Abstract
The E6 proteins from high-risk, cancer-causing types of human papillomavirus (HPV) are characterized by the presence of a PDZ (PSD95/Dlg/ZO-1) binding motif in their extreme carboxy termini, through which they interact with a number of cellular PDZ domain-containing substrates. In order to ascertain how many of these are degraded by E6 in vivo, we performed an extensive analysis of the effects of E6 ablation on the expression levels of a number of previously reported E6 PDZ substrates. Using HPV type 16 (HPV-16)-positive CaSKi cells and HPV-18-positive HeLa cells, we have found that MAGI-1 is a major degradation target of both HPV-16 and HPV-18 E6. In contrast, hDlg, hScrib, PTPN3, TIP2, FAP1, and PSD95 all exhibit various degrees of susceptibility to E6-induced degradation, and a high degree of HPV type specificity is observed for certain substrates. We also show that E6 preferentially targets MAGI-1 within the nucleus and at membrane sites. One of the direct consequences of MAGI-1 degradation is a loss of tight-junction integrity, as determined by mislocalization of the tight-junction protein ZO-1. Ablation of E6 expression restores tight junctions, and this restoration is dependent on the presence of MAGI-1. These results demonstrate that oncogenic HPV E6 proteins disrupt cellular tight junctions through the degradation of MAGI-1, and they provide further evidence of how the PDZ binding potential of E6 can contribute to HPV-induced malignancy.
Human papillomaviruses (HPVs) are the causative agent of cervical cancer, which is brought about primarily by the combined action of two viral oncoproteins, E6 and E7 (reviewed in reference 55). These proteins target, respectively, the p53 (40, 50) and pRb (5, 9) tumor suppressors; they cooperate in the induction of keratinocyte immortalization (17, 33) and tumor formation in transgenic mice (14, 24); and they are required for the continued proliferation and survival of cervical-tumor-derived cell lines (21, 53). It is clear, however, that other cellular targets of both viral proteins are necessary for their full transforming activity. In the case of the high-risk HPV E6 oncoproteins, an interesting feature is the presence of a class 1 PDZ (PSD95/Dlg/ZO-1)-binding motif (PBM) at their carboxy termini (27), which is absent from E6 proteins derived from the low-risk HPV types. This PDZ binding potential renders these E6 proteins capable of interacting with, and potentially more importantly, targeting for proteasome-mediated degradation, a subset of PDZ domain-containing cellular substrates, including the cell polarity regulators human Dlg (hDlg) (11) and human Scribble (hScrib) (35), both of which are classified as potential tumor suppressor proteins (3, 6, 12, 37, 51, 52). Other E6 PDZ domain-containing targets include the MAGI family of proteins (13, 46), which are scaffolding molecules involved in the regulation of tight-junction (TJ) assembly (34). So far, at least 10 different PDZ domain-containing substrates of E6 have been described, including PSD95 (16), PATJ (25, 43), MUPP1 (26), TIP1 (15), TIP2 (10), PTPN3 (20, 49), PTPN13 (41), and CAL (19). In most cases, these proteins have also been reported to be targets for E6-induced degradation (48). Many of these proteins are involved in diverse regulatory pathways, including the assembly of cell-cell junctions and cell attachment, and in the control of cell signaling. Potential tumor suppressor activities have also been assigned to several of these proteins. An important question that remains to be answered is whether or not all of these substrates are equally susceptible to E6-induced degradation in vivo. We know from previous studies that minor differences in the PDZ-binding motif between HPV type 16 (HPV-16) and HPV-18 E6 can significantly affect PDZ domain targeting, with HPV-18 E6 preferentially binding to hDlg, while HPV-16 E6 preferentially binds to hScrib (47). Similar results have also been reported for CAL (19), and structural studies on E6 complexed with Dlg and MAGI-1 provide some molecular explanations for these apparent differences (29, 44, 54). It is also clear, however, that while PDZ selection by E6 is highly specific, it can give potentially misleading results if assessed only under conditions of overexpression or in vitro, since at high concentrations the E6 PBM can potentially recognize any class I PDZ domain (54).
In this study we wanted to compare the patterns of expression of diverse PDZ domain-containing substrates of E6 in cells derived from cervical tumors where the expression of the E6/E7 oncoproteins is ablated using small interfering RNA (siRNA). In addition, we also analyzed the biological consequences of the targeting of one of these proteins, MAGI-1, by E6 with respect to the reestablishment of cellular TJs.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa, CaSKi, SiHa, and H1299 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin-streptomycin (100 U/ml), and glutamine (300 μg/ml). For the delivery of all siRNAs (Dharmacon), the cells were seeded on 6-cm-diameter dishes at a confluence of 1.2 × 105 and were transfected using Lipofectamine 2000 (Invitrogen) with siRNA against either luciferase, HPV-16 E6/E7 (5′UUAAAUGACAGCUCAGAGG), HPV-18 E6/E7 (5′CAUUUACCAGCCCGACGAG), E6AP, or the different PDZ proteins (relevant Dharmacon Smart Pools). For siRNA transfection followed by immunofluorescent analysis, HeLa cells were seeded at the same confluence on glass coverslips.
Antibodies.
The following antibodies were used: rabbit polyclonal anti-MAGI-1 (M5691), rabbit polyclonal anti-PTPN3 (T6453), and mouse monoclonal anti-α-tubulin (T6199) (Sigma); mouse monoclonal anti-PSD95 (6G6-1C9) and mouse monoclonal anti-p84 (5E10) (Abcam); and mouse monoclonal anti-ZO1 (ZO1-1A12) (Invitrogen). The following antibodies were purchased from Santa Cruz Biotechnology: mouse monoclonal anti-p53 (DO-1), mouse monoclonal anti-α-actinin (H-2), mouse monoclonal anti-Dlg (2D11), goat polyclonal anti-Scribble (C-20), goat polyclonal anti-TIP2 (N-19), rabbit polyclonal anti-FAP1 (H-300), rabbit polyclonal anti-E-cadherin (H-108), and mouse monoclonal anti-vimentin (V-9).
Western blotting.
Cells were lysed in 2× sodium dodecyl sulfate (SDS) sample buffer (100 mM Tris HCl [pH 6.8], 200 mM dithiothreitol [DTT], 4% SDS, 20% glycerol, 0.2% bromophenol blue), and the whole-cell extracts were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and blotted onto 0.45-μm-pore-size nitrocellulose membranes (Schleicher and Schuell). The membranes were blocked at 37°C for 1 h in 10% milk-phosphate-buffered saline (PBS), except for those probed with anti-MAGI-1, anti-PTPN3, and anti-PSD95 antibodies, which were blocked in 5% milk-PBS. The blots were incubated with the appropriate primary antibodies diluted in the washing buffer (10% milk-PBS, 0.5% Tween 20), except for the anti-MAGI-1, anti-PTPN3, and anti-PSD95 antibodies, which were diluted in 5% milk-PBS and 0.05% Tween 20. The incubation times were 2 h at room temperature for all antibodies, except for the anti-PTPN3, anti-FAP1, anti-E-cadherin, and anti-PSD95 antibodies, which were incubated overnight at 4°C. After several washes, the membranes were incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (Dako) for 1 h at room temperature. After extensive washing, the blots were developed with the enhanced chemiluminescence (ECL) or ECL Plus reagent (GE Healthcare) according to the manufacturer's instructions. Protein band intensities were quantitated where possible using the OptiQuant quantification program.
Immunofluorescence and microscopy.
Cells were fixed with 3.7% paraformaldehyde in PBS and were permeabilized with 0.1% Triton X-100 in PBS. Primary antibodies were incubated for 2 h at 37°C, washed extensively in PBS, and incubated for 20 min at 37°C with a secondary anti-rabbit or anti-mouse antibody conjugated to fluorescein or rhodamine (Molecular Probes). Samples were washed several times with water and were mounted with Vectashield mounting medium (Vector Laboratories) on glass slides. Slides were analyzed with either a Leica DMLB fluorescence microscope with a Leica photo camera (A01M871016) or a Zeiss LSM 510 confocal microscope with two lasers giving excitation lines at 480 and 510 nm. The data were collected with a 60× objective oil immersion lens.
RESULTS
MAGI-1, Dlg, and Scrib are degraded by E6 in HeLa and CaSKi cells.
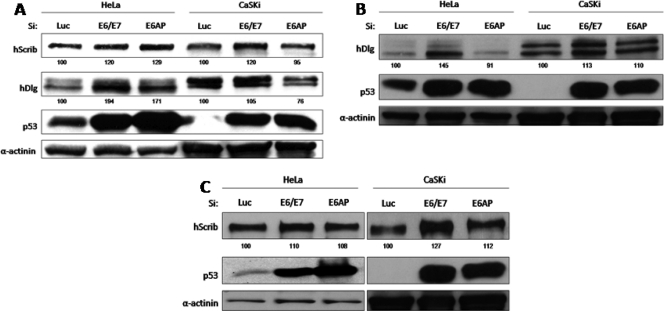
Although several studies have identified a number of PDZ domain-containing proteins as potential targets of the high-risk HPV E6 proteins, many of these proteins were analyzed in ectopic overexpression systems; moreover, each study used either different cell types or different HPV E6 proteins, making it difficult to compare directly the susceptibilities of these targets to E6-induced degradation in vivo. In an attempt to compare directly the relative susceptibilities of the various PDZ domain targets of E6 to degradation, we used siRNA to block E6/E7 expression in CaSKi (HPV-16-positive) and HeLa (HPV-18-positive) cells. Extracts from these cells were analyzed by Western blotting and were compared with those from control cells treated with siRNA to luciferase, or from cells treated with siRNA to E6AP. In the first set of assays, we analyzed changes in the levels of expression of hScrib and hDlg at 48 h and 72 h posttransfection. The results in Fig. 1 show strong increases in the levels of p53 at both time points following siRNA treatment against E6/E7 and E6AP in both CaSKi and HeLa cells. This is in agreement with the findings of previous studies (18, 39) and confirms efficient ablation of E6 expression. For hDlg, there was a modest increase in expression in HeLa cells at the 48-h and 72-h time points, confirming hDlg as a substrate for HPV-18 E6-induced degradation in HeLa cells (30, 31, 16), while there was a slight increase in the overall level of hDlg expression in CaSKi cells. It is also noteworthy that hDlg migrated somewhat differently in the two cell lines, most likely reflecting differences in the degree of phosphorylation (data not shown). Whether this might also affect its susceptibility to E6 degradation (32, 36) remains to be determined. In contrast to the pattern of hDlg expression, there was a significant increase in hScrib levels in E6/E7 siRNA-treated CaSKi cells at the 72-h time point, whereas changes in hScrib levels in HeLa cells were minimal. E6AP ablation had only modest effects, which were somewhat dependent on the time at which the assays were performed, on both hDlg and hScrib; however, there were slight increases in hDlg expression in HeLa cells (Fig. 1A) and in hScrib expression in CaSKi cells (Fig. 1C). Taken together, these results confirm that hDlg and hScrib are proteolytic substrates of HPV-18 and HPV-16 E6, respectively, in vivo.
FIG. 1.
hDlg and hScribble are degraded in HeLa and CaSKi cells according to their differential susceptibilities to HPV-16 and HPV-18 E6 oncoproteins. (A) HPV-positive HeLa and CaSKi cells were transfected with luciferase siRNA, E6/E7 siRNA, or E6AP siRNA and were grown for 48 h prior to harvesting. The expression patterns of hDlg, hScribble, p53, and α-actinin (used to monitor protein loading) were assessed by Western blot analysis. (B and C) The assay described for panel A was repeated, but HeLa and CaSKi cells were harvested at 72 h posttransfection before Western blot analysis. Numbers are percentages of band intensity for hScribble and hDlg with respect to the luciferase siRNA control (100%).
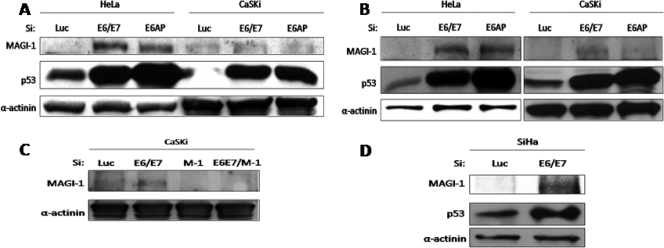
Previous structural studies had highlighted MAGI-1 as a strongly bound target of HPV-18 E6 (54), and in vitro studies had suggested that it was also a good substrate for E6-induced degradation (13, 45). A similar series of ablation experiments was therefore performed, and MAGI-1 expression was analyzed at the 48- and 72-h time points. The results in Fig. 2 demonstrate strong increases in the levels of MAGI-1 expression in HeLa cells at both time points, which were observed equally well following ablation of E6AP expression. In CaSKi cells, there were modest increases in MAGI-1 levels, though not as strong as those observed in HeLa cells. In order to confirm that the protein rescued in CaSKi cells was indeed MAGI-1, we repeated the analysis but included a siRNA against MAGI-1. As can be seen from the results in Fig. 2C, the protein that was rescued following treatment with siRNA against E6/E7 disappeared when the MAGI-1 siRNA was included. We also extended the analysis to another HPV-16-positive cell line, SiHa, and obtained similar results (Fig. 2D). Taken together, these results would suggest that while MAGI-1 is a strong substrate of HPV-18 E6, it is nonetheless also subject to HPV-16 E6-induced degradation in vivo.
FIG. 2.
MAGI-1 is efficiently rescued in HeLa and CaSKi cells upon E6/E7 ablation. (A) HeLa and CaSKi cells were transfected with luciferase siRNA, E6/E7 siRNA, or E6AP siRNA and were grown for 48 h before harvesting. Protein levels were assessed by Western blotting using an anti-MAGI-1 antibody, an anti-p53 antibody, and an anti-α-actinin antibody to monitor protein loading. Protein bands corresponding to MAGI-1 were detected using the ECL Plus system, while ECL was used to detect p53 and α-actinin expression. (B) The assay described for panel A was repeated, but cells were grown for 72 h before harvesting and Western blot analysis. (C) To confirm the identity of the band corresponding to MAGI-1, CaSKi cells were transfected with luciferase siRNA, E6/E7 siRNA, MAGI-1 (M-1) siRNA, or a combination of E6/E7 siRNA and MAGI-1 siRNA. Seventy-two hours after transfection, cells were harvested, and Western blot analysis was performed as for panels A and B. (D) The assay was repeated in SiHa cells and was processed as for panel B.
Analysis of FAP1, TIP2, PTPN3, and PSD95 expression levels following E6/E7 knockdown.
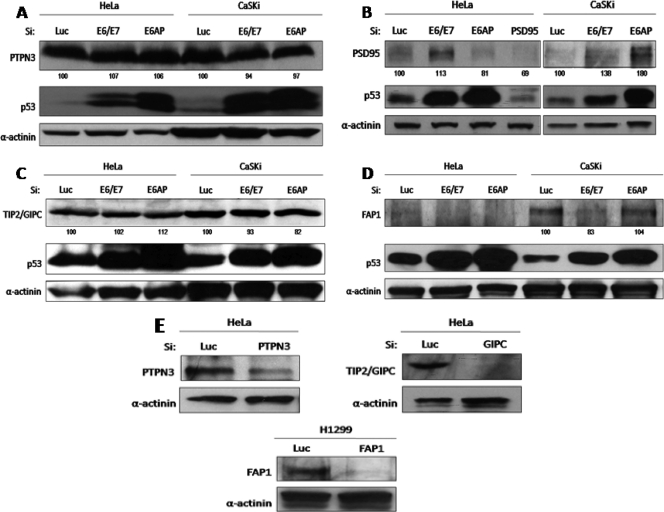
Recent studies have shown that FAP1 (PTPN13), TIP2, PTPN3, and PSD95 are also potential proteolytic substrates of E6, albeit in a variety of different experimental settings (10, 16, 20, 41, 49). Therefore, we wanted to determine whether these PDZ substrates were similarly targeted in the two cervical-tumor-derived cell lines. The cells were transfected with siRNA against E6/E7, and changes in the PDZ protein levels were ascertained by Western blot analysis. The results in Fig. 3 show no significant changes in the levels of expression of either PTPN3 (Fig. 3A) or TIP2 (Fig. 3C) following treatment with siRNA to either E6/E7 or E6AP. In contrast, there was a significant increase in the level of expression of PSD95 in HeLa cells and in CaSKi cells (Fig. 3B) following E6/E7 knockdown. Removal of E6AP had no effect on the levels of PSD95 expression in HeLa cells, while in CaSKi cells, a modest increase was obtained. These results confirm that PSD95 is a strong target for HPV-18 E6- and a weaker one for HPV-16 E6-induced degradation in vivo (16).
FIG. 3.
(A to D) Analysis of the susceptibilities of PTPN3, PSD95, TIP2/GIPC, and PTPN13/FAP1 to E6 degradation in vivo. HeLa and CaSKi cells were transfected with luciferase siRNA, E6/E7 siRNA, or E6AP siRNA and were grown for 72 h before harvesting. The expression levels of PTPN3 (A), PSD95 (B), TIP2/GIPC (C), PTPN13/FAP1 (D), p53, and α-actinin were assessed by Western blotting. Numbers are percentages of band intensity for PTPN3, PSD95, TIP2, and FAP1 relative to that for the luciferase siRNA control (100%). (E) To confirm the correct identities of the three PDZ proteins, cells were transfected with luciferase siRNA, PTPN3 siRNA, TIP2/GIPC siRNA, or PTPN13/FAP1 siRNA, and their expression patterns were revealed by Western blot analysis as in panels A, B, C, and D.
In the case of FAP1, an apparently contradictory result was obtained. Although its levels of expression in HeLa cells are very low, siRNA to E6/E7 apparently reduced FAP1 levels still further (Fig. 3D). This effect was more marked in CaSKi cells, where loss of E6/E7 expression resulted in a dramatic decrease in the levels of FAP1 expression. In addition, removal of E6AP also resulted in lower levels of FAP1 expression in HeLa cells. Since a number of these proteins were present at quite low levels, we also verified that the correct proteins were detected by the antibodies. To do this, we performed a series of Western blot analyses on cell extracts following transfection with siRNAs to each of the PDZ domain-containing proteins. The results in Fig. 3E (PTPN3, TIP2, and FAP1) and Fig. 3B (PSD95) show that the protein recognized by the relevant antibody also disappeared following transfection with the relevant siRNA.
Taken together, these results demonstrate that PTPN3 and TIP2 are not major targets of E6-induced degradation in monolayer cultures of cells derived from cervical tumors. In contrast, PSD95 appears to be a good substrate for HPV-18 E6, in agreement with previous studies (16). Finally, it would appear that, at least in cervical-tumor-derived cells in monolayer culture, E6/E7 might actually increase the levels of FAP1 expression. Whether this occurs through E6's PDZ interactions with FAP1 or through an as yet unknown function of E7 remains to be determined.
HPV E6 preferentially degrades nuclear and membrane-bound pools of MAGI-1.
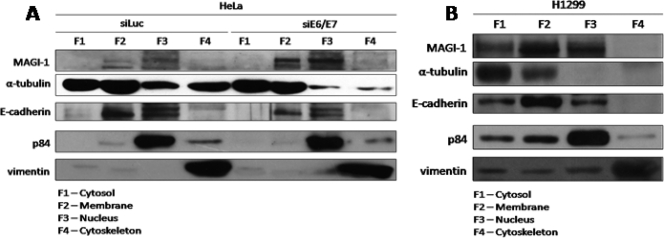
Previous studies have shown that certain cellular pools of hDlg are more susceptible than others to E6-induced degradation (31, 32, 36). Since MAGI-1 is a major common target for both HPV-16 and HPV-18 E6, we were interested in determining whether there are also certain cellular pools of MAGI-1 that are preferentially targeted by E6. To investigate this, we fractionated HeLa cells after 72 h of siRNA ablation of E6/E7 expression. For comparison, we also included H1299 cells in the analysis to determine where MAGI-1 would normally be expressed in the absence of HPV sequences. The cells were fractionated into cytosolic (F1), membrane (F2), nuclear (F3), and cytoskeletal (F4) components, and the levels of MAGI-1 expression in each fraction were ascertained by Western blot analysis. The results obtained again demonstrate that MAGI-1 is a strong substrate for HPV-18 E6-induced degradation in vivo (Fig. 4 A). Interestingly, the bulk of the MAGI-1 protein that was rescued upon ablation of E6/E7 expression resided in the membrane and nuclear fractions of the cell, and the largest pool was actually present within the nucleus. In contrast, a similar fractionation of H1299 cells (Fig. 4B) showed that the main concentration of MAGI-1 was found at membrane sites, with slightly smaller pools in the nuclear and cytosolic fractions. These studies demonstrate that the rescue of MAGI-1 from E6-induced degradation results in preferential restoration of MAGI-1 expression at membrane sites and also within the nucleus.
FIG. 4.
Subcellular fractionation reveals that MAGI-1 is degraded at membrane and nuclear locations in HPV-positive cells. (A) HeLa cells were transfected with luciferase siRNA or E6/E7 siRNA. After 72 h, cells were fractionated into 4 subcellular compartments: cytosol (F1), membrane (F2), nucleus (F3), and cytoskeleton (F4). The expression patterns of MAGI-1 and those of the four subcellular fraction markers E-cadherin, p84, α-tubulin, and vimentin were assessed by Western blotting. (B) The subcellular fractionation was repeated on H1299 cells, and the levels of MAGI-1 and the subcellular fraction markers were detected as for panel A.
HPV E6-induced degradation of MAGI-1 disrupts cellular TJs.
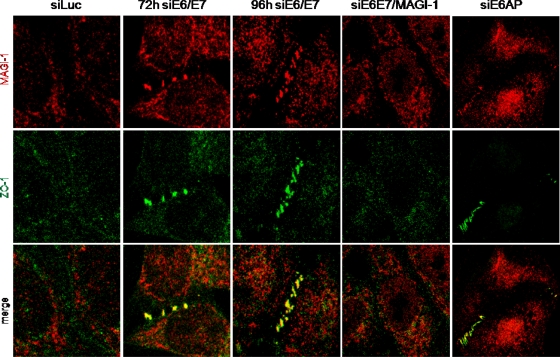
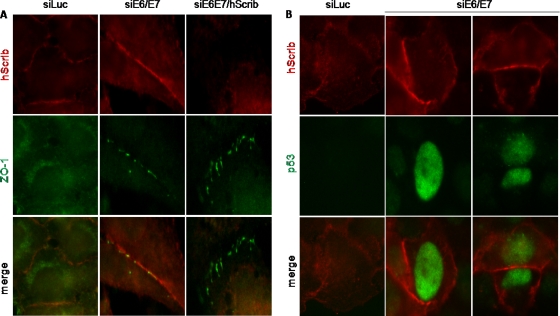
Although there is no information on the potential role of MAGI-1 in the nucleus, previous studies have implicated the membrane-bound form of MAGI-1 in the establishment of cellular TJs (34). It has also been shown that TJs are disrupted in HPV-positive cells, and a possible role for hScrib in this phenotype has been suggested (35). However, we reasoned that MAGI-1 was a more likely candidate to explain the disruption of TJs by HPV E6, and therefore, we proceeded to investigate TJ status in cells in which E6/E7 expression had been ablated. At the same time, we performed siRNA ablation of MAGI-1 and of hScrib on a subset of cells treated with siRNA to E6/E7 in order to determine whether any changes in TJs were MAGI-1 or hScrib dependent. At 72 h and 96 h after transfection of the siRNAs, HeLa cells were fixed and analyzed by immunofluorescence for MAGI-1 and a TJ marker, ZO-1 (42, 7). We focused primarily on cells that were in contact so that junctions would have the opportunity to become established, and the results for MAGI-1 are shown in Fig. 5. As can be seen, no MAGI-1 protein was detectable at sites of cell-cell contact in luciferase siRNA control cells, and ZO-1 displayed a diffuse pattern of expression and was also absent at these sites. In contrast, siRNA to E6/E7 resulted in a very marked accumulation of MAGI-1 expression at cell-cell junctions. Interestingly, this occurred in a beaded structure at the 72-h time point, and there was also a perfect colocalization with ZO-1 in these structures, suggesting the reinitiation of correct TJ formation. By the 96-h time point, this pattern was even more marked, and an intact TJ had already formed between adjacent cells. These results demonstrate that TJs can be reestablished in HeLa cells when E6/E7 expression is ablated. Interestingly, a similar pattern of staining was obtained upon ablation of E6AP expression, consistent with the results from the Western blot analyses. We also analyzed cells that had been cotransfected with a MAGI-1 siRNA. As can be seen in Fig. 5, there was a complete failure to reestablish TJs in cells treated with the MAGI-1 siRNA, as determined by the pattern of ZO-1 expression, even by the 96-h time point. In contrast, ablation of hScrib (Fig. 6 A) had no effect on the reestablishment of TJs upon ablation of E6/E7 expression. In addition, removal of E6/E7 expression in HeLa cells resulted in a marked increase in hScrib expression at the cell membrane, despite the apparent lack of significant levels of degradation seen in Fig. 1. Similar results were also obtained in CaSKi cells (Fig. 6B), suggesting that there may also be an element of relocalization of hScrib by E6, rather than just simple degradation. However, taken together, these results demonstrate that loss of TJs in an HPV-positive, tumor-derived cell line is due to the ability of E6 to induce the degradation of MAGI-1.
FIG. 5.
Rescue of MAGI-1 from E6-induced degradation restores TJs. HeLa cells were seeded on glass coverslips and were transfected with either luciferase siRNA, HPV-18 E6/E7 siRNA, a combination of HPV-18 E6/E7 siRNA and MAGI-1 siRNA, or E6AP siRNA. Cells were grown for 72 h or 96 h before being fixed, incubated with anti-MAGI-1 and anti-ZO-1 antibodies, and counterstained with rhodamine-conjugated (MAGI-1) or fluorescein-conjugated (ZO-1) secondary antibodies. Confocal images were taken at wavelengths of 480 and 510 nm.
FIG. 6.
Loss of hScrib does not affect TJ formation. HeLa cells (A) and CaSKi cells (B) were seeded onto glass coverslips and were transfected with either luciferase siRNA, HPV-18 E6/E7 siRNA, HPV-18 E6/E7 siRNA with hScrib siRNA, or HPV-16 E6/E7 siRNA as indicated (with two examples of CaSKi cells shown). Cells were grown for 96 h before fixing and staining for hScrib (red) and ZO-1 (green) in HeLa cells or for hScrib (red) and p53 (green) in CaSKi cells.
DISCUSSION
The presence of a class I PDZ-binding motif located on the extreme carboxy terminus of the high-risk HPV E6 proteins would, in theory, render all the cellular class I PDZ domain-containing proteins vulnerable as potential interacting partners and putative proteolytic substrates for these viral oncoproteins. However, from the data presented above and elsewhere (44, 47), it is clear that proteolytic degradation by E6 of its potential PDZ domain-containing substrates is highly protein specific. This is particularly true for MAGI-1, whose levels of expression are low in HPV-positive cells and increase dramatically following removal of E6/E7 expression in both HPV-16- and HPV-18-containing cell lines. Most importantly, this restoration of MAGI-1 expression correlates with the formation of TJs in these HPV-positive cells. The role of E6AP in this activity of E6 remains unclear. Thus, in HeLa cells, degradation of MAGI-1 appears also to require the presence of E6AP, while in CaSKi cells, removal of E6AP has a minimal effect. The reasons for these differences are not clear, but they continue to demonstrate important differences in how different substrates are targeted by E6 proteins derived from different HPV types.
In the case of the other potential PDZ substrates of E6, we found some marked differences in how they are targeted by HPV-16 and HPV-18 E6. Thus, hDlg appears more susceptible to degradation by HPV-18 E6 than by HPV-16 E6, and the reverse is true for hScrib. These results are in agreement with those of previously reported interaction studies in vitro (47). Similarly, PSD95 appears to be similar to hDlg in that it is efficiently targeted by HPV-18 E6 but less so by HPV-16 E6, and this is in agreement with previous reports (16). Of the remaining PDZ substrates of E6 that we were able to analyze, we failed to obtain conclusive evidence that FAP1, TIP2, or PTPN3 was targeted for degradation, either by HPV-16 E6 or by HPV-18 E6, in monolayer cultures of cells derived from cervical tumors. Indeed, the loss of E6/E7 expression may actually decrease the levels of FAP1 expression. The underlying mechanisms behind this are unclear but merit further investigation. However, these studies were done using ablation of both E6 and E7 expression, and we cannot exclude the possibility that some effects may be related to E7 depletion. In addition, these studies do not rule out the possibility that these PDZ domain-containing proteins may be degradation substrates of E6 in other biological settings: during different stages of the normal viral life cycle where the cells are subject to terminal differentiation, or at an earlier stage of tumor development. This might reflect differences in the phosphorylation status of the target protein, which could influence accessibility to E6 and subsequent targeting (32, 36). Finally, we should also emphasize that although degradation has been proposed as a major mechanism by which E6 exerts its function, it is possible that some of these substrates may be only bound by E6 and that blocking of a certain PDZ substrate-ligand interaction might be sufficient for modulation of the function of that particular cellular PDZ domain-containing protein by E6. Alternatively, E6 might also alter the localization of the substrate (as may be the case with HPV-18 E6 and hScribble in HeLa cells) and thereby alter its function.
Since biochemical data suggest that MAGI-1 is the strongest interacting partner of HPV E6 (45, 54), we focused on further defining the relevance of MAGI-1 degradation to HPV-induced malignancy. Using differential cell fractionation, we found that abolition of E6/E7 expression restores MAGI-1 at two main locations within the cell: the cell membrane and the cell nucleus. This suggests that whatever functions these two pools of MAGI-1 perform, the removal of one or both has advantages for the virus. No studies that could offer an explanation of the function of MAGI-1 in the nucleus are currently available. Sequence analysis of MAGI-1 reveals a strong bipartite nuclear localization signal in the carboxy terminus of the protein (8), consistent with the notion that a pool of MAGI-1 is normally resident within the nucleus. Further studies will aim at more fully defining the functions of this form of the protein.
In contrast, the membrane-bound form of MAGI-1 has been implicated in the control of TJs (34), which are lost in HPV-positive cells (25, 35, 43). The results reported here show that this loss is indeed a result of E6 directing the degradation of MAGI-1. Using ZO-1 as a marker of TJ integrity, we have confirmed that these junctions are largely absent in HPV-18-positive HeLa cells and that ablation of E6 expression results in a clear reaccumulation of MAGI-1 at the cell membrane, accompanied by an accumulation of ZO-1 at the same cellular location. Interestingly, this appears to be a slow process, in which a bead-like structure, indicative of the early stages of junction formation (22), is apparent at 72 h after transfection with E6/E7 siRNA, and more-complete junctions are visible by 96 h. To verify that the restoration of TJs depends on the rescue of MAGI-1 from E6-induced degradation, we cotransfected a MAGI-1 siRNA with the E6/E7 siRNA, after which there was no evidence of TJ formation. In contrast, cotransfection of siRNA to hScrib with the E6/E7 siRNA had no deleterious effects on TJ reassembly, confirming the specificity of the results with MAGI-1. These results demonstrate that the loss of TJs in HPV-18-positive HeLa cells is a direct consequence of the ability of E6 to direct the degradation of MAGI-1; thus, they provide a biological explanation of why this protein is targeted by the virus during the life cycle and by E6 in malignancy. TJs play an important role in differentiation: their correct assembly promotes exit from the cell cycle and contributes to keratinocyte differentiation (1, 4, 38). Loss of TJs can therefore be expected to delay the differentiation process. In addition, TJs participate directly in the regulation of cell proliferation by modulating signaling cascades such as mitogen-activated protein kinase (MAPK), PKB/Akt, and RhoA signaling (1, 23, 28). Interestingly, ZO-1 is also believed to be involved in controlling cell proliferation by binding and sequestering the transcription factor ZONAB/DbpA at the cell membrane (2). Taken together, these studies suggest that the degradation of MAGI-1 by HPV E6 and the consequent perturbation of TJ assembly can have pleiotropic effects, both with respect to the virus life cycle and with respect to HPV-induced malignancy.
Acknowledgments
We are grateful to Miranda Thomas for critical comments on the manuscript and to all the members of the Banks lab for advice and support.
This work was supported in part by a research grant from the Associazione Italiana per la Ricerca sul Cancro.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1.Aijaz, S., F. D'Atri, S. Citi, M. S. Balda, and K. Matter. 2005. Binding of GEF-H1 to the tight junction-associated adaptor cingulin results in inhibition of Rho signaling and G1/S phase transition. Dev. Cell 8:777-786. [DOI] [PubMed] [Google Scholar]
- 2.Balda, M. S., M. D. Garrett, and K. Matter. 2003. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J. Cell Biol. 160:423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilder, D., M. Li, and N. Perrimon. 2000. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289:113-116. [DOI] [PubMed] [Google Scholar]
- 4.Bordin, M., F. D'Atri, L. Guillemot, and S. Citi. 2004. Histone deacetylase inhibitors up-regulate the expression of tight junction proteins. Mol. Cancer Res. 2:692-701. [PubMed] [Google Scholar]
- 5.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 6.Cavatorta, A. L., et al. 2004. Differential expression of the human homologue of Drosophila discs large oncosuppressor in histologic samples from human papillomavirus-associated lesions as a marker for progression to malignancy. Int. J. Cancer 111:373-380. [DOI] [PubMed] [Google Scholar]
- 7.Denker, B. M., and S. K. Nigam. 1998. Molecular structure and assembly of the tight junction. Am. J. Physiol. 274:F1-F9. [DOI] [PubMed] [Google Scholar]
- 8.Dobrosotskaya, I., R. K. Guy, and G. L. James. 1997. MAGI-1, a membrane-associated guanylate kinase with a unique arrangement of protein-protein interaction domains. J. Biol. Chem. 272:31589-31597. [DOI] [PubMed] [Google Scholar]
- 9.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 10.Favre-Bonvin, A., C. Reynaud, C. Kretz-Remy, and P. Jalinot. 2005. Human papillomavirus type 18 E6 protein binds the cellular PDZ protein TIP-2/GIPC, which is involved in transforming growth factor beta signaling and triggers its degradation by the proteasome. J. Virol. 79:4229-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardiol, D., et al. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 18:5487-5496. [DOI] [PubMed] [Google Scholar]
- 12.Gardiol, D., A. Zacchi, F. Petrera, G. Stanta, and L. Banks. 2006. Human discs large and scrib are localized at the same regions in colon mucosa and changes in their expression patterns are correlated with loss of tissue architecture during malignant progression. Int. J. Cancer 119:1285-1290. [DOI] [PubMed] [Google Scholar]
- 13.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griep, A. E., et al. 1993. Tumorigenicity by human papillomavirus type 16 E6 and E7 in transgenic mice correlates with alterations in epithelial cell growth and differentiation. J. Virol. 67:1373-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hampson, L., C. Li, A. W. Oliver, H. C. Kitchener, and I. N. Hampson. 2004. The PDZ protein Tip-1 is a gain of function target of the HPV16 E6 oncoprotein. Int. J. Oncol. 25:1249-1256. [PubMed] [Google Scholar]
- 16.Handa, K., et al. 2007. E6AP-dependent degradation of DLG4/PSD95 by high-risk human papillomavirus type 18 E6 protein. J. Virol. 81:1379-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawley-Nelson, P., K. H. Vousden, N. L. Hubbert, D. R. Lowy, and J. T. Schiller. 1989. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 8:3905-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 13:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong, K. W., H. Z. Kim, S. Kim, Y. S. Kim, and J. Choe. 2007. Human papillomavirus type 16 E6 protein interacts with cystic fibrosis transmembrane regulator-associated ligand and promotes E6-associated protein-mediated ubiquitination and proteasomal degradation. Oncogene 26:487-499. [DOI] [PubMed] [Google Scholar]
- 20.Jing, M., J. Bohl, N. Brimer, M. Kinter, and S. B. Vande Pol. 2007. Degradation of tyrosine phosphatase PTPN3 (PTPH1) by association with oncogenic human papillomavirus E6 proteins. J. Virol. 81:2231-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonson, A. L., L. M. Rogers, S. Ramakrishnan, and L. S. Downs, Jr. 2008. Gene silencing with siRNA targeting E6/E7 as a therapeutic intervention in a mouse model of cervical cancer. Gynecol. Oncol. 111:356-364. [DOI] [PubMed] [Google Scholar]
- 22.Kimura, R., T. Ishida, M. Kuriyama, K. Hirata, and Y. Hayashi. 2010. Interaction of endothelial cell-selective adhesion molecule and MAGI-1 promotes mature cell-cell adhesion via activation of RhoA. Genes Cells 15:385-396. [DOI] [PubMed] [Google Scholar]
- 23.Kotelevets, L., et al. 2005. Implication of the MAGI-1b/PTEN signalosome in stabilization of adherens junctions and suppression of invasiveness. FASEB J. 19:115-117. [DOI] [PubMed] [Google Scholar]
- 24.Lambert, P. F., et al. 1993. Epidermal cancer associated with expression of human papillomavirus type 16 E6 and E7 oncogenes in the skin of transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 90:5583-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latorre, I. J., et al. 2005. Viral oncoprotein-induced mislocalization of select PDZ proteins disrupts tight junctions and causes polarity defects in epithelial cells. J. Cell Sci. 118:4283-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74:9680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. S., R. S. Weiss, and R. T. Javier. 1997. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. U. S. A. 94:6670-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, D., and R. J. Mrsny. 2000. Oncogenic Raf-1 disrupts epithelial tight junctions via downregulation of occludin. J. Cell Biol. 148:791-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, Y., G. D. Henry, R. S. Hegde, and J. D. Baleja. 2007. Solution structure of the hDlg/SAP97 PDZ2 domain and its mechanism of interaction with HPV-18 papillomavirus E6 protein. Biochemistry 46:10864-10874. [DOI] [PubMed] [Google Scholar]
- 30.Mantovani, F., P. Massimi, and L. Banks. 2001. Proteasome-mediated regulation of the hDlg tumour suppressor protein. J. Cell Sci. 114:4285-4292. [DOI] [PubMed] [Google Scholar]
- 31.Massimi, P., N. Gammoh, M. Thomas, and L. Banks. 2004. HPV E6 specifically targets different cellular pools of its PDZ domain-containing tumour suppressor substrates for proteasome-mediated degradation. Oncogene 23:8033-8039. [DOI] [PubMed] [Google Scholar]
- 32.Massimi, P., N. Narayan, A. Cuenda, and L. Banks. 2006. Phosphorylation of the discs large tumour suppressor protein controls its membrane localisation and enhances its susceptibility to HPV E6-induced degradation. Oncogene 25:4276-4285. [DOI] [PubMed] [Google Scholar]
- 33.Münger, K., et al. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata, M., et al. 2005. Tight junction protein MAGI-1 is up-regulated by transfection with connexin 32 in an immortalized mouse hepatic cell line: cDNA microarray analysis. Cell Tissue Res. 319:341-347. [DOI] [PubMed] [Google Scholar]
- 35.Nakagawa, S., and J. M. Huibregtse. 2000. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 20:8244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narayan, N., V. K. Subbaiah, and L. Banks. 2009. The high-risk HPV E6 oncoprotein preferentially targets phosphorylated nuclear forms of hDlg. Virology 387:1-4. [DOI] [PubMed] [Google Scholar]
- 37.Navarro, C., et al. 2005. Junctional recruitment of mammalian Scribble relies on E-cadherin engagement. Oncogene 24:4330-4339. [DOI] [PubMed] [Google Scholar]
- 38.Saitou, M., et al. 2000. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol. Biol. Cell 11:4131-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 40.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 41.Spanos, W. C., et al. 2008. The PDZ binding motif of human papillomavirus type 16 E6 induces PTPN13 loss, which allows anchorage-independent growth and synergizes with Ras for invasive growth. J. Virol. 82:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson, B. R., J. D. Siliciano, M. S. Mooseker, and D. A. Goodenough. 1986. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 103:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storrs, C. H., and S. J. Silverstein. 2007. PATJ, a tight junction-associated PDZ protein, is a novel degradation target of high-risk human papillomavirus E6 and the alternatively spliced isoform 18 E6. J. Virol. 81:4080-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas, M., J. Dasgupta, Y. Zhang, X. Chen, and L. Banks. 2008. Analysis of specificity determinants in the interactions of different HPV E6 proteins with their PDZ domain-containing substrates. Virology 376:371-378. [DOI] [PubMed] [Google Scholar]
- 45.Thomas, M., B. Glaunsinger, D. Pim, R. Javier, and L. Banks. 2001. HPV E6 and MAGUK protein interactions: determination of the molecular basis for specific protein recognition and degradation. Oncogene 20:5431-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas, M., et al. 2002. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 21:5088-5096. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, M., P. Massimi, C. Navarro, J. P. Borg, and L. Banks. 2005. The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 24:6222-6230. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, M., et al. 2008. Human papillomaviruses, cervical cancer and cell polarity. Oncogene 27:7018-7030. [DOI] [PubMed] [Google Scholar]
- 49.Töpffer, S., A. Muller-Schiffmann, K. Matentzoglu, M. Scheffner, and G. Steger. 2007. Protein tyrosine phosphatase H1 is a target of the E6 oncoprotein of high-risk genital human papillomaviruses. J. Gen. Virol. 88:2956-2965. [DOI] [PubMed] [Google Scholar]
- 50.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 51.Wodarz, A. 2000. Tumor suppressors: linking cell polarity and growth control. Curr. Biol. 10:R624-R626. [DOI] [PubMed] [Google Scholar]
- 52.Woodhouse, E., E. Hersperger, and A. Shearn. 1998. Growth, metastasis, and invasiveness of Drosophila tumors caused by mutations in specific tumor suppressor genes. Dev. Genes Evol. 207:542-550. [DOI] [PubMed] [Google Scholar]
- 53.Yoshinouchi, M., et al. 2003. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by E6 siRNA. Mol. Ther. 8:762-768. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., et al. 2007. Structures of a human papillomavirus (HPV) E6 polypeptide bound to MAGUK proteins: mechanisms of targeting tumor suppressors by a high-risk HPV oncoprotein. J. Virol. 81:3618-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]