Abstract
Resolving the nonicosahedral components in large icosahedral viruses remains a technical challenge in structural virology. We have used the emerging technique of Zernike phase-contrast electron cryomicroscopy to enhance the image contrast of ice-embedded herpes simplex virus type 1 capsids. Image reconstruction enabled us to retrieve the structure of the unique portal vertex in the context of the icosahedral capsid and, for the first time, show the subunit organization of a portal in a virus infecting eukaryotes. Our map unequivocally resolves the 12-subunit portal situated beneath one of the pentameric vertices, thus removing uncertainty over the location and stoichiometry of the herpesvirus portal.
Herpes simplex virus type 1 (HSV-1) is an enveloped mammalian virus approximately 200 nm in diameter, consisting of a 125-nm icosahedral capsid surrounded by tegument and envelope (17). The HSV-1 genome is packaged into the capsid through a unique structure called the portal, which is present at one of the 12 pentameric vertices (15, 19). The location of the portal with respect to the capsid shell and its oligomeric structure have not been unambiguously defined. Previous studies have suggested positions both outside and inside the capsid (2, 3, 19) and that in vitro-assembled HSV-1 portals can form as multimers of 11 to 14 subunits with roughly equal frequency (19). To date, no study has definitively determined the location and quaternary structure of the HSV-1 portal in its native capsid environment.
Single-particle virus reconstructions are typically generated with icosahedral symmetry imposed, meaning that any features that are not icosahedrally arranged will be lost. As the portal is believed to be located at just one of the 12 5-fold vertices in the capsid, it cannot be definitively resolved in icosahedral reconstructions, necessitating the use of a nonicosahedral, symmetry-free approach. Unfortunately, reconstructions of viruses free of icosahedral enforcement, which are ideal for resolving portal structures, are limited by difficulties in identifying the orientation of the unique vertex (2, 3). In the case of the HSV-1 capsid, there are no sentinel markers for the location of the portal, such as those provided by the external tail structures in some double-stranded DNA bacteriophages (1, 4, 9, 10, 20). The lack of a nonicosahedrally arranged protein density that protrudes from the surface of the HSV-1 capsid presents a computational challenge in determining, from the noisy electron cryomicroscopy (cryo-EM) image, which of the 12 vertices is the portal vertex. An additional complication is that the portal is approximately the same size (∼814 to 1,036 kDa in total, assuming 11 to 14 copies of UL6 [19]), as the pentons (745 kDa) that occupy the other 5-fold vertices and, consequently, does not show up as an obvious excess or absence of density in projection images of capsids. Even though the HSV-1 capsid lacks a tail density, a feature that has proven to be an efficient marker of the portal vertex in bacteriophages (4, 10, 13), we posited that recent advances in Zernike phase-contrast EM (ZPC-EM), which produces dramatic low-resolution contrast enhancement (5, 6), would prove useful in helping to identify this unique vertex. Furthermore, when used in conjunction with an advanced reconstruction algorithm (12), our ability to identify this vertex should be greatly enhanced. Identification of this vertex would then make it possible to reconstruct HSV-1 capsids to a moderate resolution using single-particle cryo-EM without enforcing any symmetry (14).
ZPC-EM relies on using a thin carbon film with a small hole in the middle, suspended at the back focal plane of the microscope's objective lens, to retard the phase of scattered electrons by π/2 with respect to the unscattered beam (7). The resulting shift in phase is manifested in dramatically enhanced low-resolution image contrast compared to that of conventional imaging methods.
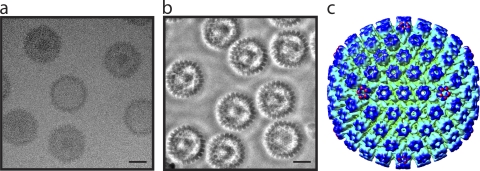
HSV-1 B capsids, purified by sucrose gradient sedimentation (16), were applied to thin-carbon-film-coated Quantifoil grids (14) and plunge-frozen in liquid ethane using a Vitrobot (FEI, Eindhoven). The grids were kept at 100K in a JEM2200FSC field-emission electron microscope (JEOL, Tokyo, Japan) operating at 200 kV, equipped with an in-column energy filter and a Zernike-type phase plate. Images were recorded on a Tietz 4,000- by 4,000-pixel SlowScan charge-coupled-device (CCD) camera (TVIPS, Germany) at ×56,000 detector magnification, with a total dose of 20 e/Å2. The ZPC-EM image was targeted at close to zero defocus in order to obtain maximum contrast enhancement. The contrast of the resulting images is substantially higher than that of conventional cryo-EM images (Fig. 1 a and b). The white halo around each particle in Fig. 1b is a consequence of the phase-contrast optics and is characteristic of ZPC-EM images (6, 14).
FIG. 1.
Zernike phase-contrast electron microscopy (ZPC-EM) of HSV-1 B capsids. (a) Image of ice-embedded HSV-1 B capsids collected using conventional cryo-EM. Bar = 50 nm. (b) Equivalent image collected using ZPC-EM. Bar = 50 nm. (c) Radially colored icosahedral reconstruction of the HSV-1 B capsid showing the characteristic T=16 capsid shell.
The particle images were boxed with the e2boxer.py program (18) and reconstructed using the Multi-Path Simulated Annealing algorithm (11-13). From 353 CCD frames, 6,033 single-particle images of HSV-1 B capsids were collected. Under ZPC-EM conditions, we were able to reconstruct an icosahedral map (Fig. 1c) without the need for contrast transfer function correction (6, 14). The resolved icosahedral orientation of each particle was subsequently used to initiate the process of determining its asymmetric orientation (12). In effect, the algorithm compares an iteratively refined de novo model of the portal density to specific regions of the raw data in order to determine which one of the 60 equivalent icosahedral orientations is most likely to be the “true” asymmetric orientation. This procedure does not require a priori knowledge of the portal structure or its location in the capsid (12). To enhance the specificity of this selection process, we chose only those particles where the mean score for one vertex was statistically higher (P value < 0.05) than the score for the other 11 vertices. The asymmetric model was refined until no further improvements were observed. The 2,308 particles whose asymmetric orientations satisfied these statistical conditions were used for reconstructing the final asymmetric map, estimated at ∼25 Å by the 0.5 Fourier Shell Correlation criterion (Fig. 2 a). The seemingly low resolution, in comparison to the total number of particles used (14), is a reflection of the fact that the B capsid's portal vertex is not readily apparent in the raw images, and as a result, the asymmetric orientations for some of the particle images might be incorrectly assigned.
FIG. 2.
Reconstruction of the HSV-1 B capsid without symmetry enforcement. (a) Radially colored asymmetric reconstruction of the HSV-1 B capsid showing a large density situated beneath one of the 12 5-fold vertices (pink). The internal scaffold density is shown in white. (b) Axial view of a 5-fold vertex (red arrow in panel a) showing a typical penton density. (c) Axial view of the unique vertex (orange arrow in panel a) showing the lack of obvious penton density (compare to panel b). (d) Axial slice of the asymmetric map (violet slicing plane in panel a) showing the 12-fold symmetry of the portal complex (pink).
Both the icosahedral and the asymmetric reconstruction of the HSV-1 B capsid (Fig. 1c and 2a) show the characteristic T=16 capsid shell. The asymmetric reconstruction shows a large density underneath one of the 12 vertices (Fig. 2a). In addition, the capsid shell at this vertex is markedly different from the other 11 vertices and lacks the typical penton density (Fig. 2b and c) seen there. This finding is consistent with the tomographic analysis of chemically treated HSV-1 B capsids, which concluded that the outer surface of the capsid at this position is formed by the top of the portal (3). The relatively weak capsid shell density seen at this vertex may arise due to difficulty in accurately assigning the asymmetric orientations, which would result in the inclusion of some penton density at this vertex. Even though the portal sits beneath the proteins that comprise the capsid shell, a portion of it is externally exposed and would be accessible to other viral or cellular proteins, as well as to antibodies specific to the portal protein UL6 (2, 15).
Figure 2d shows a cross-section of the capsid passing through the region of density beneath the unique vertex. The cross-section clearly shows that this density (Fig. 3 a) has an apparent 12-fold symmetry, as confirmed by rotational correlation analysis (Fig. 3b). It is important to note that no symmetry was imposed during the generation of this map and that both the 5-fold nature of the surrounding capsid shell and the 12-fold symmetry of the internal density arise directly from the data. The simultaneous visualization of these two symmetry-mismatched features is only possible if they are not free to adopt different rotational positions but are fixed relative to each other. As the B capsids used in this study contained cleaved scaffolding proteins, we cannot eliminate the possibility that these proteins contribute to the density we see. However, the presence of this structure at only one vertex and its 12-fold nature, which is consistent with previously described portal structures for tailed bacteriophages (4, 10, 13), gives us confidence that we are detecting the HSV-1 portal. Although a 12-subunit structure was one of several forms found for isolated in vitro-assembled HSV-1 portals (19), this is the first definitive demonstration that the portal in a virus of eukaryotes shares the same stoichiometry as that of bacteriophages.
FIG. 3.
12-Fold symmetry of the HSV-1 B capsid portal. (a) External view of the portal density extracted from the c1 map (Fig. 2a) showing 12 radially arranged densities. (b) Rotational correlation of an annulus of data from the c1 reconstruction shown in panel a confirms the 12-fold arrangement of the portal. (c) The c12 average of the portal density shown in panel a sitting within the HSV-1 B capsid shell. Side, external, and oblique views are shown.
To assess the general structural features and arrangement of the HSV-1 portal subunits, we applied c12 symmetry to our asymmetric map and extracted the 12-fold-averaged portal density (Fig. 3c). The organization and overall dimensions of our portal structure resemble those previously published for the 12-subunit form of the in vitro-assembled HSV-1 portal. However, comparison with the published figures appears to show subtle differences, for example, in the dimensions and features of the peripheral flanges (19). It is not clear whether this reflects genuine variation between the two structures in biochemically isolated and capsid-resident states or differences in the methods used for the two reconstructions.
Previous studies, which have reported two incompatible positions for the portal complex, one inside (3, 8) and the other outside the floor of the capsid shell (2), cast uncertainty over its true location. These studies, which used cryoelectron tomography to examine the nonicosahedral features of the capsid, were unable to resolve the subunit organization and provide unambiguous identification of the portal, leading to the confusion over its true location. The description of the portal in HSV-1 B capsids presented here resolves this uncertainty. The 12-fold symmetry of the internal density (Fig. 3) convincingly identifies it as the portal and establishes the location inside the capsid shell as being correct.
Protein structure accession number.
The asymmetric capsid reconstruction and 12-fold-averaged portal of HSV-1 have been deposited in EMDB (accession number EMD-5255).
Acknowledgments
We thank David McNab of the MRC Virology Unit for excellent technical assistance and Michael F. Schmid at the NCMI for his numerous helpful discussions.
This work was supported by the NIH (grants R01AI0175208 and P41RR002250), the Robert Welch Foundation (grant Q1242), and funding from the Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Corporation to K.N. R.H.R. is supported by NIH training grants (grant GM07330 through the MSTP and grant T15LM007093 through the Gulf Coast Consortia). F.J.R. is funded by the United Kingdom MRC.
Footnotes
Published ahead of print on 24 November 2010.
REFERENCES
- 1.Agirrezabala, X., et al. 2005. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 24:3820-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardone, G., et al. 2007. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology 361:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, J., M. Schmid, F. Rixon, and W. Chiu. 2007. Electron cryotomography reveals the portal in the herpesvirus capsid. J. Virol. 81:2065-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, J., P. Weigele, J. King, W. Chiu, and W. Jiang. 2006. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14:1073-1082. [DOI] [PubMed] [Google Scholar]
- 5.Danev, R., R. Glaeser, and K. Nagayama. 2009. Practical factors affecting the performance of a thin-film phase plate for transmission electron microscopy. Ultramicroscopy 109:312-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danev, R., and K. Nagayama. 2008. Single particle analysis based on Zernike phase contrast transmission electron microscopy. J. Struct. Biol. 161:211-218. [DOI] [PubMed] [Google Scholar]
- 7.Danev, R., and K. Nagayama. 2001. Transmission electron microscopy with Zernike phase plate. Ultramicroscopy 88:243-252. [DOI] [PubMed] [Google Scholar]
- 8.Deng, B., C. M. O'Connor, D. H. Kedes, and Z. H. Zhou. 2007. Direct visualization of the putative portal in the Kaposi's sarcoma-associated herpesvirus capsid by cryoelectron tomography. J. Virol. 81:3640-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fokine, A., et al. 2004. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. U. S. A. 101:6003-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, W., et al. 2006. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, X., W. Jiang, J. Jakana, and W. Chiu. 2007. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J. Struct. Biol. 160:11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, X., R. H. Rochat, and W. Chiu. 15 June 2010. Reconstructing cyano-bacteriophage P-SSP7 structure without imposing symmetry. Nat. Protoc. doi: 10.1038/nprot.2010.96. [DOI]
- 13.Liu, X., et al. 2010. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat. Struct. Mol. Biol. 17:830-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murata, K., et al. 2010. Zernike phase contrast cryo-electron microscopy and tomography for structure determination at nanometer and subnanometer resolutions. Structure 18:903-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newcomb, W. W., et al. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts, A., et al. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subak-Sharpe, J. H., and D. J. Dargan. 1998. HSV molecular biology: general aspects of herpes simplex virus molecular biology. Virus Genes 16:239-251. [DOI] [PubMed] [Google Scholar]
- 18.Tang, G., et al. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38-46. [DOI] [PubMed] [Google Scholar]
- 19.Trus, B., et al. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang, Y., et al. 2006. Structural changes of bacteriophage phi29 upon DNA packaging and release. EMBO J. 25:5229-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]