Abstract
Human parainfluenza virus type 1 (HPIV1) is an important respiratory pathogen in young children, the immunocompromised, and the elderly. We found that infection with wild-type (WT) HPIV1 suppressed the innate immune response in human airway epithelial cells by preventing not only phosphorylation of interferon regulatory factor 3 (IRF3) but also degradation of IκBβ, thereby inhibiting IRF3 and NF-κB activation, respectively. Both of these effects were ablated by a F170S substitution in the HPIV1 C proteins (F170S) or by silencing the C open reading frame [P(C−)], resulting in a potent beta interferon (IFN-β) response. Using murine knockout cells, we found that IFN-β induction following infection with either mutant relied mainly on melanoma-associated differentiation gene 5 (MDA5) rather than retinoic acid-inducible gene I (RIG-I). Infection with either mutant, but not WT HPIV1, induced a significant accumulation of intracellular double-stranded RNA (dsRNA). These mutant viruses directed a marked increase in the accumulation of viral genome, antigenome, and mRNA that was coincident with the accumulation of dsRNA. In addition, the amount of viral proteins was reduced compared to that of WT HPIV1. Thus, the accumulation of dsRNA might be a result of an imbalance in the N protein/genomic RNA ratio leading to incomplete encapsidation. Protein kinase R (PKR) activation and IFN-β induction followed the kinetics of dsRNA accumulation. Interestingly, the C proteins did not appear to directly inhibit intracellular signaling involved in IFN-β induction; instead, their role in preventing IFN-β induction appeared to be in suppressing the formation of dsRNA. PKR activation contributed to IFN-β induction and also was associated with the reduction in the amount of viral proteins. Thus, the HPIV1 C proteins normally limit the accumulation of dsRNA and thereby limit activation of IRF3, NF-κB, and PKR. If C protein function is compromised, as in the case of F170S HPIV1, the resulting PKR activation and reduction in viral protein levels enable the host to further reduce C protein levels and to mount a potent antiviral type I IFN response.
Human parainfluenza virus type 1 (HPIV1) is an important and uncontrolled respiratory pathogen that causes a significant burden of disease, mainly in young children, the immunocompromised, and the elderly (13, 16, 21, 27, 46, 52). HPIV1 is a single-stranded, negative-sense, nonsegmented RNA virus in the Paramyxoviridae family. The viral genome, 15.6 kb in length, consists of six genes (3′-N-P/C-M-F-HN-L-5′) that encode the nucleoprotein (N), phosphoprotein (P), C proteins, matrix (M) protein, fusion (F) protein, hemagglutinin (HA)-neuraminidase (HN) protein, and the large polymerase (L) protein. Each gene encodes a single major protein, with the exception of the P/C gene, which encodes the P protein in one open reading frame (ORF) and a nested set of four carboxy-coterminal C proteins (C′, C, Y1, and Y2) expressed from individual start sites in a second open reading frame. The C proteins play a critical role in HPIV1 virulence by inhibiting apoptosis, regulating type I interferon (IFN) production and signaling, and controlling the transcription of a large number of host genes (6, 8, 9, 64). The C proteins of Sendai virus (SeV), or murine PIV1, have considerable sequence conservation with the HPIV1 C proteins. However, the P/C gene organization of SeV differs from that of HPIV1 in that SeV expresses, in addition to the C proteins, a second accessory protein, the V protein, that also exerts an inhibitory role on the host innate antiviral response (2). The SeV V protein has been reported to inhibit IFN-β induction via inhibition of MDA5 signaling and to inhibit IFN signaling and apoptosis induction (2). In addition, some of the immune evasion activities of SeV and HPIV1 are species specific (9, 11) and the C deletion mutants of SeV, such as 4C−, do express the SeV V protein, hindering clear separation of V-specific and C-specific effects (28, 35). For these reasons, we believe that a careful examination of the innate immune response to HPIV1 is warranted and may yield important clues about the pathogenesis of this human pathogen that cannot be inferred from observations made with the murine homologue SeV.
Type I IFN is a central mediator of antiviral innate immunity. The induction of IFN synthesis following virus infection depends on a number of pattern recognition receptors that recognize conserved pathogen-associated molecular patterns and initiate downstream signaling cascades (31). The presence of double-stranded RNA (dsRNA), an intermediate of RNA viral replication, is recognized as a pathogen-associated molecular pattern by Toll-like receptor 3 (TLR3) and two caspase recruitment domain (CARD)-containing RNA helicases, retinoic acid-inducible gene I (RIG-I) and melanoma associated-differentiation gene 5 (MDA5), which act as intracytoplasmic sensors of dsRNA (1, 26, 50, 58, 72, 74). Whereas TLR3 mainly senses extracellular dsRNA on antigen-presenting cells, RIG-I and MDA5 are constitutively present and detect intracellular dsRNA (1, 74). TLR3 signals through an adaptor called TIR domain-containing adaptor inducing IFN-β (TRIF), while RIG-I and MDA5 recruit another CARD-containing adaptor called mitochondrial antiviral signaling protein (MAVS; also referred to as IPS-1, Cardif, or VISA) to relay the signal to the kinases TBK1 and IKKɛ, which phosphorylate interferon regulatory factor 3 (IRF3), and to IKKβ, which activates the NF-κB pathway (15, 33, 48, 54, 69). Once activated, IRF3 translocates into the nucleus and binds the positive regulatory domain III (PRDIII) of the IFN-β promoter. RIG-I and MDA5 can distinguish between different RNA viruses (32). It was originally reported that IFN production in response to infection with negative-sense RNA viruses, including SeV, was dependent on RIG-I expression, whereas the response to infection with positive-sense RNA viruses was dependent on MDA5 expression (32, 66). This model was in agreement with earlier reports that found a substantial accumulation of cytoplasmic dsRNA during infection with positive-sense RNA viruses but not with negative-sense RNA viruses (67). However, a more recent study suggested that MDA5 does contribute in vivo to sustaining the IFN response and restricting SeV infection (20). After the host cell recognizes and signals the presence of foreign RNA, IRF-3 and NF-κB, together with ATF-2/c-Jun (AP-1), form an enhanceosome that binds to the IFN-β promoter, inducing transcription of the IFN-β gene. Once type I IFN is produced and secreted, it binds to the type I IFN receptor on the surface of the infected and neighboring cells to amplify a robust antiviral response (41, 60). One IFN-inducible gene product, protein kinase R (PKR), is best known for its ability to bind dsRNA and subsequently phosphorylate the α subunit of eukaryotic initiation factor 2 (eIF2α), thereby inhibiting translation initiation during viral infection (24). PKR has also been implicated in contributing to NF-κB activation, IRF1 activation, and IFN induction by poly(I:C), independent of its kinase activity, as another mechanism of innate antiviral host defense (3, 7, 12, 34, 70, 77).
We generated a number of HPIV1 mutants to investigate C protein function as well as to generate novel live vaccines that would be attenuated by disturbing the IFN antagonistic function of the C proteins, a strategy that has been shown to result in attenuation of replication in vivo (4, 5, 64). Previously, a phenylalanine-to-serine substitution of amino acid 170 of SeV was shown to significantly attenuate a highly virulent SeV strain, increasing the 50% lethal dose (LD50) 20,000-fold in mice (19). This CF170S mutation was introduced into the P/C gene of recombinant HPIV1 by reverse genetics (30, 47). Whereas wild-type (WT) HPIV1 infection prevented IRF3 dimerization, its nuclear translocation, and IFN-β production, infection with HPIV1 containing the CF170S mutation (F170S virus) led to IRF3 activation and IFN-β production (64). Not unexpectedly, the F170S mutant was attenuated for replication in the respiratory tracts of hamsters and African green monkeys (AGMs) (4, 64). A stabilized version of this mutation is present in a live-attenuated investigational HPIV1 vaccine that is currently in phase 1 trials in children (ClinicalTrials.gov ID no. NCT00641017). We also constructed an HPIV1 mutant, referred to as P(C−), that does not express any of the four C proteins due to point mutations that silence the C open reading frame and which was even more attenuated than the F170S strain in vivo (6).
In the present study, we found that the C proteins of HPIV1 profoundly suppress the innate immune response in epithelial cells by preventing the activation of both IRF3 and NF-κB. We found that mutation or silencing of the C gene ablated both of these inhibitory effects. In contrast to the WT, infection with both F170S and P(C−) HPIV1 resulted in the accumulation of significant amounts of cytoplasmic dsRNA with kinetics that were more rapid for the P(C−) virus. The accumulation of dsRNA coincided with an increase in the accumulation of viral genomic/antigenomic RNA or mRNA and a decrease in N protein levels, which could lead to incomplete encapsidation. The enhanced induction of IFN-β observed with these mutants relied mainly on MDA5 rather than on RIG-I. PKR activation coincided with dsRNA accumulation, contributed to IFN-β induction, and restricted viral C protein expression. Thus, PKR activation provides a mechanism that allows the host cell to eliminate the very protein responsible for evading virus detection.
MATERIALS AND METHODS
Cells, viruses, and DNA transfection.
A549 human respiratory epithelial cells were maintained in F-12 medium supplemented with 0.1 mg/ml gentamicin sulfate, 4 mM l-glutamine, and 10% fetal bovine serum (FBS). Human embryonic kidney 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and gentamicin sulfate. Mouse embryonic fibroblasts (MEFs) were prepared from RIG-I+/− MDA5+/−, RIGI−/−, and MDA5−/− mice and maintained in DMEM with 10% FBS and gentamicin sulfate, as described previously (32). Stable short hairpin RNA-containing PKR (shPKR) knockdown and control (shCON) 293A cells were maintained in DMEM with 10% FBS and gentamicin sulfate supplemented with 1 μg/ml puromycin as described previously (75). Doxycycline-inducible, stable cells that express WT C′ protein tagged at the C terminus with a c-Myc epitope were generated using Flp-In T-REx 293 cells grown in 100 μg/ml hygromycin and 15 μg/ml blasticidin (Invitrogen). C protein expression was induced by incubation for 24 h with 1 μg/ml doxycycline (Sigma). Sucrose gradient-purified recombinant WT, F170S, and P(C−) HPIV1 strains were prepared and the titers were determined by limiting dilution assay as reported previously (8). All viral infections were carried out using a multiplicity of infection (MOI) of 5 tissue culture infective doses (TCID50)/ml at 32°C in medium containing 1.2% trypsin TrypLE Select (Invitrogen). For type I IFN neutralization, 5,000 neutralizing antibody units (NU)/ml anti-IFN-α and 2,000 NU/ml anti-IFN-β (PBL InterferonSource) were added at 12 h postinfection (p.i.). For transient transfections, 3 μl Lipofectamine 2000 (Invitrogen) per μg total DNA was added for every 400,000 293T cells for 48 h at 37°C. For poly(I:C)-induced IFN-β expression, 3 μl Lipofectamine 2000 per μg of poly(I:C) (Sigma) was added for every 400,000 293T cells for 6 h at 37°C. For the preparation of cellular extracts, cells were suspended in lysis buffer (1% Triton X-100, 50 mM Tris HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA) with protease inhibitor cocktail (Sigma) at a concentration of 2,000 cells/μl for 30 min at 4°C and centrifuged at 15,000 × g for 10 min. Halt protease and phosphatase inhibitor cocktail (Pierce) were added to the lysis buffer whenever phosphorylation states were examined. After centrifugation, the supernatant was used for Western blotting and immunoprecipitation (IP).
Real-time RT-PCR.
Intracellular RNA or virion-associated RNA was extracted and analyzed by real-time reverse transcription (RT)-PCR as described previously (8). The relative levels of expression of mRNA for human and murine IFN-β and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were determined by using TaqMan gene expression assays (Applied Biosystems) for human IFN-β (Hs01077958_s1), human GAPDH (Hs99999905_m1), mouse IFN-β (Mm00439546_s1), and mouse GAPDH (Mm99999915_g1), with the RT performed on intracellular RNA with random hexamers. Experiments were performed in triplicate. Combined genomic and antigenomic RNA, genomic RNA, antigenomic RNA, and IFN-β expression in shPKR 293 cells was measured in triplicate and confirmed in two independent experiments. Student's t test was used to determine statistical significance. To measure the relative copy numbers of HPIV1 genome plus antigenome in sucrose-purified virus stocks, RT was performed on extracted virion RNA with random hexamers and PCR was performed to amplify genome nucleotides 15502 to 15600, corresponding to the downstream end of the L gene and the adjoining trailer region (forward primer ACCAGACAAGAGTTTAAGAAATATCGA, reverse primer AGAAATCCCTTTAACTGACTCATAAAAACATAGT, and probe CAACAGACAAGAGTATTAATAAT). To distinguish between genomic and antigenomic RNAs in intracellular RNA, RT was performed with a positive-sense primer representing genome nucleotides 15502 to 15519 (AGAAATCCCTTTAACTGA) to measure the genome or with a negative-sense primer representing genome nucleotides 15583 to 15600 (CTTAAACTCTTGTCTGGT) to measure the antigenome. PCR was then performed to amplify nucleotides 15502 to 15600 of the genome or its complement, using the primer-probe set described above. To measure the relative copy numbers of the N and P mRNAs in intracellular RNA, RT was performed with oligo(dT) primer and PCR was performed to amplify, in the case of N, genome nucleotides 917 to 1014 (forward primer AGCATCTTTCATGAACACCATCAAGT, reverse primer GGCTTCTCAATTTGTTTATATCTGGTCTCA, and probe CCGCCCTGACACTAT) and, in the case of P, genome nucleotides 2624 to 2685 (forward primer GAAGACCACAACACCAAAACCA, reverse primer GCTGCTGACTCTTCGTTCTTTG, and probe ACGGTGGAACTTTTG).
Western blotting and antibodies.
Cells were lysed and clarified as described above for the preparation of cellular extracts, and the resulting extracts (15 μl) were denatured and reduced in sample loading buffer (0.3 M Tris HCl, 5% SDS, 50% glycerol, 0.1 M dithiothreitol [DTT]) at 95°C for 5 min, separated by SDS-PAGE on 4 to 20% Tris-glycine gels (Invitrogen), and transferred to 0.45-μm-pore polyvinylidene difluoride (PVDF) membranes (Invitrogen). Membranes were blocked overnight with 5% milk powder and 1% bovine serum albumin (BSA) or whenever probing with phospho-specific antibodies, with 5% BSA in 0.1% Tween-phosphate-buffered saline (PBS). Rabbit polyclonal antiserum was raised against a synthetic peptide corresponding to an internal sequence (amino acids 78 to 92; TITTKTEQSQRRPK) in the HPIV1 C proteins shared by C′, C Y1, and Y2 by ProSci (Poway, CA), against a synthetic peptide corresponding to an internal sequence (amino acid residues 485 to 499; RRLADRKQRLSQANN) in the HPIV1 N protein by ProSci, and against a synthetic peptide corresponding to a N-terminal sequence (amino acids 10 to 24; RDPEAEGEAPRKQES) in the HPIV1 P protein by Spring Valley Laboratories (Woodbine, MD). Membranes were probed with rabbit antiserum at a 1:1,000 dilution with 5% milk and 1% BSA in 0.1% Tween-PBS for 1 h. The antibodies used included rabbit polyclonal antibodies to human pIRF3 (S396) (#07-582; Millipore), IRF3 (sc-9082; Santa Cruz Biotechnology), IκB-β (sc-945; Santa Cruz Biotechnology), ATF2 (sc-187; Santa Cruz Biotechnology), pP38 (T180/Y182) (sc-101759; Santa Cruz Biotechnology), P38 (sc-7149; Santa Cruz Biotechnology), PKR (sc-707; Santa Cruz Biotechnology), pEIF2α (S51) (BML-SA405; Enzo Life Sciences), eIF2α (E0157; Sigma), MDA5 (ALX-210-935; Alexis Biochemicals), Flag epitope tag (F7425; Sigma), hemagglutinin (HA) epitope tag (H6908; Sigma), Myc epitope tag (C3956; Sigma), and rabbit monoclonal antibody to human pPKR (T451) (#2283-1; Epitomics) or mouse monoclonal antibodies to human pATF2 (T71) (sc-8398; Santa Cruz Biotechnology), GAPDH (G9295; Sigma), and V5 epitope tag (R960-25; Invitrogen). Horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Santa Cruz Biotechnology) was used at a dilution of 1:5,000, and horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Kirkegaard & Perry Laboratories) was used at a dilution of 1:2,400 for 1 h. Membranes were washed three times for 5 min with 0.1% Tween-PBS after primary and secondary antibody incubations. Enhanced chemiluminescent substrate (Pierce) was used to visualize proteins on BIOMAX MR film (Kodak). Relative band intensity was determined by densitometry using ImageJ.
Immunoprecipitation.
Cell lysates extracted from transfected 293T cells were mixed for 16 h at 4°C with anti-Myc agarose (Pierce), washed extensively with wash buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl), eluted with 0.1 M glycine (pH 2.8), and separated by SDS-PAGE.
Immunofluorescence.
A549 cells were fixed with 2% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 10% goat serum and 0.02% BSA for 2 h. Mouse monoclonal J2 anti-dsRNA antibody (English and Scientific Consulting) at a 1:200 dilution or rabbit polyclonal anti-HPIV1 P antibody at a dilution of 1:100 was added in 1% goat serum and 0.002% BSA for 1 h at 37°C. Texas Red-conjugated goat anti-rabbit and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse secondary antibodies (Jackson ImmunoResearch) were added at a dilution of 1:250 for 45 min at 37°C. Cells were mounted in ProLong Gold reagent with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) and visualized on a Leica SP5 confocal microscope.
Plasmid constructs.
A cDNA for HPIV1 C was generated by PCR (forward primer CATCGGTACCGTGGATACCTCAGCATCCAAAACTCTCCTTCCC and reverse primer ATCACATGCGGCCGCTTACAGATCTTCTTCAGAAATAAGTTTTTGTTCTTCTTGTACTATGTGTGCTGCTAGTTTCC) of the full-length antigenomic cDNA for WT HPIV1 and constructed with the insertion of a C-terminal c-Myc tag and KpnI and EagI restriction sequences to allow cloning into pcDNA3.1 (Invitrogen). The nucleotide sequence was confirmed by using BigDye terminator and a DNA analyzer 3730 (Applied Biosystems). pEF-IRES V5-TRIM25 was kindly provided by Michaela Gack and Jae Jung (18). pEF-BOS HA-Riplet was kindly provided by Hiroyuki Oshiumi and Tsukasa Seya (49). pRK5 Flag-TRAF3 and Myc-TRADD3 were kindly provided by Jurg Tschopp (44). pCMV2 Flag-TRAF6 was kindly provided by Masashi Muroi and Ken-ichi Tanamoto (45). pEF-BOS Flag-RIGI and Flag-MDA5 were kindly provided by Takashi Fujita (73, 74). pcDNA3 Flag-TBK1 was kindly provided by Katherine Fitzgerald and Makoto Nakanishi (15, 63). pCMV2 Flag-IPS1 has been previously described (33). pcDNA3 Flag-BIRC2 and Flag-BIRC4 were kindly provided by Temesgen Samuel (53). pFlag-CMV2 Flag-FADD was kindly provided by Andrew Thorburn (62).
RESULTS
HPIV1 activation of the interferon induction pathway.
We found previously that, in stark contrast to WT HPIV1, HPIV1 with an F170S point mutation in the C proteins or in which the C gene was silenced by point mutations induced the transcription of a broad array of cellular genes—in particular genes involved in type I IFN production and the subsequent antiviral response (8, 64). To investigate the mechanism of C-mediated antagonism of the host innate immune response, we used three cell types. A549 cells were used to model infection in human respiratory epithelial cells, 293 human kidney cells were used for transfection and knockdown experiments, and mouse embryonic fibroblasts were used for knockout experiments. We found that the HPIV1-induced IFN phenotypes were comparable between the three cell types: specifically, in each cell type, F170S and P(C−) HPIV1 induced similar, abundant levels of IFN-β mRNA expression, whereas WT HPIV1 did not induce significant IFN-β expression (data not shown).
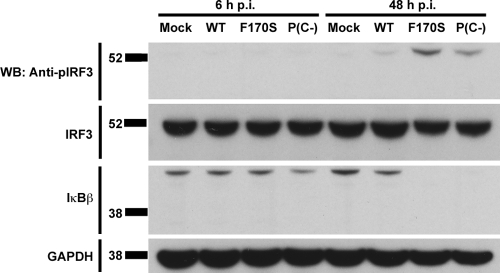
IFN-β induction is transcriptionally controlled by the coordinate assembly of an enhanceosome consisting of IRF3, NF-κB, and AP-1 at the IFN-β promoter (40, 71). We examined the activation of the three components of the IFN-β enhanceosome during HPIV1 infection and found that both F170S and P(C−) HPIV1 induced IRF3 phosphorylation and IκBβ degradation, which would permit nuclear translocation of IRF3 and NF-κB, respectively, whereas WT HPIV1 effectively blocked IRF3 phosphorylation and IκBβ degradation (Fig. 1). On the other hand, WT, F170S, and P(C−) HPIV1 did not induce detectable phosphorylation of P38 or ATF2 (see Fig. S1 in the supplemental material).
FIG. 1.
Activation of the IRF3 and NF-κB components of the IFN-β enhanceosome by infection with WT and mutant HPIV1. A549 cells were infected with WT, F170S, or P(C−) HPIV1 at an MOI of 5 for 6 or 48 h. Phosphorylated IRF3 (Ser 396), total IRF3, and total IκBβ were detected by Western blotting (WB).
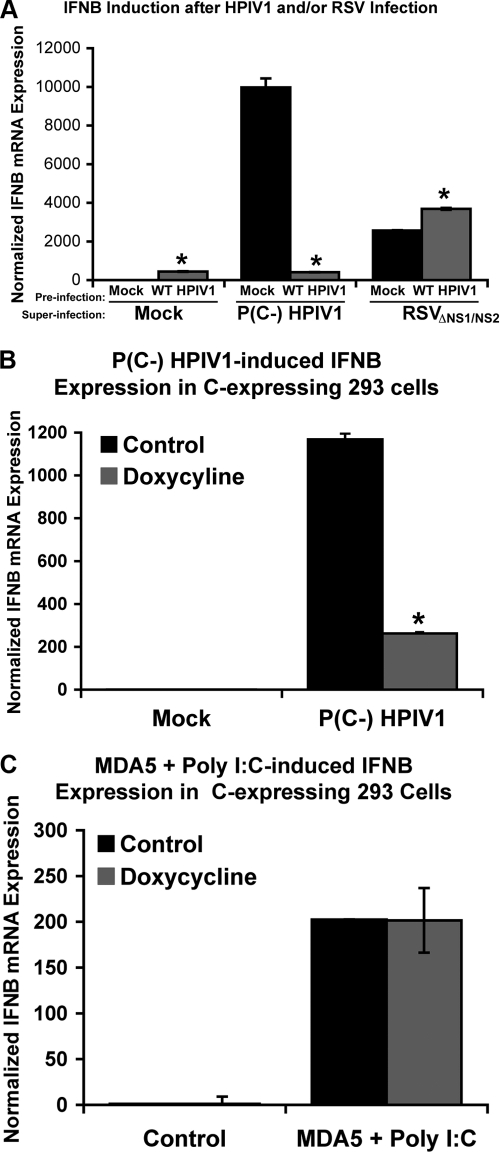
To investigate the basis for the observed antagonism of the host response by the WT HPIV1 C proteins, a selective search for cellular C-interacting proteins was performed among RIG-I/MDA5 pathway components. Cells transfected with expression plasmids expressing a tagged C′ protein and various tagged proteins of the RIG-I/MDA5 pathway were lysed and analyzed in coimmunoprecipitation experiments under various conditions in an attempt to sensitively detect any interactions. However, we were unable to detect any cellular C-interacting proteins among those tested, including TRIM25, Riplet, RIGI, MDA5, IPS1, TBK1, TRAF3, TRAF6, BIRC2, BIRC4, TRADD, and FADD, even though, under the same conditions, TRAF3 was immunoprecipitated with TRADD, a known binding partner of TRAF3 (data not shown). In addition, yeast two-hybrid screens using HPIV1 C proteins as bait did not detect any C-interacting proteins in the RIG-I/MDA5 pathway, and neither did immunoprecipitation (IP)/mass spectroscopy screens (data not shown). Since we could not identify any cellular C-interacting proteins, we investigated whether supplying the HPIV1 C proteins in trans could block the IFN-β induction pathway triggered by infection with respiratory syncytial virus (RSV) or by transfection with poly(I:C), which mainly involves activation of RIG-I and MDA5, respectively (38, 55). Infection with WT HPIV1, as a source of C proteins, strongly suppressed IFN-β induction in response to superinfection with P(C−) HPIV1, but failed to block IFN-β induction in response to superinfection with an RSV mutant that expresses green fluorescent protein (GFP) from an added gene and lacks the NS1 and NS2 RSV IFN antagonist genes (ΔNS1/NS2 RSV) (Fig. 2 A). As a control, analysis of GFP expression showed that the efficiency of infection by ΔNS1/NS2 RSV was not affected by preinfection with WT HPIV1 (not shown). To test whether the HPIV1 C proteins could interfere with IFN production triggered through MDA5, a doxycycline-inducible stable human cell line that expresses the HPIV1 C proteins was generated. In this system, expression of the HPIV1 C proteins in trans strongly suppressed IFN-β induction by infection with P(C−) HPIV1 (Fig. 2B), confirming the efficient expression of functional C protein. In contrast, expression of the HPIV1 C proteins had no effect on IFN-β induction in response to transfection with poly(I:C) (Fig. 2C). In this last experiment, the cells had also been transfected with a plasmid expressing human MDA5 to provide the homologous sensor for dsRNA prior to the transfection of poly(I:C). Taken together, these results indicate that the HPIV1 C proteins supplied in trans can efficiently suppress IFN induction by HPIV1 mutants, but not by heterologous inducers.
FIG. 2.
IFN-β induction in A549 cells by heterologous RIG-I and MDA5 stimuli is not affected by expression of the HPIV1 C proteins in trans. (A) A549 cells were preinfected with WT HPIV1 at an MOI of 5 for 24 h and then superinfected with either P(C−) HPIV1 or GFP-expressing ΔNS1/NS2 RSV at a MOI of 5 for another 24 h. IFN-β expression was measured by real-time PCR and was normalized to the mock-preinfected, mock-superinfected sample. *, P < 0.01 compared to mock-preinfected samples. (B) C protein expression was induced in stable C protein-expressing 293 cells with 1 μg/ml doxycycline for 24 h. Cells were then mock infected or infected with P(C−) HPIV1 at an MOI of 5 for another 24 h. IFN-β expression was measured by real-time PCR and was normalized to the control treated, mock-infected sample. *, P < 0.01 compared to control treated cells. (C) C protein expression was induced in stable C protein-expressing 293 cells with 1 μg/ml doxycycline for 24 h, transfected with 2.5 μg MDA5 expression plasmid for another 18 h, and then transfected with 10 μg poly(I:C) for 6 h. IFN-β expression was measured by real-time PCR and was normalized to the control treated, control transfected sample.
HPIV1-induced IFN-β expression is mediated mostly by MDA5.
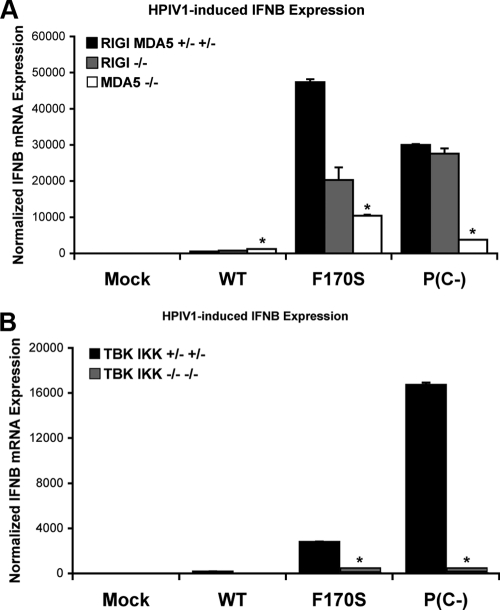
Since we were unable to obtain either direct or indirect evidence that the C proteins blocked IFN production by interacting with a host factor of the RIG-I/MDA5 pathway, we evaluated the involvement of this pathway in IFN-β induction by HPIV1, using knockout MEFs. In contrast to WT HPIV1, F170S and P(C−) HPIV1 strongly induced IFN-β expression in control MEFs (Fig. 3A, black bars). RIG-I knockout led to a slight decrease (2.3-fold) in IFN-β expression in F170S-infected cells, but no effect was observed in P(C−)-infected cells (Fig. 3A). MDA5 knockout led to a greater reduction in IFN-β expression in both F170S- and P(C−)-infected cells (4.5-fold and 8.0-fold, respectively) (Fig. 3A). TBK1 and IKKɛ double knockout led to a dramatic reduction in IFN-β expression in both F170S- and P(C−)-infected cells, confirming the requirement for TBK1 and IKKɛ in IFN induction during infection with F170S or P(C−) HPIV1 (Fig. 3B). The observation that knockouts of TBK1 and IKKɛ led to almost complete inhibition of IFN-β mRNA synthesis, whereas knockout of MDA5 led to only a partial inhibition, raised the possibility that factors in addition to MDA5 contribute to the induction of IFN-β mRNA via TBK1 and IKKɛ.
FIG. 3.
IFN-β expression in RIG-I/MDA5 pathway knockout MEFs infected with WT or mutant HPIV1. RIG-I and MDA5 knockout MEFs (A) and TBK1 and IKKɛ double-knockout MEFs (B) were infected with WT, F170S, or P(C−) HPIV1 at a MOI of 5 for 48 h. IFN-β expression was measured by real-time PCR and normalized to mock-infected control MEFs. *, P < 0.01 compared to control MEFs infected with the same virus.
Intracellular dsRNA accumulates in cells infected with C mutant HPIV1 viruses.
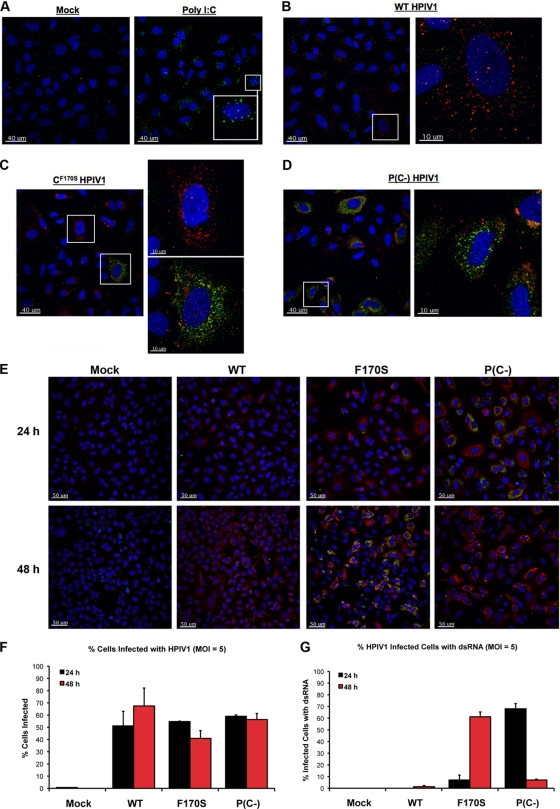
Since knockout of the dsRNA sensor MDA5 strongly reduced IFN-β expression during infection with F170S or P(C−) HPIV1, we looked for the presence of dsRNA in HPIV1-infected cells by immunofluorescence using a monoclonal antibody against dsRNA. Transfection of poly(I:C) was used as a positive control, demonstrating the ability of the dsRNA-specific monoclonal antibody to detect intracellular dsRNA (Fig. 4A). dsRNA was not detected in WT-infected cells at either 24 or 48 h (Fig. 4B and E), similar to observations previously reported for a number of negative-sense RNA viruses, including influenza, La Crosse, Sendai, and Newcastle disease viruses (67). In contrast, dsRNA was readily detected in cells infected with either the F170S or P(C−) virus; however, the kinetics of accumulation differed. In the case of F170S HPIV1, less than 10% of infected cells were positive for dsRNA at 24 h, whereas over 60% of infected cells were positive at 48 h (Fig. 4C, E, and F). In contrast to F170S HPIV1, the accumulation of dsRNA in response to P(C−) HPIV1 occurred more rapidly, with approximately 70% of infected cells containing detectable dsRNA at 24 h. However, the accumulation of dsRNA in response to the P(C−) virus was also transient: by 48 h, less than 10% of P(C−)-infected cells contained dsRNA (Fig. 4D to F). This difference did not appear to be due to differences in the kinetics or efficiencies of infection: the percentages of cells infected with WT, F170S, and P(C−) HPIV1 were similar at 24 and 48 h, as assessed by immunofluorescence using polyclonal antibodies specific to the HPIV1 P protein (Fig. 4G).
FIG. 4.
Immunofluorescence of dsRNA in A549 cells infected with WT or mutant HPIV1. A549 cells were transfected with poly(I:C) (0.02 μg/μl) (A) or infected with WT (B), F170S (C), or P(C−) (D) HPIV1 at an MOI of 5 for 24 h. HPIV1 P protein (red) was detected using a rabbit polyclonal anti-P antibody and a Texas Red-conjugated secondary anti-rabbit antibody. dsRNA (green) was detected with a mouse monoclonal anti-dsRNA antibody and an FITC-conjugated secondary anti-mouse antibody. (E) dsRNA and the HPIV1 P protein were examined as in panels A to D in A549 cells infected with WT, F170S, or P(C−) HPIV1 at an MOI of 5 for 24 and 48 h. (F and G) Quantitation of the number of cells in panel D with and without detectable dsRNA (F) or HPIV1 P protein (G) at 24 and 48 h. dsRNA levels are representative of two independent experiments.
One possible explanation for the increased accumulation of intracellular dsRNA in cells infected with the F170S and P(C−) viruses was that these virus preparations contained disproportionately high levels of defective interfering particles, which, for nonsegmented negative-strand RNA viruses, usually have copy-back genomes that can form base-paired panhandles. To investigate this possibility, we assayed a sucrose gradient-purified preparation of each virus for (i) the number of infectious particles, measured by limiting dilution assay; and (ii) the relative copy number of genomic plus antigenomic RNA, measured by real time RT-PCR with primers specific to the trailer region that would be present in both standard and defective genomes. We then calculated the ratio of infectious particles to genome/antigenome copies for each virus and then normalized these ratios to that of the F170S virus, which was assigned a value of 1.0. The resulting values were as follows: F170S, 1.0; WT, 0.28; and P(C−), 0.09. In other words, F170S HPIV1 had the highest ratio of infectious particles to genome copies, and P(C−) had the lowest, with WT HPIV1 being intermediate. This lack of a consistent trend (i.e., the F170S virus had a higher ratio of infectious particles to genome/antigenome copies than the WT virus, and yet the F170S virus led to dsRNA accumulation, whereas the WT virus did not) suggests that the dsRNA produced during infection with the F170S and P(C−) viruses probably cannot be attributed to the presence of a disproportionately high content of defective interfering particles in the initial inoculum.
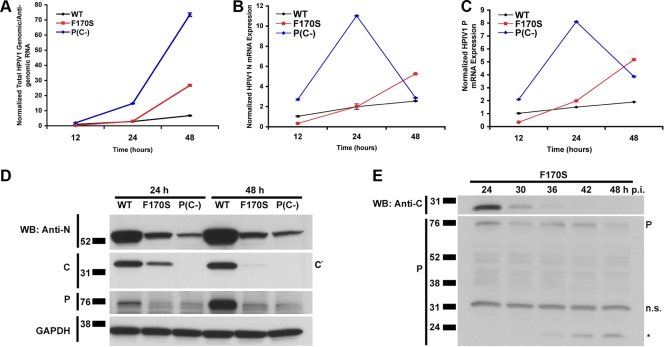
Mutation or deletion of the HPIV1 C proteins results in increased viral RNA synthesis and decreased accumulation of viral proteins.
Another potential source for intracellular dsRNA could arise from annealing between incompletely encapsidated negative-sense genomic RNA and either positive-sense antigenomic RNA or mRNA, which might occur if mutation or deletion of the C proteins perturbed viral RNA production. We therefore used real-time RT-PCR to monitor viral RNA accumulation in A549 cells infected with the WT, F170S, or P(C−) viruses. The combined amount of genomic plus antigenomic RNA was quantified using primers/probes specific for the trailer region (Fig. 5A), whereas the N and P mRNAs (Fig. 5B and C, respectively) were quantified using oligo(dT) for reverse transcription followed by gene-specific primers/probes for PCR. In cells infected with WT HPIV1, the levels of genome/antigenome, N mRNA, and P mRNA rose gradually over the time course of the experiment (Fig. 5A to C). For the F170S mutant, the levels of these RNAs were similar to those of WT HPIV1 at 24 h, but then increased substantially by 48 h. A very different pattern was observed during infection with P(C−) virus. The amount of genome/antigenome was larger than those for the other two viruses at 24 h and continued to increase to a very high level at 48 h (Fig. 5A). In contrast, the levels of N and P mRNA were extremely high at 24 h but thereafter dropped sharply (Fig. 5B). In addition, the relative levels of intracellular genomic versus antigenomic HPIV1 RNA were measured by reverse transcription with positive- or negative-sense primers, to prime genomic or antigenomic RNA, respectively, followed by real-time PCR (see Fig. S2 in the supplemental material). This showed that the ratios of genomic RNA to antigenomic RNA (average ratio, 17:1) were similar for each virus at each time point (see Fig. S2 in the supplemental material) and thus were not detectably perturbed by mutation/deletion of the C protein.
FIG. 5.
Viral RNA and protein expression over time during infection with WT and mutant HPIV1. A549 cells were infected with WT, F170S, or P(C−) HPIV1 at an MOI of 5 for 12, 24, or 48 h. Relative amounts of HPIV1 (A) combined genomic and antigenomic RNA, (B) N mRNA, and (C) P mRNA were measured by real-time PCR. The values were normalized to the 12-h time point for WT HPIV1, which was assigned a value of 1.0. (D) HPIV1 N, C, and P proteins were detected by Western blotting from samples collected in parallel to the samples in panels A to C. Shown is a representative of two independent experiments with similar results. (E) A549 cells were infected with F170S HPIV1 at an MOI of 5 for 24, 30, 36, 42, or 48 h. The C and P proteins were detected by Western blotting. P denotes the position of the P protein, n.s. indicates a nonspecific band, and the asterisk indicates a potential P cleavage or degradation product detected by the polyclonal anti-P antibody.
We also monitored the accumulation of cell-associated viral proteins in the time course experiment described above by Western blot analysis (Fig. 5D). WT-infected cells contained higher levels of N and P protein at both 24 and 48 h compared to the mutant viruses, which was surprising since WT-infected cells contained the lowest levels of N and P mRNA (Fig. 5D). In contrast, although P(C−)-infected cells contained the highest levels of genomic/antigenomic RNA at both time points and very high levels of N and P mRNAs at 24 h, they contained the lowest levels of N and P protein at both time points (Fig. 5A to D). Similarly, F170S-infected cells contained much less N and P protein than WT-infected cells despite higher levels of N and P mRNA. Lysates from WT-infected A549 cells also contained a major band of C′ protein, which is the most abundant of the HPIV1 C proteins. As expected, no C proteins were detected with P(C−) HPIV1 infection. The C′ protein also was detected in lysates from F170S-infected cells at 24 h but, as with the N and P proteins, in lower abundance compared to WT HPIV1. Interestingly, however, the C′F170S protein was not detected at 48 h. We further investigated the kinetics of accumulation of the C′F170S protein in additional time course experiments. This showed that the C′F170S protein was present at 12 and 24 h postinfection (data not shown). However, beginning at 30 h postinfection, the level of C′F170S protein began to decline until it was no longer detectable by 42 h (Fig. 5E). In this experiment, the level of the P protein expressed by F170S HPIV1 also decreased with time, coincident with the appearance of a smaller P-specific band that might be a degradation product (Fig. 5E).
These observations indicated that HPIV1 RNA synthesis indeed was perturbed by mutation and, in particular, deletion of the C protein. These observations also indicate that viral protein synthesis was inhibited in cells infected with F170S and P(C−) HPIV1. The increased production of viral RNA by F170S and P(C−) HPIV1, in the context of reduced levels of N and P protein necessary for encapsidation, could provide for the accumulation of incompletely encapsidated genome capable of annealing with positive-sense RNA to form the observed dsRNA.
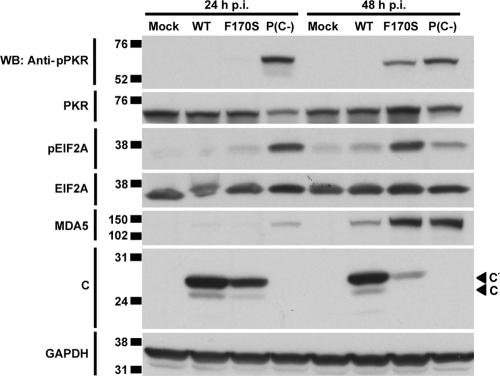
HPIV1 infection and dsRNA accumulation induce PKR activation.
Since dsRNA was highly abundant during infection with P(C−) and F170S HPIV1 at 24 and 48 h, respectively, we examined PKR activation at the same time points. When bound to dsRNA, PKR is activated by autophosphorylation. Activated PKR can phosphorylate eIF2α, resulting in an inhibition of translational initiation (24) that potentially could contribute to the reduced accumulation of viral proteins in cells infected with F170S and P(C−) HPIV1. Consistent with this idea, we found that both F170S and P(C−) HPIV1 induced PKR autophosphorylation and eIF2α phosphorylation, with kinetics mirroring those of dsRNA accumulation (Fig. 4E and F and Fig. 6). Specifically, P(C−) HPIV1 infection induced activation of PKR by 24 h, and F170S HPIV1 infection induced activation of PKR by 48 h. In addition, infection with either mutant strongly induced the expression of MDA5, a highly IRF- and IFN-inducible gene (Fig. 6). The C′ and C proteins were expressed at 24 and 48 h in cells infected with WT HPIV1 and not in cells infected with P(C−) HPIV1, as expected. Consistent with the results in Fig. 5D, both species were present in F170S-infected cells at 24 h, and their accumulation greatly decreased by 48 h (Fig. 6). Thus, infection with F170S and P(C−) HPIV1 activated PKR and phosphorylated eIF2α, which might contribute to the reduced accumulation of viral proteins. However, this may not be a complete explanation for the reduced accumulation of viral proteins in the case of F170S HPIV1, since there was little activation of PKR and eIF2α at 24 h (Fig. 6), a time when the reduction in protein accumulation was already evident (Fig. 5D and 6).
FIG. 6.
PKR activation during infection with WT and mutant HPIV1. A549 cells were infected with WT, F170S, or P(C−) HPIV1 at an MOI of 5 for 24 or 48 h. PKR phosphorylation, total PKR, eIF2α (EIF2A) phosphorylation, total eIF2α, total MDA5, and C proteins were detected by Western blotting.
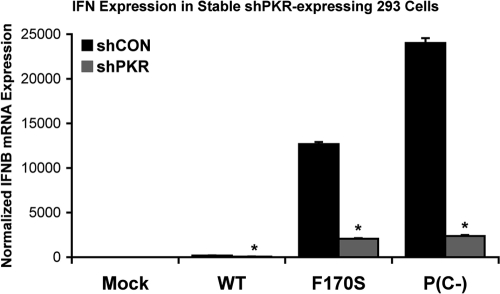
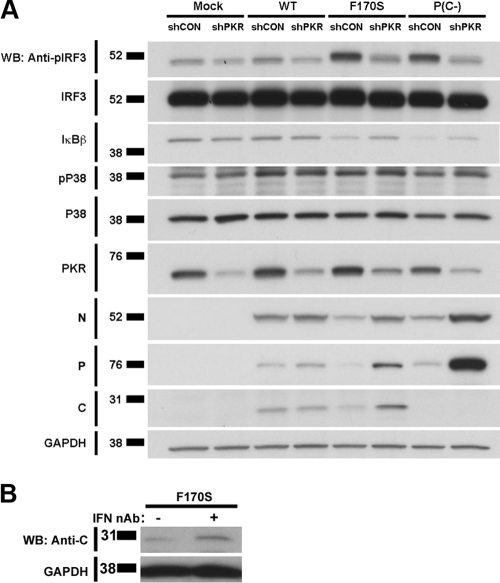
PKR contributes to HPIV1-induced IFN induction as well as the reduced accumulation of viral proteins.
We examined the effects of PKR activation using stable PKR knockdown 293 cells, in which expression of PKR expression was strongly reduced, although not completely blocked, due to the constitutive expression of an shRNA specific to PKR. PKR knockdown significantly reduced IFN-β expression in F170S- and P(C−)-infected cells by 6.2- and 10.1-fold, respectively (Fig. 7). This reduction in IFN-β expression corresponded with a reduction in IRF3 phosphorylation (3.1- and 3.4-fold, respectively) (Fig. 8A; see Fig. S3a in the supplemental material for quantitation). IκBβ degradation was only slightly reduced (1.3- and 1.6-fold, respectively) in PKR knockdown cells (Fig. 8A; see Fig. S3b in the supplemental material for quantitation). P38 phosphorylation was similar in control and PKR knockdown cells (Fig. 8A; see Fig. S3c in the supplemental material for quantitation). In the mock-infected cells, the expression of PKR in the knockdown cells was 25% that of the control knockdown cells (Fig. 8A; see Fig. S3d in the supplemental material for quantitation). Interestingly, PKR knockdown also reversed the decrease in the accumulation of the C′F170S, N, and P proteins during infection with the C mutant/deletion viruses, an effect that was not observed during infection with the WT HPIV1 (Fig. 8A; see Fig. S3e to g in the supplemental material for quantitation). These findings indicated that the reduction in viral protein synthesis in normal cells infected with the C mutant viruses (e.g., Fig. 5D and E and Fig. 6) indeed is linked to PKR activity. Since PKR is an IFN-inducible gene product, the induction of additional PKR due to IFN-mediated signaling could contribute to the loss of viral protein expression during infection with the mutant HPIV1 viruses. Indeed, we found that the decrease in C′F170S protein expression could be partially blocked by IFN-neutralizing antibodies (Fig. 8B).
FIG. 7.
IFN-β expression induced by infection with WT and mutant HPIV1 during PKR knockdown. Stable 293 cells expressing the shPKR and control shCON small hairpin RNAs were infected with WT, F170S, or P(C−) HPIV1 at an MOI of 5 for 48 h. IFN-β expression was measured by real-time PCR and normalized to mock-infected shCON cells. *, P < 0.01 compared to control 293A cells infected with the same virus.
FIG. 8.
Effect of PKR on activation of the components of the IFN-β enhanceosome and accumulation of the C protein in HPIV1 WT and mutant virus-infected cells. (A) Stable shPKR-expressing and control shCON 293A cells were infected with WT, F170S, or P(C−) HPIV1 at an MOI of 5 for 48 h. Phosphorylated IRF3, total IRF3, total IκBβ, phosphorylated P38, total P38, and total PKR, as well as viral N, P, and C proteins were detected by Western blotting. (B) Type I IFN affects C′ protein expression during HPIV1 infection. A549 cells were infected with F170S HPIV1 at an MOI of 5. At 12 h postinfection, neutralizing antibodies against IFN-α (5,000 NU/ml) and IFN-β (2,000 NU/ml) were added. C protein was detected by Western blotting at 30 h.
DISCUSSION
Infection with C mutant HPIV1s [F170S and P(C−)], but not WT HPIV1, effectively stimulates IFN-β production in epithelial A549 cells (64). In addition, F170S, but not WT, HPIV1 trigger IRF3 dimerization and nuclear translocation, indicating that the wild-type C proteins block the activation of IRF3 (64). In the present study, the mechanisms by which the C proteins prevent IFN-β induction were explored in more detail. IFN-β production is optimally induced by the assembly of an enhanceosome comprised of IRF3, NF-κB, and AP-1 at the IFN-β promoter. Here, we found that the F170S and P(C−) mutants were unable to inhibit IRF3 and NF-κB activation, whereas WT HPIV1 inhibited their activation, suggesting an interaction of the HPIV1 C proteins with the RIG-I/MDA5 pathway upstream of IRF3 and NF-κB activation. Numerous examples of viral proteins that actively inhibit this pathway are known (10). For instance, the influenza A virus NS1 protein inhibits RIG-I activation by binding to TRIM25 and the hepatitis C NS3/4A protease cleaves and inactivates MAVS (17, 37). In this context, we hypothesized that the HPIV1 C proteins acted to inhibit IFN-β production by binding to or otherwise interfering with one or more components of the RIG-I/MDA5 pathway. However, four observations led us to reject this hypothesis: (i) none of the known members of the RIG-I/MDA5 pathway that we examined could be immunoprecipitated with the C proteins; (ii) there was no significant decrease in the abundance or shift in the gel mobility of any of these host proteins indicative of C protein-mediated modification or degradation; (iii) the yeast two-hybrid assay and immunoprecipitation in conjunction with mass spectrometry failed to detect interactions; and (iv) supplying the C proteins in trans failed to block IFN-β induction by RSV and poly(I:C), known RIG-I and MDA5 stimuli, respectively. This inability of the HPIV1 C proteins to prevent heterologous MDA5- and RIG-I-mediated IFN-β induction suggested that they do not antagonize the components of the IFN production pathway at or downstream of RIG-I and MDA5 but rather prevent the activation of these pathways.
Knockout of TBK1 and IKKɛ completely ablated the IFN-β response to F170S and P(C−) HPIV1 infection, confirming that the RIG-I/MDA5 pathway was necessary for IFN-β induction by these two viruses. Surprisingly, and in contrast to earlier suggestions that paramyxovirus sensing depended mainly on RIG-I (23, 32, 39, 57), we found that IFN-β induction in murine cells relied more strongly on MDA5 than on RIG-I. However, these results are consistent with more recent studies using SeV that indicated that both RIG-I and MDA5 could contribute to IFN-β production in vitro and resistance in vivo (20, 76). However, these findings cannot be readily extrapolated to HPIV1, because SeV, in contrast to HPIV1, also expresses a V protein, a known inhibitor of MDA5 (2, 11, 20, 57, 76). Since MDA5 primarily recognizes long dsRNA molecules (or, more specifically, webs of dsRNA and single-stranded RNA [ssRNA] [51]), its involvement in IFN-β expression during infection with F170S and P(C−) HPIV1 suggested that accumulation of dsRNA might occur, and this was indeed found to be the case. Although earlier studies had reported dsRNA accumulation after infection with positive-strand RNA viruses, double-stranded RNA viruses, and DNA viruses but not with negative-strand RNA viruses (36, 56, 67, 68), more recent studies also detected dsRNA in cells infected with a C deletion mutant of SeV (59). Our findings of dsRNA accumulation in cells infected with C protein mutants but not in WT HPIV1 suggest that the HPIV1 C proteins are indeed required to prevent the accumulation of dsRNA that would otherwise trigger a potent host innate antiviral response.
The appearance of dsRNA was temporally associated with phosphorylation of PKR and eIF2α and decreased accumulation of the N, P, and C proteins in F170S-infected cells. PKR knockdown studies confirmed that PKR is a major contributor to the loss of the N, P, and CF170S proteins. In addition to its role in inhibiting translation during viral infection, PKR has also been implicated in NF-κB activation, IRF1 activation, and IFN induction by poly(I:C) via MAVS, effects that are independent of its kinase activity and that provide another mechanism of innate antiviral host defense (3, 7, 12, 34, 42, 70, 77). Our data indicate that PKR contributes significantly to IRF3 phosphorylation and induction of IFN-β following infection with HPIV1 C protein mutants that generated dsRNA. These finding are consistent with a previous report on a C-protein-deficient measles virus mutant that required PKR for maximal IFN-β induction (43). Thus, both MDA5 and PKR contribute to the generation of IFN-β during infection with C mutant HPIV1s, and both are activated by dsRNA.
Quantitation of viral RNA levels showed that accumulation of genomic and antigenomic RNA, but also viral mRNA, was more pronounced in P(C−)- and F170S-infected A549 cells than in WT HPIV1-infected cells. Concomitant with the increase in viral RNA species, N and P protein levels decreased, which led us to speculate that this imbalance could give rise to unencapsidated RNAs that could anneal with each other or with excess viral mRNA to form dsRNA. However, we have not yet confirmed the identity of the dsRNA species detected in A549 cells. The current model of genome replication by nonsegmented negative-sense RNA viruses involves N protein encapsidation of the template. In the absence of concurrent encapsidation, naked replication products are made that terminate prematurely (65). These RNAs could stimulate the innate immune system.
For SeV, the C proteins have previously been implicated in inhibiting viral RNA synthesis and regulating the balance between genome and antigenome levels (14, 28, 29). Interestingly, one study reported that infection of primate LLC-MK2 and Vero cells with a SeV C deletion strain led to a predominance of antigenomic RNA compared to genomic RNA and to an increase in viral protein synthesis (29). Our results differ from these in that the accumulation of viral proteins was decreased rather than increased, and deletion of the HPIV1 C proteins did not alter the ratio of viral genomic RNA to antigenomic RNA. Our results provide an alternative explanation for the observation that a recombinant SeV strain expressing HPIV1 C in place of SeV C failed to block IRF3 activation in murine cells (11). This originally was interpreted to indicate that the respirovirus C proteins, like the rubulavirus V protein, are determinants of host range (11). However, it may be that the HPIV1 C proteins do not function efficiently in the context of the heterologous SeV N, P, and L proteins to prevent the accumulation of dsRNA and subsequent IRF3 activation. We (unpublished data) and others (22, 25) have observed an association between the HPIV1 C proteins and proteins of the homologous replication complex, and this association might be virus specific.
This study has a number of limitations that need to be considered when interpreting our results. First of all, the experimental models used (A549, MEF, and 239 cells) have their own limitations. For example, although the A549 cell line is an accepted model for human respiratory epithelial cells, it is an immortalized cell line, and as such its signaling pathways might be altered. To address this concern, we have previously used primary human airway epithelial cells to confirm that the F170S mutant readily induced type I IFN induction in this organized epithelium. However, the P(C−) mutant was too restricted in replication in this organized epithelium to permit a valid comparison to WT HPIV1 (6). Another limitation is the uncertainty as to whether PKR- and IFN-β-induced inhibition of protein synthesis can fully account for the reduced accumulation of viral proteins during infection with the HPIV1 mutants. For example, in cells infected with the F170S mutant, the levels of N and P protein were reduced compared to those in WT-infected cells even at 24 h, a time point before the activation of PKR was detected. Also, while the rapid accumulation of genomic and antigenomic RNA with the P(C−) mutant could be imagined to outpace the synthesis of N and P protein and thus initiate dsRNA formation, this proposed mechanism seems less convincing for the F170S mutant with its more gradual dsRNA accumulation. Thus, there may be additional host inhibitory effects on viral protein accumulation that are activated in C mutant-infected cells or, alternatively, the C proteins may have a positive effect on viral protein synthesis. Further studies to explore these alternative mechanisms of regulating viral protein synthesis are clearly needed.
To our surprise, we were unable to detect a physical interaction between wild-type C proteins and any of the examined members of the RIG-I/MDA5 pathway, either by co-IP or by yeast two-hybrid assay or mass spectroscopy. Rather, the C proteins seem to regulate viral RNA synthesis to prevent the production of dsRNA that would otherwise activate MDA5 and PKR which, in turn, would activate IRF3 and NF-κB, result in the induction of IFN-β, and inhibit protein synthesis. We propose that the following series of events occur during infection with P(C−) HPIV1. dsRNA is generated early, within the first 24 h of infection, due to the combined rapid increase of mRNA, genomic RNA, and antigenomic RNA. The dsRNA activates PKR and also induces IFN-β via the RIG-I/MDA5 pathway. PKR activation and IFN-β induction decrease viral protein synthesis, thereby maintaining low levels of N protein. IFN-β might also inhibit protein synthesis by the induction of inhibitors such as IFIT1 and IFIT2 (61). In addition, apoptosis is activated early in P(C−)-infected cells, but the mechanism underlying this induction remains undefined (6). The combined effects of PKR activation, IFN-β induction, and the initiation of apoptosis are likely responsible for the decline in viral macromolecular synthesis that starts between 24 and 48 h after infection with P(C−) HPIV1. The events occurring during infection with the F170S mutant are delayed but could be similar to those in P(C−) HPIV1-infected cells. The kinetics of dsRNA accumulation and decreased expression of mutant C proteins described here suggest that a race between the evasion of host antiviral responses and the elimination of viral antagonists of dsRNA accumulation is key in determining the outcome of HPIV1 infections.
Supplementary Material
Acknowledgments
We thank Shirin Munir and Christine Winter for providing the sucrose-purified GFP-tagged ΔNS1/NS2 RSV. We are grateful to Owen Schwartz, Juraj Kabat, Lily Koo, and Steven Becker (NIAID Biological Imaging Core) for assistance with confocal microscopy.
This research was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases. This research was also supported in part by a cooperative research and development agreement (CRADA) between NIH and MedImmune, LLC, for the development of PIV and RSV vaccines.
Footnotes
Published ahead of print on 1 December 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 2.Andrejeva, J., et al. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltzis, D., S. Li, and A. E. Koromilas. 2002. Functional characterization of pkr gene products expressed in cells from mice with a targeted deletion of the N terminus or C terminus domain of PKR. J. Biol. Chem. 277:38364-38372. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett, E. J., et al. 2006. Introducing point and deletion mutations into the P/C gene of human parainfluenza virus type 1 (HPIV1) by reverse genetics generates attenuated and efficacious vaccine candidates. Vaccine 24:2674-2684. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett, E. J., et al. 2005. Human parainfluenza virus type I (HPIV1) vaccine candidates designed by reverse genetics are attenuated and efficacious in African green monkeys. Vaccine 23:4631-4646. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett, E. J., et al. 2008. Human parainfluenza virus type 1 C proteins are nonessential proteins that inhibit the host interferon and apoptotic responses and are required for efficient replication in nonhuman primates. J. Virol. 82:8965-8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet, M. C., R. Weil, E. Dam, A. G. Hovanessian, and E. F. Meurs. 2000. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol. Cell. Biol. 20:4532-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boonyaratanakornkit, J. B., et al. 2009. The C proteins of human parainfluenza virus type 1 (HPIV1) control the transcription of a broad array of cellular genes that would otherwise respond to HPIV1 infection. J. Virol. 83:1892-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bousse, T., R. L. Chambers, R. A. Scroggs, A. Portner, and T. Takimoto. 2006. Human parainfluenza virus type 1 but not Sendai virus replicates in human respiratory cells despite IFN treatment. Virus Res. 121:23-32. [DOI] [PubMed] [Google Scholar]
- 10.Bowie, A. G., and L. Unterholzner. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8:911-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers, R., and T. Takimoto. 2009. Host specificity of the anti-interferon and anti-apoptosis activities of parainfluenza virus P/C gene products. J. Gen. Virol. 90:1906-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu, W. M., et al. 1999. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity 11:721-731. [DOI] [PubMed] [Google Scholar]
- 13.Counihan, M. E., D. K. Shay, R. C. Holman, S. A. Lowther, and L. J. Anderson. 2001. Human parainfluenza virus-associated hospitalizations among children less than five years of age in the United States. Pediatr. Infect. Dis. J. 20:646-653. [DOI] [PubMed] [Google Scholar]
- 14.Curran, J., J. B. Marq, and D. Kolakofsky. 1992. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology 189:647-656. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald, K. A., et al. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 16.Forster, J., et al. 2004. Prospective population-based study of viral lower respiratory tract infections in children under 3 years of age (the PRI.DE study). Eur. J. Pediatr. 163:709-716. [DOI] [PubMed] [Google Scholar]
- 17.Gack, M. U., et al. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gack, M. U., et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916-920. [DOI] [PubMed] [Google Scholar]
- 19.Garcin, D., M. Itoh, and D. Kolakofsky. 1997. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology 238:424-431. [DOI] [PubMed] [Google Scholar]
- 20.Gitlin, L., et al. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6:e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb, J., et al. 2009. Community-acquired respiratory viral infections in lung transplant recipients: a single season cohort study. Transplantation 87:1530-1537. [DOI] [PubMed] [Google Scholar]
- 22.Grogan, C. C., and S. A. Moyer. 2001. Sendai virus wild-type and mutant C proteins show a direct correlation between L polymerase binding and inhibition of viral RNA synthesis. Virology 288:96-108. [DOI] [PubMed] [Google Scholar]
- 23.Hausmann, S., J. B. Marq, C. Tapparel, D. Kolakofsky, and D. Garcin. 2008. RIG-I and dsRNA-induced IFNbeta activation. PLoS One 3:e3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershey, J. W. 1991. Translational control in mammalian cells. Annu. Rev. Biochem. 60:717-755. [DOI] [PubMed] [Google Scholar]
- 25.Horikami, S. M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornung, V., et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 27.Hui, D. S., et al. 2008. Influenza-like illness in residential care homes: a study of the incidence, aetiological agents, natural history and health resource utilisation. Thorax 63:690-697. [DOI] [PubMed] [Google Scholar]
- 28.Irie, T., N. Nagata, T. Igarashi, I. Okamoto, and T. Sakaguchi. 2010. Conserved charged amino acids within Sendai virus C protein play multiple roles in the evasion of innate immune responses. PLoS One 5:e10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irie, T., N. Nagata, T. Yoshida, and T. Sakaguchi. 2008. Paramyxovirus Sendai virus C proteins are essential for maintenance of negative-sense RNA genome in virus particles. Virology 374:495-505. [DOI] [PubMed] [Google Scholar]
- 30.Itoh, M., Y. Isegawa, H. Hotta, and M. Homma. 1997. Isolation of an avirulent mutant of Sendai virus with two amino acid mutations from a highly virulent field strain through adaptation to LLC-MK2 cells. J. Gen. Virol. 78:3207-3215. [DOI] [PubMed] [Google Scholar]
- 31.Janeway, C. A., Jr. 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54:1-13. [DOI] [PubMed] [Google Scholar]
- 32.Kato, H., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 33.Kawai, T., et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, A., et al. 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 16:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurotani, A., et al. 1998. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells 3:111-124. [DOI] [PubMed] [Google Scholar]
- 36.Lee, J. Y., J. A. Marshall, and D. S. Bowden. 1994. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology 200:307-312. [DOI] [PubMed] [Google Scholar]
- 37.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 102:17717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, P., et al. 2007. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 81:1401-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loo, Y. M., et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maniatis, T., et al. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb. Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 41.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McAllister, C. S., and C. E. Samuel. 2009. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J. Biol. Chem. 284:1644-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McAllister, C. S., et al. 2010. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J. Virol. 84:380-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michallet, M. C., et al. 2008. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity 28:651-661. [DOI] [PubMed] [Google Scholar]
- 45.Muroi, M., and K. Tanamoto. 2008. TRAF6 distinctively mediates MyD88- and IRAK-1-induced activation of NF-kappaB. J. Leukoc. Biol. 83:702-707. [DOI] [PubMed] [Google Scholar]
- 46.Murphy, B. R., et al. 1988. Current approaches to the development of vaccines effective against parainfluenza and respiratory syncytial viruses. Virus Res. 11:1-15. [DOI] [PubMed] [Google Scholar]
- 47.Newman, J. T., et al. 2004. Generation of recombinant human parainfluenza virus type 1 vaccine candidates by importation of temperature-sensitive and attenuating mutations from heterologous paramyxoviruses. J. Virol. 78:2017-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161-167. [DOI] [PubMed] [Google Scholar]
- 49.Oshiumi, H., M. Matsumoto, S. Hatakeyama, and T. Seya. 2009. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J. Biol. Chem. 284:807-817. [DOI] [PubMed] [Google Scholar]
- 50.Pichlmair, A., et al. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 51.Pichlmair, A., et al. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761-10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed, G., P. H. Jewett, J. Thompson, S. Tollefson, and P. F. Wright. 1997. Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children < 5 years old. J. Infect. Dis. 175:807-813. [DOI] [PubMed] [Google Scholar]
- 53.Santoro, M. M., T. Samuel, T. Mitchell, J. C. Reed, and D. Y. Stainier. 2007. Birc2 (cIap1) regulates endothelial cell integrity and blood vessel homeostasis. Nat. Genet. 39:1397-1402. [DOI] [PubMed] [Google Scholar]
- 54.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 55.Spann, K. M., K. C. Tran, and P. L. Collins. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J. Virol. 79:5353-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stollar, B. D., and V. Stollar. 1970. Immunofluorescent demonstration of double-stranded RNA in the cytoplasm of Sindbis virus-infected cells. Virology 42:276-280. [DOI] [PubMed] [Google Scholar]
- 57.Strahle, L., et al. 2007. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J. Virol. 81:12227-12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahasi, K., et al. 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 29:428-440. [DOI] [PubMed] [Google Scholar]
- 59.Takeuchi, K., T. Komatsu, Y. Kitagawa, K. Sada, and B. Gotoh. 2008. Sendai virus C protein plays a role in restricting PKR activation by limiting the generation of intracellular double-stranded RNA. J. Virol. 82:10102-10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniguchi, T., and A. Takaoka. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14:111-116. [DOI] [PubMed] [Google Scholar]
- 61.Terenzi, F., D. J. Hui, W. C. Merrick, and G. C. Sen. 2006. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 281:34064-34071. [DOI] [PubMed] [Google Scholar]
- 62.Thomas, L. R., A. Henson, J. C. Reed, F. R. Salsbury, and A. Thorburn. 2004. Direct binding of Fas-associated death domain (FADD) to the tumor necrosis factor-related apoptosis-inducing ligand receptor DR5 is regulated by the death effector domain of FADD. J. Biol. Chem. 279:32780-32785. [DOI] [PubMed] [Google Scholar]
- 63.Tojima, Y., et al. 2000. NAK is an IkappaB kinase-activating kinase. Nature 404:778-782. [DOI] [PubMed] [Google Scholar]
- 64.Van Cleve, W., et al. 2006. Attenuating mutations in the P/C gene of human parainfluenza virus type 1 (HPIV1) vaccine candidates abrogate the inhibition of both induction and signaling of type I interferon (IFN) by wild-type HPIV1. Virology 352:61-73. [DOI] [PubMed] [Google Scholar]
- 65.Vidal, S., and D. Kolakofsky. 1989. Modified model for the switch from Sendai virus transcription to replication. J. Virol. 63:1951-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, Q., et al. 2009. Role of double-stranded RNA pattern recognition receptors in rhinovirus-induced airway epithelial cell responses. J. Immunol. 183:6989-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu, L. G., et al. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 70.Yang, Y. L., et al. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yie, J., K. Senger, and D. Thanos. 1999. Mechanism by which the IFN-beta enhanceosome activates transcription. Proc. Natl. Acad. Sci. U. S. A. 96:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoneyama, M., and T. Fujita. 2007. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 282:15315-15318. [DOI] [PubMed] [Google Scholar]
- 73.Yoneyama, M., et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 74.Yoneyama, M., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 75.Yoon, C. H., E. S. Lee, D. S. Lim, and Y. S. Bae. 2009. PKR, a p53 target gene, plays a crucial role in the tumor-suppressor function of p53. Proc. Natl. Acad. Sci. U. S. A. 106:7852-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yount, J. S., L. Gitlin, T. M. Moran, and C. B. Lopez. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai virus defective interfering particles. J. Immunol. 180:4910-4918. [DOI] [PubMed] [Google Scholar]
- 77.Zamanian-Daryoush, M., T. H. Mogensen, J. A. DiDonato, and B. R. Williams. 2000. NF-kappaB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-kappaB-inducing kinase and IkappaB kinase. Mol. Cell. Biol. 20:1278-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.