Abstract
Glucose transport into mammalian cells is mediated by a group of glucose transporters (GLUTs) on the plasma membrane. Human cytomegalovirus (HCMV)-infected human fibroblasts (HFs) demonstrate significantly increased glucose consumption compared to mock-infected cells, suggesting a possible alteration in glucose transport during infection. Inhibition of GLUTs by using cytochalasin B indicated that infected cells utilize GLUT4, whereas normal HFs use GLUT1. Quantitative reverse transcription-PCR and Western analysis confirmed that GLUT4 levels are greatly increased in infected cells. In contrast, GLUT1 was eliminated by a mechanism involving the HCMV major immediate-early protein IE72. The HCMV-mediated induction of GLUT4 circumvents characterized controls of GLUT4 expression that involve serum stimulation, glucose concentration, and nuclear functions of ATP-citrate lyase (ACL). In infected cells the well-characterized Akt-mediated translocation of GLUT4 to the cell surface is also circumvented; GLUT4 localized on the surface of infected cells that were serum starved and had Akt activity inhibited. The significance of GLUT4 induction for the success of HCMV infection was indicated using indinavir, a drug that specifically inhibits glucose uptake by GLUT4. The addition of the drug inhibited glucose uptake in infected cells as well as viral production. Our data show that HCMV-specific mechanisms are used to replace GLUT1, the normal HF GLUT, with GLUT4, the major glucose transporter in adipose tissue, which has a 3-fold-higher glucose transport capacity.
Human cytomegalovirus (HCMV), a herpesvirus, is an enveloped double-stranded DNA virus which relies on host cells to synthesize large amounts of viral proteins as well as viral DNA. This requires a significant amount of energy and building blocks for the synthesis of biomolecules. These are supplied by increased glucose and glutamine utilization in infected cells (9, 28, 27). With respect to glucose utilization, it is known that many DNA viruses increase glucose uptake in infected cells (3, 21, 37). Although this has been known for HCMV-infected human fibroblasts for over 2 decades (21), the mechanism for increased glucose uptake has remained unclear.
Glucose is a polar molecule which does not readily diffuse across the hydrophobic plasma membrane; therefore, specific carrier molecules exist to mediate its uptake. The glucose transporters (GLUTs) are facilitative transporters that carry hexose sugars across the membrane without requiring energy. GLUTs comprise a family of at least 13 members, GLUT 1 to 12 plus the proton (H+)-myoinositol cotransporter (HMIT) (16, 40). These transporters belong to a family of proteins called solute carrier family 2 (gene symbol SLC2A). A common structural feature of the SLC2A family proteins is the presence of 12 transmembrane domains with both the amino- and carboxyl-terminal ends located on the cytosolic side and a unique N-linked oligosaccharide side chain located on either the first or fifth extracellular loop. Structurally, the GLUTs can be divided into three classes: GLUT1 to -4 (class I), GLUT5, -7, -9, and -11 (class II), and GLUT6, -8, -10, and -12 and HMIT (class III) (16, 40).
Class I is comprised of the most-well-characterized glucose transporters, GLUT1 to GLUT4. GLUT1 is ubiquitously distributed in various tissues with different levels of expression in different cell types. It is most abundant in fibroblasts, erythrocytes, and endothelial cells, with low levels of expression in muscle, liver, and adipose tissue (6, 11, 32). GLUT2 is a low-affinity transporter for glucose and is found primarily in the intestines, pancreatic β-cells, kidney, and liver (38). GLUT3 mRNA expression is almost ubiquitous in human tissues, although the protein distribution is restricted to brain and testis (12). GLUT3 transports glucose with high affinity (it has the lowest Km of the GLUTs), which allows neurons to have enhanced access to glucose, especially under conditions of low blood glucose. GLUT4 is the major glucose transporter in adipose tissue, as well as in skeletal and cardiac muscle. Insulin stimulation leads to Akt activation, which mediates the rapid translocation of GLUT4 from intracellular vesicles to the cell surface, resulting in an increase in cellular glucose transport activity (5, 22-24).
In the studies presented here we show that HCMV-specific mechanisms are used to replace GLUT1, the normal human fibroblast (HF) GLUT, with GLUT4, which has a 3-fold-higher glucose transport capacity. The HCMV-mediated induction of GLUT4 circumvents characterized controls of GLUT4 expression, as well as the Akt-mediated translocation of GLUT4 to the cell surface. Treatment of infected cultures with indinavir, a drug which specifically inhibits glucose uptake by GLUT4, inhibited glucose transport and severely inhibited viral production, indicating the significance of GLUT4 induction for the success of an HCMV infection.
MATERIALS AND METHODS
Cell culture, viruses, drugs, and plasmids.
Life-extended human foreskin fibroblasts (HFs) (7), 293T cells, and ihfie1.3 cells (25) were propagated and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM GlutaMAX (all reagents were obtained from Invitrogen, Carlsbad, CA). HCMV infections (multiplicity of infection [MOI], 3) were performed in serum-starved HFs by using the Towne strain of HCMV modified to contain green fluorescent protein (GFP) under the control of the simian virus 40 (SV40) early promoter. For immunofluorescence studies, the wild-type Towne strain of HCMV without the GFP expression cassette was used. The following drugs were used at concentrations discussed below in Results: cytochalasin B (catalog number C2743; Sigma); AKTi (Akt inhibitor VIII; catalog number 124018; Calbiochem); indinavir sulfate (8145; obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH). Plasmids pRSV72 and pRSV86 containing cDNAs encoding IE72 and IE86, respectively, were previously described (43); plasmid pCD-MIE capable of expressing all HCMV major immediate-early (MIE) proteins was constructed by cloning a 5.3-kb NdeI-SalI fragment from pRL43a (34) into pCDNA3 to replace the 2.7-kb NdeI-BsmI fragment; plasmid pHA-GLUT4-GFP was previously described (10, 20); lentiviral expression plasmid encoding a short hairpin RNA (shRNA) for ACL (shACL) was purchased from OpenBiosystems (RHS3979-97066582); and a similar lentiviral expression plasmid encoding a control shRNA targeting firefly luciferase RNA was constructed by cloning oligos CCGGGTTCGTCACATCTCATCTACCCTCGAGGGTAGATGAGATGTGACGAACTTTTTG (sense) and AATTCAAAAAGTTCGTCACATCTCATCTACCCTCGAGGGTAGATGAGATGTGACGAAC (antisense) into the pLKO.1 plasmid (41) between the AgeI and EcoR I sites.
Glucose uptake assay.
Glucose uptake was assayed using a modified protocol (4). Briefly, HFs in 12-well plates were serum starved for 24 h and then either mock infected or HCMV infected in serum-free DMEM for 48 h. The cells were then washed twice using 1 ml of HEPES-buffered saline (HBS; 140 mM NaCl, 20 mM HEPES [pH 7.4], 5.0 mM KCl, 2.5 mM MgSO4, 1.0 mM CaCl2). A 0.4-ml volume of transport solution (4.0 μCi/ml d-[6-3H]glucose, 100 μM glucose in HBS), with or without drug, was added to each well and incubated for 5 min at room temperature. The assay was stopped by adding 0.5 ml ice-cold stop solution (50 mM glucose in 0.9% NaCl), quickly washed three more times with 1.0 ml of ice-cold stop solution, and aspirated to dryness. Cells were lysed by adding 0.3 ml of 0.1 N NaOH; the activity in a 150-μl aliquot was determined by liquid scintillation counting using an LKB 1209 RACKBETA counter. Glucose concentrations in cell culture media were measured using a Nova Biomedical Flex Analyzer as previously described (39).
Immunofluorescence.
Subconfluent HFs were either mock infected or infected with wild-type HCMV. At 2 h postinfection (hpi), the cells were trypsinized and electroporated with 5.0 μg of pHA-GLUT4-GFP per million cells by using an Amaxa Nucleofactor II according to the manufacturer's recommendations with the basic nucleofector kit for primary fibroblasts (catalog number VP1-1002; Lonza). The cells were then plated on glass coverslips in six-well plates and cultured for 2 days. At 48 hpi, both mock- and HCMV-infected cells were refed with serum-free DMEM for 4 h and then either left untreated or treated with insulin and AKTi. For AKTi treatment, AKTi was added to the DMEM to a final concentration of 1 μM during the 4 h of serum starvation; for insulin stimulation, cells were refed with DMEM containing 100 nM insulin for 10 min after the 4 h of serum starvation at 37°C. After treatment, coverslips were washed in phosphate-buffered saline (PBS) and fixed in 0.4% paraformaldehyde at room temperature. The cells were not permeabilized but were blocked in Tris-buffered saline (TBS) containing 5% bovine serum albumin and 0.5% Tween 20, followed by incubation with 5.0 μg/ml of anti-hemagglutinin (anti-HA) antibody (12CA5; Roche Applied Science) in blocking buffer for 2 h at room temperature. Coverslips were washed three times with PBS and then incubated with Alexa Fluor 594 donkey anti-mouse secondary antibody (Invitrogen) at a 1:120 dilution for 1 h at room temperature. Coverslips were finally washed with PBS and mounted using VectaShield (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole (DAPI). Fluorescent images were captured using a Nikon Eclipse E600 microscope.
Quantitative RT-PCR.
Total RNA was isolated using the SV total RNA isolation system (Promega) according to the manufacturer's protocol. cDNA was synthesized from 1.5 μg of total RNA using the SuperScript first-strand synthesis system for reverse transcription-PCR (RT-PCR; Invitrogen). Quantitative real-time PCR was performed on equal volumes of cDNA product by using the TaqMan Universal PCR master mix (Applied Biosystems) on the ABI 7000 system. Gene expression data were normalized to peptidyl prolyl isomerase A (PPIA) mRNA levels (35). The following primer sets from Applied Biosystems were used in this study: GLUT1 (assay ID Hs00197884_m1), GLUT4 (assay ID Hs00168966_m1), and PPIA (assay ID Hs99999904_m1).
shRNA lentiviruses.
The procedures for the growth and utilization of shRNA-encoding lentiviruses have been previously described (44).
Western analysis.
Whole-cell extracts were made by lysing cells in lysis buffer (20 mM Tris-Cl [pH 7.4], 1.0% NP-40, 0.1% SDS, 10 mM NaF, 1.0 mM EDTA, 0.2 mM Na3VO4, 1.0 mM phenylmethylsulfonyl fluoride, 1.0 μg/ml aprotinin, 1.0 μg/ml leupeptin). Western blotting was performed by following the procedures described previously (45). The following antibodies were used in this study: anti-ACL (42), anti-actin (MAB1501; Chemicon), anti-ex2/3, which detects all known HCMV major immediate-early proteins (43), anti-GLUT1 (NB300-666; Novus Biologicals), anti-GLUT4 (ab65976; Abcam), anti-phospho-ACL S454 (catalog number 4331; Cell Signaling Technology), anti-phospho-Akt S473 (catalog number 9271; Cell Signaling Technology), anti-pp28 (sc-56975; Santa Cruz Biotechnology), anti-pp52 (sc-69744; Santa Cruz Biotechnology), and anti-pp65 (sc-52401; Santa Cruz Biotechnology).
Virus growth curves.
Confluent HFs in 60-mm dishes were serum starved for 24 h and then infected with HCMV in serum-free DMEM at an MOI of 3. After a 2-hour adsorption period, the cells were washed once with serum-free DMEM and then refed with serum-free DMEM containing either no drug or 200 μM indinavir. For 24-hour drug treatments, indinavir was added to the medium (200 μM) 24 h prior to each harvest time point. At 0, 24, 48, 72, 96, and 120 hpi, viruses were harvested and viral titration were performed as previously described (19). The experiment was set up in duplicate dishes. One set of dishes was used to harvest viruses; a parallel set of dishes was used to make whole-cell extracts to check expression of viral proteins.
RESULTS
HCMV infection causes reduced GLUT1 expression and increased GLUT4 expression.
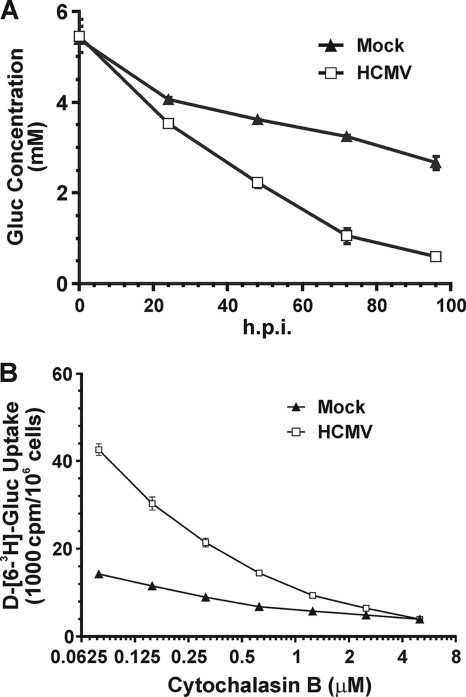
Figure 1 A shows that the glucose concentration is depleted much faster in cell culture medium of HCMV-infected HFs than in mock-infected HFs. In addition, at late times in infection, when the glucose concentration in the cell culture medium was very low (<1 mM), infected cells continued transporting glucose. Thus, glucose uptake appears to be facilitated in infected cells, possibly through the induction of a GLUT with a higher glucose affinity than GLUT1, the normal GLUT present in uninfected HFs. To test this, we used cytochalasin B, a chemical inhibitor that inhibits glucose transport by class I and III GLUTs (40), to determine the 50% inhibitory concentration (IC50) in HCMV-infected and mock-infected cells at 48 hpi. Cells were treated with increasing concentrations of cytochalasin B, resulting in progressive inhibition of glucose uptake in both mock- and HCMV-infected cells (Fig. 1B). The IC50 for mock-infected cells was approximately 0.5 μM, similar to that for GLUT1 (0.44 μM) (40); however, the IC50 for infected cells was 0.2 μM, suggesting that an alteration in the GLUTs had occurred during infection. The 0.2 μM IC50 for infected cells is similar to the reported IC50 for GLUT4 (0.1 to 0.2 μM) (40).
FIG. 1.
(A) Glucose (Gluc) consumption is increased in HCMV-infected cells. Serum-starved HFs were either mock treated or HCMV infected (MOI of 3) in serum-free DMEM. Culture medium was collected at the indicated time points, and the glucose concentration in the medium was assayed as described in Materials and Methods. (B) Cytochalasin B inhibition of glucose transport in mock- and HCMV-infected cells. HFs were infected as described for panel A; at 48 hpi, glucose uptake was assayed in the presence of increasing concentrations of cytochalasin B, as described in Materials and Methods.
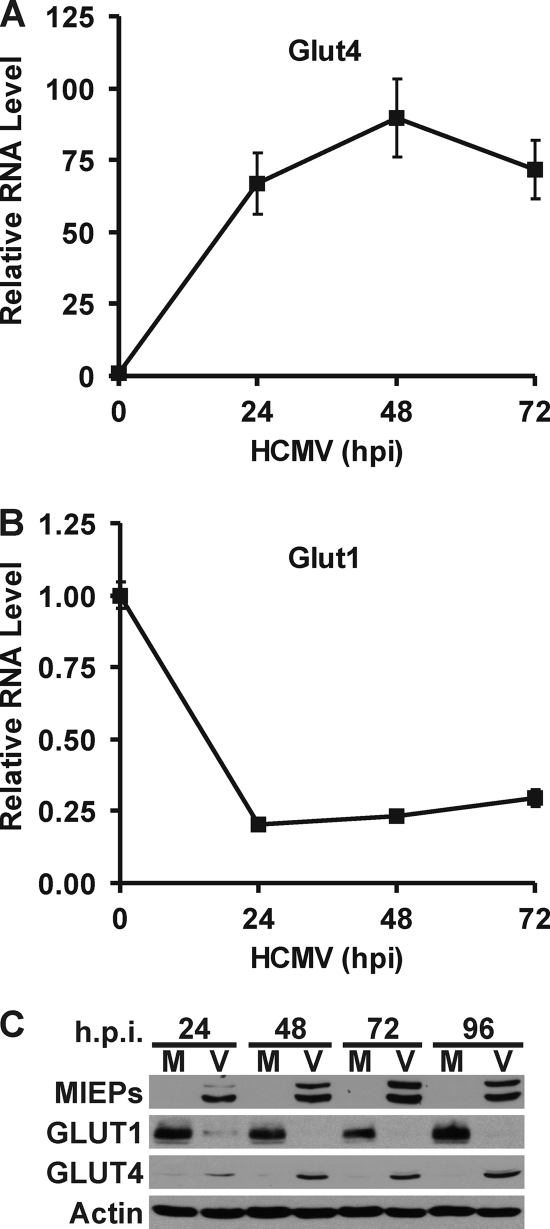
To determine if GLUT4 expression is increased in HCMV-infected cells, total RNA was isolated and GLUT4 mRNA levels were measured using quantitative RT-PCR. Figure 2 A shows that HCMV-infected cells dramatically induce GLUT4 mRNA levels. Surprisingly, the level of mRNA for GLUT1, the normal HF GLUT, was significantly reduced in HCMV-infected cells (Fig. 2B). Figure 2C shows that these changes in mRNA levels were reflected at the protein level. Mock-infected cells had a high GLUT1 protein level, while in HCMV-infected cells the GLUT1 protein level decreased to a very low level by 24 hpi and was nearly undetectable after 48 hpi. In contrast, the very low level of GLUT4 in mock-infected cells dramatically increased in infected cells by 24 hpi and kept increasing throughout the infection. The expression of the HCMV MIE proteins showed that an HCMV infection was established.
FIG. 2.
HCMV-infected cells have increased GLUT4 expression and decreased GLUT1 expression. Total RNA was isolated at 0, 24, 48, and 72 hpi. GLUT4 (A) and GLUT1 (B) mRNA levels were measured by quantitative RT-PCR and normalized to PPIA mRNA levels. (C) Whole-cell extracts were prepared from mock-infected (M) and HCMV-infected (V) infected HFs at 24, 48, 72, and 96 hpi. GLUT1, GLUT4, MIEPs and actin protein levels were determined by Western analysis.
HCMV IE72 reduces GLUT1 expression.
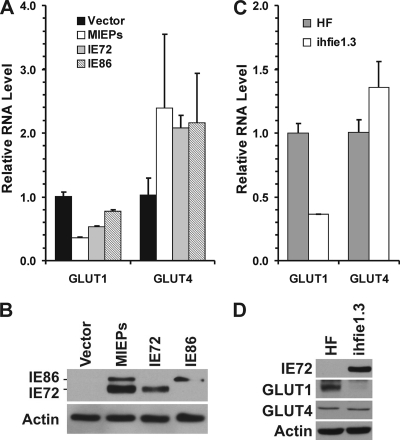
The very rapid drop in GLUT1 mRNA and protein levels by 24 hpi suggested that a very early viral mechanism was mediating it, possibly involving a MIE protein. For this reason, we tested GLUT1 mRNA levels in HFs electroporated with plasmids expressing the MIE proteins. Figure 3 A shows the RT-PCR results, comparing cells electroporated with a control vector, a plasmid which expresses the genomic MIE gene from HCMV, capable of producing all the MIE proteins, and plasmids that express cDNAs for the individual MIE proteins IE72 (IE1) and IE86 (IE2). Figure 3B shows the expression of IE72 and IE86 in each transfection experiment. Expression of the entire MIE gene or IE72 caused a significant decrease in GLUT1 expression (Fig. 3A); IE86 also decreased GLUT1 mRNA levels, but not as much as IE72.
FIG. 3.
GLUT1 expression is inhibited by IE72. (A) GLUT1 mRNA levels decreased in HFs transiently transfected with HCMV IE72. HFs were electroporated with pRSV72, pRSV86, pCD-MIE, or a control vector (see Materials and Methods). At 48 h after electroporation, the cells were refed with serum-free DMEM. Total RNA was isolated at 72 h postelectroporation. GLUT1 and GLUT4 mRNA levels were measured by quantitative RT-PCR and normalized to PPIA mRNA levels. (B) Cultures prepared in parallel with those reported in panel A were harvested for protein level and analyzed by Western blotting for MIE proteins and actin. (C and D) Both the GLUT1 mRNA level and protein levels were decreased in ihfie1.3 cells, which stably express HCMV IE72.
The lowering of GLUT1 levels by IE72 was also tested in the ihfie1.3 human fibroblast cell line, which stably expresses IE72 (25). A comparison of GLUT1 mRNA levels between these cells and normal HFs showed that ihfie1.3 cells expressed only one-third as much GLUT1 mRNA as normal HFs (Fig. 3C). Further, Western analysis showed that GLUT1 protein levels were very low in ihfie1.3 cells (Fig. 3D) compared to normal HFs, suggesting that IE72 alone can reduce GLUT1 expression. IE86 may also be able to repress GLUT1 RNA levels, as suggested by the transfection data in Fig. 3A; however, a reliable cell line expressing only IE86 is not available.
Figure 3A shows that GLUT4 mRNA levels only modestly increased, approximately 2-fold, in transfected cells expressing IE72, IE86, or both. This was reiterated in the studies with ihfie1.3 cells (Fig. 3C and D). These results suggest that the MIE proteins when expressed by themselves cannot account for the great increase in GLUT4 mRNA seen in infected cells (Fig. 2A). Thus, another immediate-early protein, or an early protein, may mediate GLUT4 transcription. This is further examined below in Fig. 4.
FIG. 4.
(A) GLUT4 expression is resistant to glucose concentration in HCMV-infected cells. HFs were mock treated or HCMV infected as described in the text. At 2 hpi the cells were refed with serum-free medium containing 1, 5, or 25 mM glucose. At 48 hpi, total RNA was isolated, and GLUT4 mRNA levels were determined by quantitative RT-PCR. (B) ACL is activated in HCMV infection. Western analysis was performed to determine the levels of total and phosphorylated (S454) ACL in mock-infected (M) or HCMV-infected (V) cell extracts harvested at 48 hpi; MIEPs and actin were also analyzed. (C) ACL is not required for the activation of GLUT4 expression in HCMV infection. Confluent HFs were mock infected or infected with lentiviral vectors expressing a control shRNA (shCTRL) or an shRNA specific for ACL (shACL). Two days after lentiviral infection, the cells were refed with serum-free DMEM. One day later the cells were either mock treated or HCMV infected; at 48 hpi total RNA was isolated and GLUT4 mRNA levels were determined by quantitative RT-PCR. (D) Time course of GLUT4 mRNA levels during HCMV infection. Total RNA was prepared from HCMV-infected HFs at 0, 4, 8, 12, 24, 48, 72, and 96 hpi. GLUT4 mRNA levels were determined by RT-PCR as described in the text.
Induction of GLUT4 by HCMV infection is significantly resistant to glucose concentration.
It has been reported that GLUT4 expression can be regulated in a glucose-dependent manner (2, 13). For example, in rats with induced diabetes, circulating glucose increases and GLUT4 mRNA levels are markedly decreased in fat cells and skeletal muscle cells (18). Therefore, we cultured mock-infected and HCMV-infected HFs in DMEM with different concentrations of glucose. At 2 hpi, the medium was changed to medium containing 1, 5, or 25 mM glucose; the cells were harvested at 48 hpi and the GLUT4 mRNA levels were determined (Fig. 4 A). In mock-infected cells, the very low levels of GLUT4 mRNA were not affected by the different glucose concentrations. In HCMV-infected cells, the virally induced levels of GLUT4 mRNA were moderately decreased at higher glucose concentrations; however, even at 25 mM glucose the HCMV-mediated induction of GLUT4 mRNA was very high, suggesting that in HCMV-infected cells GLUT4 mRNA levels can be significantly increased by a mechanism that is, in large part, resistant to inhibition by high glucose concentrations.
GLUT4 activation does not involve ATP-citrate lyase.
Recent studies have suggested that ACL, the enzyme that converts citrate to acetyl coenzyme A (CoA) and oxaloacetate, plays a critical role in determining the total amount of histone acetylation in mammalian cells. ACL-dependent production of acetyl-CoA in the nucleus contributes to increased histone acetylation during the cellular response to growth factor stimulation and during adipocyte differentiation (42). In this regard, the experiments showed that ACL-dependent histone acetylation contributes to the elective regulation of genes involved in glucose metabolism, including GLUT4. In HCMV-infected HFs, we observed a moderate increase in total ACL protein levels in infected cells at 48 hpi (Fig. 4B), as well as a significant increase in the phosphorylation of ACL at Ser454 (Fig. 4B), which has been reported to activate ACL (33). While this activation of ACL is important for acetyl-CoA production for fatty acid synthesis that is critical for HCMV infection (28), it is also possible that ACL's nuclear activity may participate in GLUT4 expression. To test this we depleted ACL using shRNAs and found that at 48 hpi GLUT4 levels increased identically in control and ACL-depleted, HCMV-infected cells (Fig. 4C). These results suggest that a virus-specific, ACL-independent mechanism mediates GLUT4 expression in HCMV-infected cells.
Our data thus far suggest that the HCMV-induced increase of GLUT4 levels is independent of ACL function in histone acetylation and is significantly resistant to glucose concentration; thus, it is nutrient independent. In addition, the data in Fig. 3A and C suggest that independent expression of the MIE proteins has only a modest effect on GLUT4 mRNA levels. Thus, as suggested above, it is likely that an early protein may be involved in GLUT4 activation. The involvement of an early protein is supported by the more detailed evaluation of GLUT4 mRNA levels during the time course of an HCMV infection in HFs (Fig. 4D). These data show that GLUT4 RNA levels increased very little during the immediate-early period (4 to 12 hpi), which correlates with the modest activation mediated by transfected MIE proteins (Fig. 3A). A more significant increase in GLUT4 mRNA was seen by 24 hpi, and this was substantially increased between 24 and 48 hpi. These data suggest that early viral protein synthesis is needed for the full activation of GLUT4 expression in infected cells.
Translocation of GLUT4 in HCMV-infected cells bypasses Akt signaling.
Under conditions when increased glucose uptake is not needed, GLUT4 resides primarily in intracellular vesicles. Upon signaling for increased glucose uptake, GLUT4 translocates to the plasma membrane; for example, insulin stimulation activates Akt, which mediates the rapid translocation of GLUT4 to the cell surface (5). HCMV infection is able to activate Akt signaling (15, 19), which was shown by the increase of Akt phosphorylation at Ser473 (Fig. 5 A). Treatment with AKTi abolished S473 phosphorylation in infected cells (Fig. 5A). When we compared untreated and AKTi-treated HCMV-infected cells, there was no difference in total GLUT4 protein levels in whole-cell extracts (Fig. 5), indicating that the increase in GLUT4 expression in infected cells does not involve Akt signaling.
FIG. 5.
Translocation of GLUT4 can bypass Akt signaling in HCMV-infected cells. (A) Akt signaling is not involved in the activation of GLUT4 expression in HCMV-infected cells. Serum-starved HFs were pretreated with 1 μM AKTi for 1 h. Then, the cells were either mock treated or infected with HCMV in serum-free medium in the presence or absence of 1 μM AKTi. Whole-cell extracts were prepared at 48 hpi, and GLUT4, phospho-Akt S473, MIEPs, and actin protein levels were determined by Western analysis. (B) Translocation of GLUT4 can bypass Akt signaling in HCMV-infected cells. Results of fluorescence microscopy analysis of the cellular localization of HA-GLUT4-GFP in HFs are shown. At 2 hpi, mock- or HCMV-infected HFs were electroporated with the HA-GLUT4-GFP reporter plasmid. At 48 hpi the cells were serum starved for 4 h and then either left untreated or treated with insulin and AKTi as described in detail in Materials and Methods. GFP fluorescence (green) is a measure of total GLUT4 expression. Immunofluorescence staining for the HA tag (red) is a measure of GLUT4 on the cell surface. The nuclei were stained with DAPI (blue).
The data above show that AKTi was effective in inhibiting Akt activation in infected cells; therefore, we used it to determine whether Akt activity was necessary for GLUT4 translocation to the cell surface in HCMV-infected HFs. At 2 hpi, mock- or HCMV-infected cells were electroporated with the HA-GLUT4-GFP reporter plasmid. The doubly tagged GLUT4 construct is GFP-tagged at the C terminus and HA tagged in the first extracellular loop (10). Fluorescence microscopic analysis of nonpermeabilized cells expressing this protein shows total GLUT4 by GFP fluorescence (green) regardless of it cellular location; however, immunofluorescent detection of the HA tag (red) can only occur if the GLUT4 is on the cell surface, where the HA tag is outside the cell and accessible by anti-HA antibody. At 48 hpi the cells were serum starved for 4 h and then either left untreated or treated with insulin and AKTi as described in Materials and Methods. In Fig. 5B, immunofluorescence microscopy shows that GFP fluorescence stayed in the cytoplasm in mock-infected, serum-starved cells. As expected, upon insulin stimulation a significant amount of GLUT4 translocated to the cell surface, as indicated by HA surface staining. However, the presence of AKTi inhibited GLUT4 surface localization. In contract, in infected cells the HA staining of cell surface GLUT4 was readily detected in serum-starved cells. This was neither increased by insulin treatment nor inhibited by treatment with AKTi. These data show that GLUT4 protein translocation in infected cells does not require insulin stimulation and is insensitive to Akt inhibition. Thus, HCMV infection induces translocation of GLUT4 to the cell surface by a mechanism that bypasses Akt signaling. Previous studies have suggested that overexpression of GLUT4 alone, similar to that seem in HCMV-infected cells, can increase GLUT4 levels on the cell surface (8).
Indinavir impairs HCMV growth by reducing glucose uptake.
In order to assess the significance of GLUT4 during HCMV infection, it would be ideal to use shRNA depletion. However, after testing the available shRNAs for GLUT4, we found none that could significantly deplete GLUT4 levels in infected cells (data not shown). Thus, we turned to the inhibitor indinavir, an HIV protease inhibitor which is used in combination with other drugs in AIDS antiviral therapy. Many patients being treated with indinavir develop insulin resistance (29). It has been determined that insulin resistance results from indinavir's selective inhibition of the glucose transport activity of GLUT4, but not GLUT1, at physiologic concentrations of indinavir (30). Therefore, we used indinavir to show that GLUT4-mediated glucose uptake is necessary for viral growth.
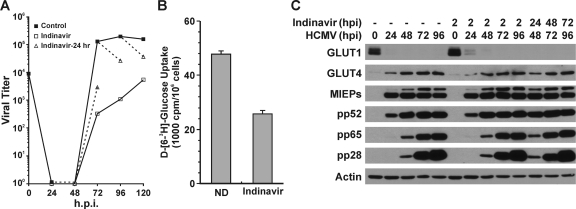
Figure 6 A shows a normal growth curve of HCMV in HFs. The dashed lines indicate the changes in growth caused by a 24-h treatment with 200 μM indinavir from 48 to 72, 72 to 86, and 96 to 120 hpi. In all cases the indinavir treatment slowed infectious virion formation; the most significant effect was in the 48-to-72-hpi period, during which the titer was reduced by 1.5 logs. Treatment with indinavir from the beginning of the infection (drug added at 2 hpi) had a more sever effect on viral growth (Fig. 6A).
FIG. 6.
Inhibition of GLUT4 by indinavir results in reduced HCMV growth. (A) Indinavir inhibits production of infectious HCMV virions. HCMV virus growth curves were generated in the presence of indinavir or water (drug solvent, Control). HFs were infected with HCMV as described in the text. The black line with solid squares indicates the control growth curve in which water was added at 2 hpi. The black line with open squares indicates the indinavir-treated cultures where 200 μM indinavir was added at 2 hpi and staged in the infection until harvesting. The dashed line with open triangles indicates that indinavir was added for 24 h prior to harvest, i.e., 24 to 48, 48 to 72, 72 to 96, or 96 to 120 hpi. (B) Glucose uptake was inhibited by indinavir in HCMV-infected cells. Glucose uptake was measured in HCMV-infected cells in the presence or absence of 200 μM indinavir as described in Materials and Methods. ND, no drug. (C) Virus protein levels in control and indinavir-treated cultures. Whole-cell extracts were prepared from HCMV-infected cultures treated with indinavir as described for panel A and evaluated by Western analysis.
Inhibition of glucose uptake by indinavir was assayed at 48 hpi; the cells were either mock treated or treated with 200 μM indinavir for 1 h before glucose uptake analysis. The results showed that the 1-h indinavir treatment inhibited glucose uptake in HCMV-infected HFs by at least 50% (Fig. 6B). Figure 6C shows Western analysis results of proteins in normal and indinavir-treated cells. As described above, HCMV-infected cells without indinavir treatment had reduced GLUT1 and elevated GLUT4 protein levels, and this was not altered by indinavir treatment (Fig. 6C). We also noted that temporal expression and levels of viral proteins representative of the immediate-early (MIEPs), early (pp52), and late (pp65 and pp28) classes of HCMV genes were not altered by indinavir treatment from 2 hpi or during the last 24 h of infection before harvest. The lack of effects of indinavir on viral protein levels suggests that the indinavir inhibitory effects on viral growth are at the level of infectious virion formation. This is likely, since the inhibition of glucose consumption would limit the ability of infected cells to make fatty acids, which are necessary for virion maturation (28), but may not affect viral protein synthesis and accumulation.
DISCUSSION
Metabolic flux analysis of HCMV-infected cells, compared to mock-infected cells, shows global metabolic upregulation in infected cells (27, 28). This includes greatly increased glycolysis, in which the vast majority of glucose-derived acetyl-CoA supports fatty acid synthesis to make membranes needed by the virus (28). As a result there is a great decrease in glucose-derived carbon entering the tricarboxylic acid (TCA) cycle. This presents the potential problem of limiting TCA cycle intermediates that are used biosynthetically and whose synthesis is the source of NADH for oxidative phosphorylation and the production of ATP. However, we have recently demonstrated that HCMV-infected cells are not as dependent on glucose for energy as are uninfected cells; in infected cells glutamine is used for the TCA cycle (anaplerosis) and is a major source of energy (9). This occurs in infected cells due to the induction of enzymes needed to convert glutamine to α-ketoglutarate, which enters the TCA cycle (9). For these alterations in metabolism to support the production of HCMV, it is key that the uptake of both glutamine and glucose increase significantly during infection. We have previously shown that glutamine uptake is increased (9), and it has long been known that glucose uptake increases in HCMV-infected cells (3, 21, 37).
In this study we analyzed the means by which HCMV increases glucose uptake. Our data show a significant alteration in the glucose transporters on the surface of HCMV-infected HFs. GLUT1, the normal glucose transporter of HFs, is a housekeeping transporter residing constitutively in the plasma membrane and is responsible for basal glucose transport (26). We showed that GLUT1 is eliminated from infected cells and replaced with GLUT4, which has a 3-fold-higher glucose transport capacity than GLUT1 (31). GLUT4 is normally an acute insulin-sensitive isoform that can quickly boost glucose transport in response to insulin stimulation in adipocytes and muscle cells (14, 17). We showed that HCMV infection increases GLUT4 expression in HFs, as well as its location on the cell surface, in order to elevate glucose uptake. This utilization of GLUT4 is critical for the success of the HCMV infection, since inhibition of GLUT4 transport function using indinavir significantly inhibited glucose uptake and HCMV growth. One caveat for the indinavir experimental findings must be mentioned: while the ability of the drug to inhibit GLUT4 is well established, indinavir is also an HIV protease inhibitor. In this regard, we do not know whether there are HCMV proteases that may be inhibited by the drug and thus complicate our virus growth inhibition results (Fig. 6A). However, Fig. 6B does show that the drug significantly inhibited glucose uptake in infected cells, as predicted by its inhibitory effects on GLUT4.
Our data suggest that the HCMV MIE proteins, particularly IE72 (IE1), mediate the reduction in GLUT1 mRNA levels in infected cells. However, why GLUT1 is eliminated in HCMV-infected cells is not clear; there is no apparent reason why the presence of GLUT1 with GLUT4 should interfere with glucose transport in HCMV-infected cells, unless there are limited sites for glucose transporters on the cell surface and the elimination of GLUT1 enables the insertion of GLUT4. The mechanism for increasing GLUT4 mRNA and protein levels is also unclear. Our data suggest that the MIE proteins by themselves have a very modest effect on GLUT4 mRNA levels; temporally, it appears that the expression of an early protein is needed for significant increases in GLUT4 mRNA.
The HCMV-mediated induction of GLUT4 appears to circumvent characterized controls of GLUT4 expression and localization. Our data show that activation of GLUT4 expression is neither nutrient dependent nor ACL dependent. As discussed above, acetyl-CoA generated by ACL is required for metabolism gene expression, including GLUT4, in response to growth factor (serum) stimulation (42). Our studies showed that GLUT4 mRNA and protein levels are increased in the absence of serum stimulation and when ACL is depleted by using shRNAs. Thus, HCMV appears to utilize a virus-specific mechanism for GLUT4 induction. Although ACL is not involved in GLUT4 upregulation, it is important in infected cells for the production of acetyl-CoA for de novo lipogenesis (28).
The HCMV-mediated induction of GLUT4 also circumvents the well-characterized Akt-mediated translocation and occurs in the absence of insulin stimulation. It has been reported that overexpression of GLUT4 can bypass insulin signaling and dramatically increase the levels of GLUT4 on the plasma membrane as well as increase glucose transport activity (1, 8, 36). Thus, we suggest that the Akt-independent localization of GLUT4 to the cell surface in infected cells is, at least in part, due to the significant increase in GLUT4 levels in infected cells.
In these studies we found that GLUT4 overproduction and localization to the cell surface is very important for the success of an HCMV infection in HFs. However, GLUT4 is not the only GLUT with a high glucose affinity; others include GLUT3, GLUT7, GLUT8, and GLUT10. HFs had no detectable GLUT7 expression in either mock- or HCMV-infected cells (data not shown). However, GLUT3, GLUT8, and GLUT10 are represented in HFs; GLUT3 and -8 mRNA levels increase modestly, 2- to 3-fold on average, over a 72-h infection time course, while GLUT10 mRNA levels decreased (see Fig. S1 in the supplemental material). Western analysis showed no change in GLUT3 or GLUT10 protein levels between mock-treated and infected cells at 48 hpi (see Fig. S2 in the supplemental material); GLUT8 could not be tested because there is no suitable antibody for Western analysis. Thus, other GLUTs may contribute to glucose uptake in HCMV-infected cells; however, the most significant and dramatic alteration in GLUT expression in infected cells is the induction of GLUT4 accompanied by the loss of GLUT1.
Supplementary Material
Acknowledgments
We thank all the members of the Alwine laboratory for critical reading of the manuscript and valuable advice. We also thank Kathryn Wellen at the University of Pennsylvania and Phillip Bilan at the Hospital for Sick Children, Toronto, Canada, for sharing their reagents and helpful comments.
This work was supported by National Institutes of Health grant R01-CA028379-29 awarded to J.C.A. by the National Cancer Institute.
Footnotes
Published ahead of print on 8 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Al-Hasani, H., D. R. Yver, and S. W. Cushman. 1999. Overexpression of the glucose transporter GLUT4 in adipose cells interferes with insulin-stimulated translocation. FEBS Lett. 460:338-342. [DOI] [PubMed] [Google Scholar]
- 2.Arnoni, C. P., et al. 2009. Regulation of glucose uptake in mesangial cells stimulated by high glucose: role of angiotensin II and insulin. Exp. Biol. Med. (Maywood) 234:1095-1101. [DOI] [PubMed] [Google Scholar]
- 3.Bardell, D. 1984. Host cell glucose metabolism during abortive infection by adenovirus type 12. Microbios 39:95-99. [PubMed] [Google Scholar]
- 4.Bilan, P. J., Y. Mitsumoto, T. Ramlal, and A. Klip. 1992. Acute and long-term effects of insulin-like growth factor I on glucose transporters in muscle cells. Translocation and biosynthesis. FEBS Lett. 298:285-290. [DOI] [PubMed] [Google Scholar]
- 5.Birnbaum, M. J. 1989. Identification of a novel gene encoding an insulin-responsive glucose transporter protein. Cell 57:305-315. [DOI] [PubMed] [Google Scholar]
- 6.Birnbaum, M. J., H. C. Haspel, and O. M. Rosen. 1986. Cloning and characterization of a cDNA encoding the rat brain glucose-transporter protein. Proc. Natl. Acad. Sci. U. S. A. 83:5784-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bresnahan, W. A., G. E. Hultman, and T. Shenk. 2000. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J. Virol. 74:10816-10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho, E., et al. 2004. GLUT4 overexpression or deficiency in adipocytes of transgenic mice alters the composition of GLUT4 vesicles and the subcellular localization of GLUT4 and insulin-responsive aminopeptidase. J. Biol. Chem. 279:21598-21605. [DOI] [PubMed] [Google Scholar]
- 9.Chambers, J. W., T. G. Maguire, and J. C. Alwine. 2010. Glutamine metabolism is essential for human cytomegalovirus infection. J. Virol. 84:1867-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson, K., A. Aviles-Hernandez, S. W. Cushman, and D. Malide. 2001. Insulin-regulated trafficking of dual-labeled glucose transporter 4 in primary rat adipose cells. Biochem. Biophys. Res. Commun. 287:445-454. [DOI] [PubMed] [Google Scholar]
- 11.Fukumoto, H., et al. 1988. Sequence, tissue distribution, and chromosomal localization of mRNA encoding a human glucose transporter-like protein. Proc. Natl. Acad. Sci. U. S. A. 85:5434-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haber, R. S., S. P. Weinstein, E. O'Boyle, and S. Morgello. 1993. Tissue distribution of the human GLUT3 glucose transporter. Endocrinology 132:2538-2543. [DOI] [PubMed] [Google Scholar]
- 13.Hainault, I., E. Hajduch, and M. Lavau. 1995. Fatty genotype-induced increase in GLUT4 promoter activity in transfected adipocytes: delineation of two fa-responsive regions and glucose effect. Biochem. Biophys. Res. Commun. 209:1053-1061. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, S. A., J. M. Buxton, B. M. Clancy, and M. P. Czech. 1990. Insulin regulation of hexose transport in mouse 3T3-L1 cells expressing the human HepG2 glucose transporter. J. Biol. Chem. 265:20106-20116. [PubMed] [Google Scholar]
- 15.Johnson, R. A., X. Wang, X. L. Ma, S. M. Huong, and E. S. Huang. 2001. Human cytomegalovirus up-regulates the phosphatidylinositol 3-kinase (PI3-K) pathway: inhibition of PI3-K activity inhibits viral replication and virus-induced signaling. J. Virol. 75:6022-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joost, H. G., et al. 2002. Nomenclature of the GLUT/SLC2A family of sugar/polyol transport facilitators. Am. J. Physiol. Endocrinol. Metab. 282:E974-E976. [DOI] [PubMed] [Google Scholar]
- 17.Kern, M., et al. 1990. Insulin responsiveness in skeletal muscle is determined by glucose transporter (Glut4) protein level. Biochem. J. 270:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klip, T. A., T. Tsakiridis, A. Marette, and P. A. Ortiz. 1994. Regulation of expression of glucose transporters by glucose: a review of studies in vivo and in cell cultures. FASEB J. 8:43-53. [DOI] [PubMed] [Google Scholar]
- 19.Kudchodkar, S. B., Y. Yu, T. G. Maguire, and J. C. Alwine. 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J. Virol. 78:11030-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lampson, M. A., A. Racz, S. W. Cushman, and T. E. McGraw. 2000. Demonstration of insulin-responsive trafficking of GLUT4 and vpTR in fibroblasts. J. Cell Sci. 113:4065-4076. [DOI] [PubMed] [Google Scholar]
- 21.Landini, M. P. 1984. Early enhanced glucose uptake in human cytomegalovirus-infected cells. J. Gen. Virol. 65:1229-12232. [DOI] [PubMed] [Google Scholar]
- 22.Langfort, J., M. Viese, T. Ploug, and F. Dela. 2003. Time course of GLUT4 and AMPK protein expression in human skeletal muscle during one month of physical training. Scand. J. Med. Sci. Sports 13:169-174. [DOI] [PubMed] [Google Scholar]
- 23.Li, L., H. Chen, and S. L. McGee. 2008. Mechanism of AMPK regulating GLUT4 gene expression in skeletal muscle cells. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 25:161-167. (In Chinese.) [PubMed] [Google Scholar]
- 24.Lira, V. A., et al. 2007. Nitric oxide increases GLUT4 expression and regulates AMPK signaling in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 293:E1062-E1068. [DOI] [PubMed] [Google Scholar]
- 25.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. U. S. A. 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueckler, M. 1990. Family of glucose-transporter genes. Implications for glucose homeostasis and diabetes. Diabetes 39:6-11. [DOI] [PubMed] [Google Scholar]
- 27.Munger, J., S. U. Bajad, H. A. Coller, T. Shenk, and J. D. Rabinowitz. 2006. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2:1165-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munger, J., et al. 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 26:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murata, H., P. W. Hruz, and M. Mueckler. 2002. Indinavir inhibits the glucose transporter isoform Glut4 at physiologic concentrations. AIDS 16:859-863. [DOI] [PubMed] [Google Scholar]
- 30.Murata, H., P. W. Hruz, and M. Mueckler. 2000. The mechanism of insulin resistance caused by HIV protease inhibitor therapy. J. Biol. Chem. 275:20251-20254. [DOI] [PubMed] [Google Scholar]
- 31.Palfreyman, R. W., A. E. Clark, R. M. Denton, G. D. Holman, and I. J. Kozka. 1992. Kinetic resolution of the separate GLUT1 and GLUT4 glucose transport activities in 3T3-L1 cells. Biochem. J. 284:275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pardridge, W. M., R. J. Boado, and C. R. Farrell. 1990. Brain-type glucose transporter (GLUT-1) is selectively localized to the blood-brain barrier. Studies with quantitative Western blotting and in situ hybridization. J. Biol. Chem. 265:18035-18040. [PubMed] [Google Scholar]
- 33.Pierce, M. W., J. L. Palmer, H. T. Keutmann, T. A. Hall, and J. Avruch. 1982. The insulin-directed phosphorylation site on ATP-citrate lyase is identical with the site phosphorylated by the cAMP-dependent protein kinase in vitro. J. Biol. Chem. 257:10681-10686. [PubMed] [Google Scholar]
- 34.Pizzorno, M. C., P. O'Hare, L. Sha, R. L. LaFemina, and G. S. Hayward. 1988. trans-Activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J. Virol. 62:1167-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radonic, A., et al. 2005. Reference gene selection for quantitative real-time PCR analysis in virus infected cells: SARS corona virus, yellow fever virus, human herpesvirus-6, camelpox virus and cytomegalovirus infections. Virol. J. 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren, J. M., et al. 1995. Overexpression of Glut4 protein in muscle increases basal and insulin-stimulated whole body glucose disposal in conscious mice. J. Clin. Invest. 95:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito, Y., and R. W. Price. 1984. Enhanced regional uptake of 2-deoxy-D-[14C]glucose in focal herpes simplex type 1 encephalitis: autoradiographic study in the rat. Neurology 34:276-284. [DOI] [PubMed] [Google Scholar]
- 38.Thorens, B., H. K. Sarkar, H. R. Kaback, and H. F. Lodish. 1988. Cloning and functional expression in bacteria of a novel glucose transporter present in liver, intestine, kidney, and beta-pancreatic islet cells. Cell 55:281-290. [DOI] [PubMed] [Google Scholar]
- 39.Tong, X., F. Zhao, A. Mancuso, J. J. Gruber, and C. B. Thompson. 2009. The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 106:21660-21665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uldry, M., and B. Thorens. 2004. The SLC2 family of facilitated hexose and polyol transporters. Pflugers Arch. 447:480-489. [DOI] [PubMed] [Google Scholar]
- 41.Wakiyama, M., T. Matsumoto, and S. Yokoyama. 2005. Drosophila U6 promoter-driven short hairpin RNAs effectively induce RNA interference in Schneider 2 cells. Biochem. Biophys. Res. Commun. 331:1163-1170. [DOI] [PubMed] [Google Scholar]
- 42.Wellen, K. E., et al. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324:1076-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, Y., and J. C. Alwine. 2002. Human cytomegalovirus major immediate-early proteins and simian virus 40 large T antigen can inhibit apoptosis through activation of the phosphatidylinositide 3′-OH kinase pathway and the cellular kinase Akt. J. Virol. 76:3731-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, Y., and J. C. Alwine. 2008. Interaction between simian virus 40 large T antigen and insulin receptor substrate 1 is disrupted by the K1 mutation, resulting in the loss of large T antigen-mediated phosphorylation of Akt. J. Virol. 82:4521-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu, Y., S. B. Kudchodkar, and J. C. Alwine. 2005. Effects of simian virus 40 large and small tumor antigens on mammalian target of rapamycin signaling: small tumor antigen mediates hypophosphorylation of eIF4E-binding protein 1 late in infection. J. Virol. 79:6882-6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.