no accumulation of either G3BP or caprin-1 in most VFs during WT virus infection, the quantitative accumulation of G3BP and caprin-1 in AVGs surrounding factories formed during infection with ΔE3L mutant VV is consistent with the interpretation that these proteins are sequestered in AVGs during ΔE3L mutant virus infection to block the progression of viral replication at the stage of intermediate transcription.
We find it significant that both caprin-1 and USP10 are present in AVGs and are interacting partners of G3BP. Caprin-1 is known to nucleate SGs when overexpressed and may regulate G3BP-mediated RNA transport, while USP10's deubiquitinating activity is regulated by association with G3BP (43, 44) and regulates membrane transport between the ER and the Golgi apparatus. The fact that both of these proteins are associated with AVGs suggests that one function of the AVG is to serve as a recruitment center for multiple different types of signals. A signaling center role is consistent with the apparent stability of AVGs, which may allow several different types of signals to be integrated in the vicinity of the VV replication center. Given the importance of dsRNA signaling in stimulating apoptosis in VV infection (25), it is also possible that apoptotic components are also present in AVGs.
A second potential AVG function is the recruitment of proteins with direct antiviral activity, such as TIA-1, to the vicinity of the VF. Previous work has pointed to an antiviral role for TIA-1 in HSV-1 and Sindbis virus replication (28), suggesting that the antiviral mechanism of TIA-1 is not restricted to poxviruses. However, TIA-1 also has a proviral role in the replication of flaviviruses (28), so its antiviral effect is not universal for all viruses. In unstressed cells, TIA-1 is predominantly nuclear and mediates alternative splicing. However, upon infection with ΔE3L mutant VV, TIA-1 accumulates in the cytoplasm and concentrates in the neighborhood of the replication center. Loss of TIA-1 results in increased replication of the ΔE3L mutant virus (Fig. 7), demonstrating its functional role in restricting infection. This suggests that TIA-1 may directly inhibit viral gene expression by binding viral mRNA and repressing its translation. Alternatively, TIA-1 recruitment to AVGs and consequent depletion from the nucleus might alter splicing of TIA-1 target genes and thereby allow a more robust antiviral response. Further work is needed to distinguish between these possibilities. It is of note that MEFs lacking TIAR, a close relative of TIA-1, do not rescue the replication of ΔE3L mutant VV (data not shown). Since TIA-1 and TIAR have different mRNA targets, this could indicate that TIA-1 specifically recognizes viral mRNA while TIAR does not.
Our data presented in Fig. 6 confirm previous studies showing that the dsRNA-activated kinase PKR is important in limiting the replication of the ΔE3L mutant virus (8, 11, 12, 50, 51). Our studies also suggest that multiple eIF2α kinases can contribute to eIF2 α phosphorylation in cells infected with the ΔE3L mutant virus, which slightly increases eIF2α phosphorylation even when PKR is absent. Consistent with the idea that eIF2α phosphorylation is a critical step in the restriction of ΔE3L mutant virus replication, infection of eIF2α S51Α cells incapable of phosphorylating eIF2α (but capable of activating PKR) with ΔE3L mutant virus resulted in a greater rescue of ΔE3L mutant virus replication and a more complete block of AVG formation. Unexpectedly, our experiments show that eIF2α phosphorylation is not the sole event required for VV replication. TIA-1 cells infected with the ΔE3L mutant virus supported virus replication despite an increase in eIF2α phosphorylation. Though not common, the replication of viruses that produce capped and poly(A) mRNAs in the presence of eIF2α phosphorylation has been noted previously (7).
This dissociation of eIF2α phosphorylation from VV replication argues strongly that phosphorylation of eIF2α per se does not restrict VV replication but that the induction of AVGs by this phosphorylation is required as well. The finding that cells lacking TIA-1 still form cytoplasmic granules but that the TIA-1-deficient granules do not block virus replication is in keeping with studies of virus disruption of SGs. For example, poliovirus degrades G3BP specifically and induces the formation of TIA-1-containing SGs, which do not inhibit virus replication. This suggests that virus evasion of the antiviral effects of cytoplasmic granules can take the form of blocking their formation or removing critical components (9, 28, 32, 35).
Elucidating the molecular mechanism by which TIA-1 inhibits ΔE3L mutant virus replication and fully investigating the composition and function(s) of these novel cytoplasmic granules are goals for future studies. The present study indicates that TIA-1 plays a previously uncharacterized role in the restriction of poxvirus replication. Previous work describes roles for TIA-1 and the related protein TIAR in cell homeostasis, growth, and cellular RNA metabolism, influencing mRNA splicing, translation and decay (38, 53). Previous findings suggest that TIA-1 suppresses HSV-1 and Sindbis viral replication, and interactions between TIA-1 and viral RNA transcripts during hepatitis B virus and WNV infections have been demonstrated (14, 45). Important remaining questions are whether TIA-1 restricts the replication of ΔE3L mutant virus through its interaction with viral RNA molecules during infection and whether similar granules form during infection with these other viruses.
Supplementary Material
Acknowledgments
We acknowledge Sara Johnston (USAMRIID) for help with experiments, Rachel Fearns and members of the Connor lab, for helpful critique of the manuscript, and Samantha Stewart, Sarah Tisdale, and Judy Yen for excellent technical support.
This work was supported by grants NIH R01 (AI 033600) and R01 (AR 051472) awarded to P.A. J.H.C. was supported in part by the Peter Paul Career Development Professorship.
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print on 8 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anderson, P., and N. Kedersha. 2006. RNA granules. J. Cell Biol. 172:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell. Biol. 10:430-436. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, P., and N. Kedersha. 2009. Stress granules. Curr. Biol. 19:R397-R398. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, P., and N. Kedersha. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141-150. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, P., and N. Kedersha. 2002. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backes, S., et al. 2010. Viral host-range factor C7 or K1 is essential for modified vaccinia virus Ankara late gene expression in human and murine cells, irrespective of their capacity to inhibit protein kinase R-mediated phosphorylation of eukaryotic translation initiation factor 2alpha. J. Gen. Virol. 91:470-482. [DOI] [PubMed] [Google Scholar]
- 7.Balachandran, S., and G. N. Barber. 2004. Defective translational control facilitates vesicular stomatitis virus oncolysis. Cancer Cell 5:51-65. [DOI] [PubMed] [Google Scholar]
- 8.Beattie, E., et al. 1996. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes. 12:89-94. [DOI] [PubMed] [Google Scholar]
- 9.Beckham, C. J., and R. Parker. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, H.-W., L. H. Uribe, and B. L. Jacobs. 1995. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J. Virol. 69:6605-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, H.-W., J. C. Watson, and B. L. Jacobs. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 89:4825-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, M. V., H.-W. Chang, B. L. Jacobs, and R. J. Kaufman. 1993. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J. Virol. 67:1688-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earl, P. L., B. Moss, L. S. Wyatt, and M. W. Carroll. 2001. Generation of recombinant vaccinia viruses. Curr. Protoc. Mol. Biol. Chapter 16:Unit16.17. [DOI] [PubMed] [Google Scholar]
- 14.Emara, M. M., and M. A. Brinton. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 104:9041-9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Essbauer, S., M. Pfeffer, and H. Meyer. 2010. Zoonotic poxviruses. Vet. Microbiol. 140:229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilks, N., et al. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irvine, K., R. Stirling, D. Hume, and D. Kennedy. 2004. Rasputin, more promiscuous than ever: a review of G3BP. Int. J. Dev. Biol. 48:1065-1077. [DOI] [PubMed] [Google Scholar]
- 18.Iseni, F., et al. 2002. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 21:5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsafanas, G. C., and B. Moss. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2:221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsafanas, G. C., and B. Moss. 2004. Vaccinia virus intermediate stage transcription is complemented by Ras-GTPase-activating protein SH3 domain-binding protein (G3BP) and cytoplasmic activation/proliferation-associated protein (p137) individually or as a heterodimer. J. Biol. Chem. 279:52210-52217. [DOI] [PubMed] [Google Scholar]
- 21.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 22.Kedersha, N., et al. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kedersha, N., S. Tisdale, T. Hickman, and P. Anderson. 2008. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448:521-552. [DOI] [PubMed] [Google Scholar]
- 24.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kibler, K. V., et al. 1997. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. J. Virol. 71:1992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohno, K. 2010. Stress-sensing mechanisms in the unfolded protein response: similarities and differences between yeast and mammals. J. Biochem. 147:27-33. [DOI] [PubMed] [Google Scholar]
- 27.Langland, J. O., and B. L. Jacobs. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133-141. [DOI] [PubMed] [Google Scholar]
- 28.Li, W., et al. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989-12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Y., X. Meng, Y. Xiang, and J. Deng. 2010. Structure function studies of vaccinia virus host range protein k1 reveal a novel functional surface for ankyrin repeat proteins. J. Virol. 84:3331-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig, H., et al. 2006. Double-stranded RNA-binding protein E3 controls translation of viral intermediate RNA, marking an essential step in the life cycle of modified vaccinia virus Ankara. J. Gen. Virol. 87:1145-1155. [DOI] [PubMed] [Google Scholar]
- 31.McEwen, E., et al. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925-16933. [DOI] [PubMed] [Google Scholar]
- 32.McInerney, G. M., N. L. Kedersha, R. J. Kaufman, P. Anderson, and P. Liljestrom. 2005. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montero, H., M. Rojas, C. F. Arias, and S. Lopez. 2008. Rotavirus infection induces the phosphorylation of eIF2alpha but prevents the formation of stress granules. J. Virol. 82:1496-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moss, B. 2007. Poxviridae: the viruses and their replication, p. 2905-2946. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 35.Piotrowska, J., et al. 2010. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 84:3654-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin, Q., C. Hastings, and C. L. Miller. 2009. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J. Virol. 83:11090-11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raven, J. F., and A. E. Koromilas. 2008. PERK and PKR: old kinases learn new tricks. Cell Cycle 7:1146-1150. [DOI] [PubMed] [Google Scholar]
- 38.Reyes, R., J. Alcalde, and J. M. Izquierdo. 2009. Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol. 10:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheuner, D., et al. 2003. The double-stranded RNA-activated protein kinase mediates viral-induced encephalitis. Virology 317:263-274. [DOI] [PubMed] [Google Scholar]
- 40.Scheuner, D., et al. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 41.Schütz, S., and P. Sarnow. 2007. How viruses avoid stress. Cell Host Microbe 2:284-285. [DOI] [PubMed] [Google Scholar]
- 42.Smith, G. L., J. A. Symons, A. Khanna, A. Vanderplasschen, and A. Alcami. 1997. Vaccinia virus immune evasion. Immunol. Rev. 159:137-154. [DOI] [PubMed] [Google Scholar]
- 43.Solomon, S., et al. 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27:2324-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soncini, C., I. Berdo, and G. Draetta. 2001. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 20:3869-3879. [DOI] [PubMed] [Google Scholar]
- 45.Tang, H., Y. Huang, J. Chen, C. Yu, and A. L. Huang. 2008. Cellular protein TIA-1 regulates the expression of HBV surface antigen by binding the HBV posttranscriptional regulatory element. Intervirology 51:203-209. [DOI] [PubMed] [Google Scholar]
- 46.Tourrière, H., et al. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823-831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Walsh, D., et al. 2008. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol. Cell. Biol. 28:2648-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wek, R. C., H. Y. Jiang, and T. G. Anthony. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34:7-11. [DOI] [PubMed] [Google Scholar]
- 49.White, J. P., A. M. Cardenas, W. E. Marissen, and R. E. Lloyd. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295-305. [DOI] [PubMed] [Google Scholar]
- 50.Xiang, Y., et al. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 76:5251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, P., B. L. Jacobs, and C. E. Samuel. 2008. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J. Virol. 82:840-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, P., J. O. Langland, B. L. Jacobs, and C. E. Samuel. 2009. Protein kinase PKR-dependent activation of mitogen-activated protein kinases occurs through mitochondrial adapter IPS-1 and is antagonized by vaccinia virus E3L. J. Virol. 83:5718-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, T., N. Delestienne, G. Huez, V. Kruys, and C. Gueydan. 2005. Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J. Cell Sci. 118:5453-5463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.