Abstract
Retroviruses replicate by reverse transcribing their single-stranded RNA genomes into double-stranded DNA using specific cellular tRNAs to prime cDNA synthesis. In HIV-1, human tRNA3Lys serves as the primer and is packaged into virions during assembly. The viral Gag protein is believed to chaperone tRNA3Lys placement onto the genomic RNA primer binding site; however, the timing and possible regulation of this event are currently unknown. Composed of the matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains, the multifunctional HIV-1 Gag polyprotein orchestrates the highly coordinated process of virion assembly, but the contribution of these domains to tRNA3Lys annealing is unclear. Here, we show that NC is absolutely essential for annealing and that the MA domain inhibits Gag's tRNA annealing capability. During assembly, MA specifically interacts with inositol phosphate (IP)-containing lipids in the plasma membrane (PM). Surprisingly, we find that IPs stimulate Gag-facilitated tRNA annealing but do not stimulate annealing in Gag variants lacking the MA domain or containing point mutations involved in PM binding. Moreover, we find that IPs prevent MA from binding to nucleic acids but have little effect on NC or Gag. We propose that Gag binds to RNA either with both NC and MA domains or with NC alone and that MA-IP interactions alter Gag's binding mode. We propose that MA's interactions with the PM trigger the switch between these two binding modes and stimulate Gag's chaperone function, which may be important for the regulation of events such as tRNA primer annealing.
Human immunodeficiency virus type 1 (HIV-1) virions contain two copies of single-stranded genomic RNA, about 2,000 structural Gag proteins, viral enzymes and accessory proteins, and host factors necessary for productive infection (14). One of these essential factors is human tRNA3Lys, normally used by the ribosome in translation but co-opted by the virus for use as a primer to initiate the reverse transcription of viral genomic RNA (35, 38, 57). The 18 nucleotides (nt) at the 3′ end of tRNA3Lys are complementary to viral genomic RNA at the primer binding site (PBS) found within the highly structured 5′ untranslated region (UTR), and viral chaperone proteins promote tRNA3Lys-PBS annealing. The timing of tRNA primer annealing and whether it is regulated are currently unknown.
Nucleic acid (NA) chaperone proteins facilitate NA folding by destabilizing secondary and tertiary structures and allowing formation of the thermodynamically most favored state (56). Previous in vitro studies have shown HIV-1 nucleocapsid (NC or NCp7) protein to be a very effective NA chaperone through a combination of destabilization and aggregation activities (16, 39, 58). NC also displays rapid NA binding kinetics, another important characteristic of chaperone proteins (15). NC can efficiently catalyze tRNA3Lys annealing onto the HIV-1 genome PBS (31, 53), as well as other NA refolding events critical to virus replication, such as RNA genome dimerization (37) and minus-strand transfer (39). In viruses lacking active protease, tRNA3Lys annealing to the PBS still occurs although at reduced levels, suggesting that the precursor protein Gag may act as an NA chaperone and facilitate this process in vivo (34). Indeed, GagΔp6, which lacks the C-terminal p6 domain, and other assembly-competent Gag variants can facilitate tRNA annealing and genome dimerization in vitro (21, 29, 59, 75).
The Gag polyprotein orchestrates virus assembly, and Gag alone is sufficient for formation of virus-like particles (VLPs), which in rabbit reticulocyte lysate resemble immature virions without membranes (9, 27). Gag consists of the matrix (MA), capsid (CA), spacer peptide 1 (SP1), NC, spacer peptide 2 (SP2), and p6 domains (Fig. 1 B), which are proteolyzed into their freestanding mature forms during or after budding (14). The structures of most individual domains have been solved by X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy in both free and ligand-bound states (20, 22, 44, 47), but the entire structure of Gag remains elusive (25). Each domain of Gag is multifunctional, serving interaction roles during assembly, structural roles in the mature virion, and facilitative roles during reverse transcription and integration (10, 25, 39). Gag's myristoylated MA domain interacts with the plasma membrane (PM) (5, 49), and the highly basic NC domain recognizes the ψ packaging signal on the genome, thereby specifically selecting unspliced retroviral RNA preferentially over other RNAs in the cell (3, 20, 28, 37). CA facilitates Gag-Gag interactions responsible for oligomerization and particle formation (2, 24, 47) and interacts with assembly cofactors (43), and p6 is important for final budding events involving the cellular export machinery (26, 65).
FIG. 1.
(A) Sequence and predicted secondary structure of unmodified human tRNA3Lys and ShortPBS RNA (HIV-1 NL4-3) used in this work. Shown in red are the complementary sequences that base pair during tRNA annealing. The structure of the PBS region shown here is based upon the model of Wilkinson et al. (74). The minihelix derived from the acceptor-TΨC domains of human tRNA3Lys is boxed. (B) Schematic showing the full-length HIV-1 Gag protein and GagΔp6 variants investigated in this study.
After proteolysis, MA remains bound to the viral membrane, CA forms the conical core, and NC facilitates many steps of reverse transcription (39, 68). Although the myristoyl group of MA allows it to nonspecifically bind to membranes, amino acids 17 to 33 on MA's surface target assembly to the PM by binding phosphatidylinositol-(4,5)-bisphosphate (PIP2) lipids in the PM inner leaflet (5, 11-13, 50, 60, 66). Inositol phosphates (IPs) such as PIP2 are necessary cofactors for in vitro assembly of VLPs that resemble wild-type (WT) HIV-1 particles; in their absence, the VLPs formed are small and aberrant (8). Decreasing the levels of cellular PIP2 by overexpressing a PIP2-specific phosphatase severely reduces virus assembly, which is retargeted to intracellular compartments (50). However, in assembly experiments other IPs tested also formed WT-like VLPs (9).
In this work, we evaluated how each of Gag's structural domains contributes to its chaperone activity in vitro. In addition to carrying out detailed kinetic studies of tRNA3Lys annealing to the PBS, we characterized the capabilities of WT and variant Gag proteins to bind, aggregate, and destabilize NA helices. Basic regions at both ends of Gag (i.e., MA and NC) have previously been shown to contribute to inositol hexakisphosphate (IP6) binding (19). To establish the contribution of NC and MA to Gag's chaperone function, a series of domain deletion variants and point mutants were prepared and tested, and the effect of IPs on tRNA annealing and NA interactions was investigated. These studies were also motivated, in part, by previous work showing that HIV-1 MA is capable of binding to and packaging NA via a “basic patch” that plays a known role in PM binding (4, 40, 51, 54). Intermolecular Gag-Gag interactions, which are essential for assembly, could be expected to enhance NA interactions by increasing binding cooperativity. To test this hypothesis, monomeric Gag variants were also examined. Taken together, our results support an essential role for the NC domain in HIV-1 Gag's chaperone function, which may be modulated by MA domain interaction with NA and IPs.
MATERIALS AND METHODS
Preparation of proteins and nucleic acids.
WT untagged GagΔp6 and variant proteins were purified to 90% homogeneity from Escherichia coli using established methods (18). QuikChange (Stratagene) site-directed mutagenesis was used to introduce point mutations and deletions. Unmyristoylated MA (His-tagged) (45), CA (77), and NC (30) were prepared essentially as previously described. During the preparation of MA, polyethylenimine precipitation followed by ammonium sulfate precipitation was used to remove contaminating NA (7, 42).
A 105-nt RNA construct derived from the PBS region of the HIV-1 genome (ShortPBS RNA) and human tRNA3Lys were transcribed in vitro according to established protocols (31). [32P]-radiolabeled tRNA3Lys for use in annealing assays was transcribed using the same protocol except that a mixture of [α-32P]GTP and GTP was used in the transcription mixture. Aminoacylation reactions were performed to assess the activity of tRNA3Lys prior to use in annealing assays as described previously (62). Two carboxyfluorescein (FAM)-labeled oligonucleotides were used in these studies: 5′-FAM-DNA20, 5′-FAM-CTTCTTTGGGAGTGAATTAG-3′ (20-nt DNA), obtained from TriLink Biotechnologies, and 3′-FAM-minihelixLys3, 5′-GCCCGGACAGGGUUCAAGUCCCUGUUCGGGCGCCAUU-FAM-3′ (37-nt RNA), purchased from Dharmacon.
Annealing assays.
ShortPBS RNA and tRNA3Lys were folded prior to use according to previous methods (31). Annealing reaction mixtures contained 50 mM HEPES, pH 7.4, 5 mM dithiothreitol (DTT), 1 mM MgCl2, 20 mM NaCl, 10 nM tRNA3Lys, and 25 nM ShortPBS RNA unless noted otherwise. Reaction mixtures were incubated at 37°C for 10 min and cooled to room temperature prior to the addition of chaperone protein. In single-time-point assays, protein concentrations ranged from 0 to 5 μM, and reaction mixtures were incubated for 120 min. In time course annealing assays, 0.4 μM protein was used, and annealing was allowed to proceed over 120 min at room temperature. In annealing assays with IPs, concentrations were 0.8 μM protein, 2 μM IP6 (Sigma), or 300 μM (1,4,5)-inositol trisphosphate (IP3) or dibutanoyl phosphatidylinositol-(4,5)-bisphosphate (PIP2-diC4; Echelon Biosciences, Inc.). In all annealing assays, reactions were quenched with 1% SDS (final concentration), phenol-chloroform extracted twice, mixed with loading dye (50% glycerol with dyes), and run on a polyacrylamide gel (5% stacking gel; 10% running gel). The gels were exposed to phosphor screens and visualized with a Typhoon Trio Variable Mode Imager (GE Healthcare). Single-time-point assays were fit to sigmoidal curves to establish the protein concentration at which annealing is half maximal (C1/2), and these values were used to determine K1/2 values. The K1/2 values are the nucleotide-to-protein ratio obtained by taking the total nucleotide concentration in the annealing reaction mixture and dividing by the C1/2 value. Time course assays were fit to single exponential curves to determine annealing rate, k (min−1).
Sedimentation assays.
Sedimentation assays were performed to monitor the ability of viral proteins to aggregate NA. Conditions were identical to single-time-point annealing assays except that instead of being quenched with SDS, the mixtures were centrifuged at 9,300 × g for 5 min at 4°C in a tabletop centrifuge. The amount of [32P]-labeled tRNA3Lys remaining in the supernatant was determined by scintillation counting.
Destabilization assays.
A 64-nt transactivation response region (TAR) DNA hairpin, 5′ labeled with Alexa Fluor 488 and 3′ labeled with DABCYL [4-(4′-dimethylaminophenylazo)benzoic acid] quencher was purchased from TriLink Biotechnologies (San Diego, CA). This DNA construct is complementary to the 59-nt TAR RNA hairpin and also contains 5′ T and 3′ TTTT overhangs to facilitate synthesis and prevent undesired fluorescence quenching (32). To assay the destabilization activity of viral proteins, a fluorescence lifetime-based approach was used as described previously (32). Briefly, in these assays 100 nM labeled TAR DNA hairpin was mixed with various amounts of viral proteins in 20 mM HEPES (pH 7.4), 50 mM NaCl, 1 mM MgCl2, 10 μM tris(2-carboxyethyl)phosphine, and 5 mM β-mercaptoethanol. Reaction mixtures were incubated for 30 min at room temperature prior to lifetime measurements on a Life-Spec (red) time-resolved spectrometer (Edinburg Instruments, Livingston, United Kingdom). Traces were best fit to a triple-exponential decay, and the lifetimes and relative populations were determined. The apparent free energy of destabilization was calculated as follows: ΔG = −kBT·ln(Bτ1), where kB is the Boltzmann constant, T is temperature in kelvins, and Bτ1 is the relative percent population of the open state. Apparent free energies of destabilization were compared between proteins over a range of concentrations in the absence and presence of 2 μM IP6.
Fluorescence measurements.
Fluorescence anisotropy (FA) was used to measure equilibrium binding of NC and Gag variants to NA. FA measurements were conducted using 20 nM fluorescently labeled NA (5′-FAM-DNA20 and 3′-FAM-minihelixLys3) in 50 mM NaCl, 20 mM HEPES, pH 7.4, 10 μM tris(2-carboxyethyl)phosphine, and 5 mM β-mercaptoethanol. MinihelixLys3 was refolded in the presence of Mg2+ as described previously (31), and the final buffer also contained 1 mM MgCl2. Measurements were obtained using a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA) according to established protocols (64). Binding affinities were determined by fitting the data to a 1:1 binding model with a correction for changes in fluorophore intensity due to protein binding (64). In the case of MA, FA also increased upon binding to NA; however, the binding was accompanied by a large loss in fluorescence intensity, making Kd (apparent dissociation constant) determination less reliable. Thus, fluorescence intensity rather than anisotropy was used to determine the NA binding affinity of MA. The Kd of IP6 binding to MA was determined from competition binding assays described below.
Determination of apparent dissociation constants by fluorescence intensity measurements.
MA-NA binding affinities were determined by titrating MA into fluorescently labeled NA and by fitting the fluorescence intensity of the NA substrate, I(MA), to the expression:
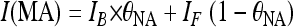 |
(1) |
Here, IB and IF are intensities of the bound and free NA, respectively, and θNA is the fraction of bound NA determined from the concentrations of MA and NA and the Kd of MA binding to NA (i.e., KdNA) using equation 2:
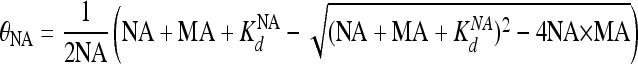 |
(2) |
Considering that the concentration of the fluorescently labeled NA is much less than the concentration of MA and KdNA, equation 2 reduces to the following:
 |
(3) |
To determine the Kd between IP6 and MA, competition binding assays were carried out by prebinding MA to NA and titrating in IP6. The recovery of fluorescence intensity curves were fit to equation 1, with θNA calculated according to the following:
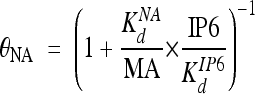 |
(4) |
Here, KdIP6 is the binding affinity of MA to IP6 in the absence of NA. Equation 4 was obtained from equation 3, but instead of using total MA concentration, the effective MA concentration, MA*, was used:
 |
(5) |
Equation 4 was obtained by substituting equation 5 into equation 3.
RESULTS
The MA domain of Gag inhibits
 annealing. The tRNA annealing activity of GagΔp6 and Gag-derived variant proteins was measured using a gel mobility shift annealing assay as previously described (31). Briefly, unmodified 76-nt human tRNA3Lys transcript was internally labeled with [α-32P]GTP and incubated with a 105-nt RNA construct (ShortPBS) derived from the PBS region of the HIV-1 genome (Fig. 1A) in the presence of recombinant HIV-1 proteins. The Gag variants used in this study (Fig. 1B) all lack the C-terminal p6 domain and also lack the N-terminal myristoyl group. Importantly, previous studies have shown that GagΔp6 (designated wild-type herein) assembles into immature VLPs that mimic authentic immature virions (8-9, 73). Deletion constructs (Gag constructs consisting of CA and NC [CANC], MA-linker-NC, and MA and CA [MACA] domains) were designed to determine the contribution of each of Gag's domains to its NA chaperone activity. Additionally, variants were tested to understand how the basic character of MA (using GagΔp6 with the mutations K30N and K32N [GagΔp6-K30,32N] and with three mutations in MA [GagΔp6-3 M]) and the ability of Gag to homodimerize (using WM-GagΔp6, WM-CANC, and WM-GagΔp6-K30,32N, where WM represents the CA mutations W316A and M317A) affect its chaperone activity (Fig. 1B). GagΔp6-3 M contains the E40R, E42L, N47K point mutations, which impart a more robust NA binding ability to HIV-1 MA (M. Sun, R. Gorelick, L. Mansky, and K. Musier-Forsyth, unpublished results).
annealing. The tRNA annealing activity of GagΔp6 and Gag-derived variant proteins was measured using a gel mobility shift annealing assay as previously described (31). Briefly, unmodified 76-nt human tRNA3Lys transcript was internally labeled with [α-32P]GTP and incubated with a 105-nt RNA construct (ShortPBS) derived from the PBS region of the HIV-1 genome (Fig. 1A) in the presence of recombinant HIV-1 proteins. The Gag variants used in this study (Fig. 1B) all lack the C-terminal p6 domain and also lack the N-terminal myristoyl group. Importantly, previous studies have shown that GagΔp6 (designated wild-type herein) assembles into immature VLPs that mimic authentic immature virions (8-9, 73). Deletion constructs (Gag constructs consisting of CA and NC [CANC], MA-linker-NC, and MA and CA [MACA] domains) were designed to determine the contribution of each of Gag's domains to its NA chaperone activity. Additionally, variants were tested to understand how the basic character of MA (using GagΔp6 with the mutations K30N and K32N [GagΔp6-K30,32N] and with three mutations in MA [GagΔp6-3 M]) and the ability of Gag to homodimerize (using WM-GagΔp6, WM-CANC, and WM-GagΔp6-K30,32N, where WM represents the CA mutations W316A and M317A) affect its chaperone activity (Fig. 1B). GagΔp6-3 M contains the E40R, E42L, N47K point mutations, which impart a more robust NA binding ability to HIV-1 MA (M. Sun, R. Gorelick, L. Mansky, and K. Musier-Forsyth, unpublished results).
Maximal chaperone activity of HIV-1 NC is known to require saturating levels of protein—that is, enough protein to coat the NA substrates. The binding site size of mature NC has been reported to be 5 to 8 nt (39). Thus, the concentration dependence of each Gag variant was first tested using a single-time-point assay in which protein concentration was varied while all other conditions remained constant (Table 1). For proteins lacking the NC domain (MA, CA, and MACA), no annealing was observed over 2 h using up to a 1:6 nucleotide-to-protein ratio. In contrast, for all proteins containing the NC domain, annealing was observed, suggesting that NC is absolutely essential for this activity. For chaperone proteins that did facilitate annealing, these single-time-point assays allowed us to determine each protein's K1/2 value, i.e., the critical nucleotide-to-protein ratio at which annealing is half maximal (Table 1). A higher K1/2 value indicates that less protein is required to facilitate tRNA annealing. The K1/2 value reflects a chaperone protein's binding affinity (Kd) and binding site size, as well as cooperativity. For NC (K1/2 of ∼7.90), about four times more protein was required to achieve half-maximal annealing relative to the amount of GagΔp6 (K1/2 of ∼30.2), consistent with previous studies comparing the two proteins (59, 75). GagΔp6, CANC (K1/2 of ∼29.9), and GagΔp6-K30,32N (K1/2 of ∼26.7) facilitate annealing at lower concentrations than their monomeric counterparts WM-GagΔp6 (K1/2 of ∼18.8), WM-CANC (K1/2 of ∼12.7), and WM-GagΔp6-K30,32N (K1/2 of ∼ 11.6) (Table 1). The more basic GagΔp6-3 M variant likewise requires less protein to facilitate annealing (K1/2 of ∼41.1), which may reflect its higher NA binding affinity (see below).
TABLE 1.
Annealing and binding parameters of Gag variantsd
| Protein variant | K1/2a | k (min−1)b | Kdminihelix (nM)c | KdssDNA (nM)c |
|---|---|---|---|---|
| NC | 7.90 ± 1.1 | 0.40 ± 0.2 | 92.2 ± 9.0 | 26.7 ± 2.0 |
| GagΔp6 | 30.2 ± 12 | 0.036 ± 0.01 | 72.8 ± 24 | 42.8 ± 10 |
| MA | — | — | 325 ± 25 | 542 ± 40 |
| CA | — | — | ND | ND |
| MACA | — | — | ND | ND |
| MA-linker-NC | 20.4 ± 1.1 | 0.097 ± 0.01 | 14.7 ± 4.0 | 12.2 ± 4.0 |
| CANC | 29.9 ± 8.2 | 0.271 ± 0.04 | 104 ± 30 | 48.7 ± 31 |
| WM-CANC | 12.7 ± 4.2 | 0.052 ± 0.01 | 62.2 ± 17 | 35.6 ± 16 |
| WM-GagΔp6 | 18.8 ± 6.8 | 0.036 ± 0.01 | 39.3 ± 3.0 | 28.0 ± 6.8 |
| WM-GagΔp6-K30,32N | 11.6 ± 0.5 | 0.101 ± 0.01 | ND | ND |
| GagΔp6-K30,32N | 26.7 ± 6.5 | 0.14 ± 0.01 | 77.0 ± 17 | 40.2 ± 23 |
| GagΔp6-3M | 41.1 ± 5.4 | 0.021 ± 0.01 | 34.6 ± 4.0 | 21.9 ± 0.7 |
K1/2 is the nucleotide-to-protein ratio at which annealing is half maximal. K1/2 values were obtained from sigmoidal fits of single-time-point annealing assays as described in Materials and Methods.
k is the annealing rate constant obtained from single exponential fits of time course annealing assays.
Kdminihelix and KdssDNA are the apparent dissociation constants for binding to 3′-FAM-minihelixLys3 and to 5′-FAM-DNA20, respectively, as determined using FA.
All experiments were performed in the presence of 1 mM MgCl2 and 20 mM NaCl with the exception of the ssDNA binding studies, which lacked MgCl2. All reported values represent the average of three or more trials with the standard deviation indicated. Dashes indicate no annealing detected. ND, not determined.
Based on the concentration dependence results, time course annealing assays were conducted using a constant nucleotide-to-protein ratio of 8.5:1. This value was chosen because all GagΔp6 variants were saturating at this concentration (Table 1); thus, under these conditions increasing the chaperone concentration would not be expected to enhance the rate of annealing. The tRNA annealing rate (k; min−1) was determined as described in Materials and Methods, and the values are reported in Table 1. Gag facilitates tRNA annealing over 10-fold more slowly than NC (0.036 ± 0.01 versus 0.40 ± 0.2 min−1) (Table 1). Higher concentrations of Gag (up to a 1:1 nucleotide-to-Gag ratio) did not result in increased rates of annealing, which is consistent with conditions of protein saturation (Fig. 2). Since NC levels are not saturating at 8.5:1 nucleotide-to-protein, its annealing rate increases to 0.82 ± 0.03 min−1 at 4.3:1 nucleotide-to-protein, or about 22-fold faster than GagΔp6. At higher concentrations of NC, the rate becomes so high that it is difficult to measure for technical reasons.
FIG. 2.
Bar graph summarizing GagΔp6-facilitated tRNA3Lys annealing rates at different nucleotide-to-GagΔp6 ratios, as shown above the bars.
Interestingly, the CANC variant facilitates tRNA annealing approximately 8-fold faster (0.271 ± 0.04 min−1) than GagΔp6. These results suggest that the presence of the basic MA domain reduces Gag's tRNA annealing efficiency. In contrast, the more basic GagΔp6-3 M variant has a slightly lower annealing rate (0.021 ± 0.01 min−1), whereas the less basic GagΔp6-K30,32N variant has an enhanced annealing rate (0.14 ± 0.01 min−1).
To test the effect of dimerization on Gag's chaperone activity, WM mutations were introduced into the GagΔp6, CANC, and GagΔp6-K30,32N variants. Monomeric WM-GagΔp6 protein facilitates tRNA annealing with the same rate as GagΔp6, suggesting that Gag multimerization does not contribute to tRNA annealing efficiency under saturating conditions. However, the monomeric WM-CANC protein shows a significantly reduced annealing rate (0.052 ± 0.01 min−1) relative to CANC. Thus, in the context of CANC, dimerization appears to have a positive effect on tRNA annealing. However, CANC-facilitated tRNA annealing is slower than that of NC, suggesting that the presence of the wild-type CA domain weakens NC's chaperone activity. Likewise, MA-linker-NC has a higher annealing rate than GagΔp6, supporting the conclusion that the CA domain reduces the chaperone activity of wild-type Gag. The dimerization-deficient WM-GagΔp6-K30,32N shows a somewhat reduced annealing rate (0.101 ± 0.01) relative to GagΔp6-K30,32N, but the effect is relatively small. Taken together, these data suggest that the effect of dimerization on tRNA annealing is highly context dependent. Despite the variable effects of CA dimerization on annealing rate, the single-time-point assays described above suggest that dimerization-competent variants are more efficient on a molar basis (i.e., have higher K1/2 values) than their monomeric counterparts (Table 1), which likely reflects their increased cooperativity.
Inositol phosphates stimulate Gag's NA chaperone activity.
We next investigated the mechanism by which MA modulates Gag's interactions with NA. Residues K30 and K32 within the MA domain of Gag have been implicated in binding to IPs in vitro (63), a result that has been corroborated by cell-based assays (50), membrane flotation experiments (12), and liposome-binding assays (4, 12). Since a primary function of MA involves membrane binding to PIP2, we investigated the effect of IPs on the chaperone activity of Gag. For these studies, we used inositol-(1,4,5)-triphosphate (IP3), a compound that resembles the head group of PIP2, as well as inositol hexakisphosphate (IP6). We also tested PIP2-diC4, a soluble analog of PIP2 which has 4-carbon acyl chains in place of the arachnidonate and stearate chains in native PIP2.
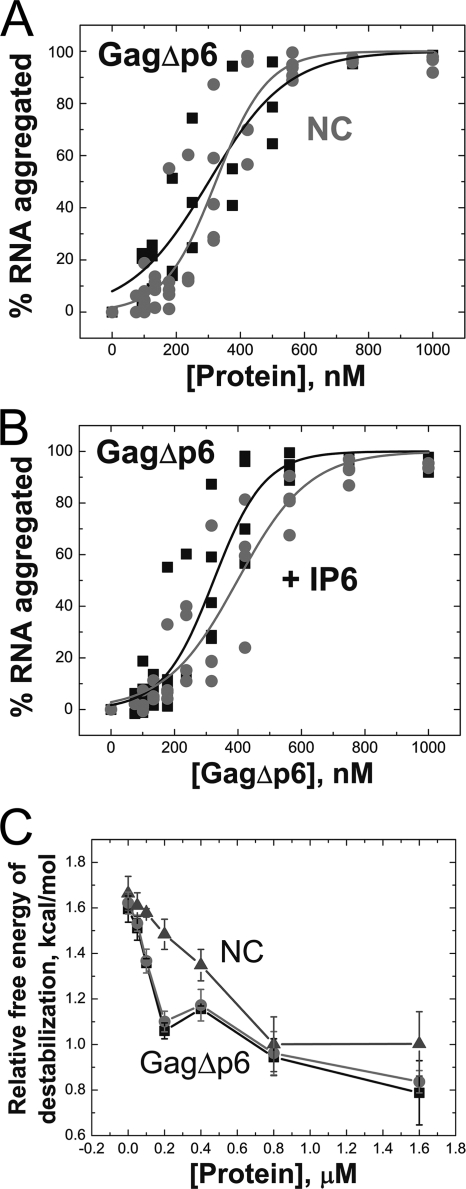
As shown in Fig. 3 A, all IPs tested enhance the ability of Gag to facilitate tRNA annealing. In the presence of 2 μM IP6 or higher concentrations of the less negatively charged IP3 or PIP2-diC4 (300 μM), up to a 13-fold enhancement in tRNA annealing is observed (Fig. 3A). The rate of tRNA annealing by GagΔp6 in the presence of IPs (0.470 ± 0.10 min−1) is even greater than that of CANC (Table 1). The need for higher concentrations of IP3 and PIP2-diC4 was expected based on in vitro assembly assays in which higher concentrations of IP3 were required to correct particle morphology (8). Further studies were conducted only with IP6. We showed that the presence of IP6 does not enhance annealing by NC or CANC, which lack the MA domain (Fig. 3B), suggesting that the observed rate increase is due to IP6-MA interactions. To confirm this hypothesis, the rate of annealing of GagΔp6-K30,32N was also tested in the absence and presence of IP6. The rate of annealing by this variant was also unaffected by IP6 addition (Fig. 3B).
FIG. 3.
The effect of IPs on tRNA3Lys annealing to ShortPBS template. (A) Annealing by GagΔp6 (filled squares) and of GagΔp6 in the presence of 2 μM IP6 (open circles), 300 μM IP3 (filled triangles), and 300 μM PIP2-diC4 (open triangles) in 20 mM NaCl. The inset shows data for annealing by GagΔp6 in the absence and presence of 20 μM IP6 in 150 mM NaCl. (B) Annealing by CANC (left), GagΔp6-K30,32N (middle), and NC (right) in the absence (solid squares) and presence (open circles) of IP6 in 20 mM NaCl. In these annealing experiments, the ratio of tRNA3Lys to ShortPBS ratio is 2.5:1, and the protein concentrations are 800 nM (4.3:1, nucleotide to protein).
As the annealing reactions described above were performed in 20 mM NaCl, the effect of IPs on Gag-facilitated tRNA annealing were also tested in the presence of physiological salt conditions (i.e., 150 mM NaCl). Under these conditions, GagΔp6 annealing is also stimulated by IP6 addition (Fig. 3A, inset) although ∼10-fold more IP6 was required to achieve a similar rate enhancement. This likely reflects the electrostatic nature of MA-IP6 interactions.
Dissecting the mechanism of IP stimulation of Gag annealing.
NA aggregation, destabilization, and rapid binding kinetics are three important properties of NA chaperone proteins such as NC. We hypothesized that IP6 stimulates Gag-facilitated tRNA annealing by enhancing one or more of these activities.
A sedimentation assay was used to assess NA aggregation (71). In this assay, 32P-labeled tRNA3Lys is incubated with GagΔp6 or NC, which leads to the formation of NA-protein aggregates that can be pelleted by centrifugation at speeds (9,300 × g) that do not pellet free protein or NA. This assay has been previously used to assess aggregation activity of a variety of retroviral NC proteins (55, 64, 69-71). GagΔp6 and NC aggregate NA with similar efficiencies (Fig. 4 A), and IP6 does not affect Gag's ability to aggregate NA (Fig. 4B).
FIG. 4.
(A) Aggregation activity of GagΔp6 (squares) compared to NC (circles). (B) GagΔp6-facilitated aggregation of NA in the presence (circles) and absence (squares) of 5:1 IP6 to GagΔp6. Data from three independent experiments are shown for each protein. (C) GagΔp6-facilitated destabilization of a fluorescently labeled TAR DNA hairpin in the absence (squares) and presence (circles) of a 5-fold excess of IP6. NC-facilitated destabilization is also shown (triangles). The relative free energy of destabilization is calculated from the percent population of TAR DNA hairpins in the open state, as described in Materials and Methods.
A time-resolved fluorescence resonance energy transfer (tr-FRET) assay was used to assess destabilization. In this assay, a DNA hairpin complementary to the HIV-1 genome's transactivation response region (TAR) RNA is labeled at the 3′ and 5′ ends with a fluorescent dye and quencher, respectively (55). This assay was previously used to characterize duplex destabilization activity of retroviral chaperone proteins (32, 55) by directly monitoring the end-to-end distance of the hairpin stem. The advantage of tr-FRET is that it allows the distribution of different populations of molecules to be observed (i.e., open, semiopen, and closed) in the absence and presence of chaperone protein. By monitoring the most open population as a function of protein concentration, we find that GagΔp6 is able to effectively destabilize the TAR hairpin and that the presence of IP6 does not affect this capability (Fig. 4C).
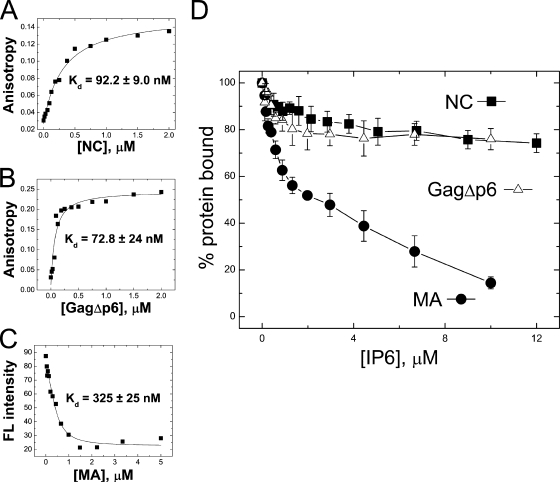
NA binding of Gag variants was next investigated using fluorescence anisotropy (FA) (Fig. 5). For these studies, binding to a 20-mer single-stranded DNA (ssDNA) oligonucleotide and a 37-nt tRNA3Lys-derived acceptor-TΨC stem minihelix was measured (Fig. 1A, boxed). In the case of MA, addition of protein was accompanied by a significant decrease in fluorescence intensity, making the use of FA less reliable. Therefore, in this case, the change in fluorescence intensity was used to determine the apparent binding affinity (Fig. 5C).
FIG. 5.
Fluorescence measurements to monitor NC (A), GagΔp6 (B), and MA (C) binding to minihelixLys3 with average binding affinities (Kd) from at least three trials indicated. For NC and GagΔp6, changes in the fluorescence anisotropy were fit to a 1:1 binding curve as described previously (60). For MA, the change in fluorescence intensity (i.e., quenching) upon binding was fit to a similar binding model described in Materials and Methods. (D) Competition binding assay wherein 20 nM 3′-FAM-labeled minihelixLys3 was preincubated with 200 nM NC, 200 nM GagΔp6, or 1 μM MA followed by titration with IP6. Fluorescence anisotropy (NC and GagΔp6) or intensity (MA) was normalized to the fraction bound.
In accord with its duplex destabilizing activity, NC preferentially binds single-stranded over double-stranded NA (39, 64). As expected, tighter binding to the ssDNA 20-mer (Kd of ∼30 nM) relative to the minihelixLys3 (Kd of ∼100 nM) is observed (Table 1). In contrast, MA binds both substrates with similar affinities, with no preference for ssDNA (Kd of ∼400 nM). All Gag variants containing NC bind strongly and with comparable affinities to both substrates (Kd of ∼35 to 100 nM), irrespective of the presence of the MA domain. Although binding of Gag variants containing both NC and MA domains is predicted to be stronger than for variants containing only NC, the relatively short NA sequences used in the FA binding studies may not support simultaneous binding of both domains to a single substrate. The only exception is the MA-linker-NC construct, which binds with substantially greater affinity (Kd of 12 to 15 nM), most likely reflecting this protein's increased flexibility compared to GagΔp6. Taken together, these results confirm that Gag interacts with NA most strongly with its NC domain but that the MA domain also binds NA with moderate affinity.
To determine whether IP6 alters the NA binding properties of NC and MA, competition assays were performed. In these experiments, the proteins were prebound to minihelixLys3, and the complexes were titrated with IP6. As shown in Fig. 5D, the fraction of MA-bound complexes decreases with increasing IP6, suggesting that MA is readily competed off the NA (∼85%) (Fig. 5D). Similar competition studies performed with free NC or GagΔp6 showed that these NA-protein complexes are only partially displaced with increasing IP6 concentration (∼20%) (Fig. 5D). These findings demonstrate that ∼2 μM IP6 is sufficient to compete with NA for binding to the MA domain of Gag, but even a 10-fold higher concentration of IP6 cannot effectively compete off the NC domain. Using these competition curves and the Kd of MA binding to the minihelix, we estimate that the Kd of MA binding to IP6 is 245 ± 60 nM (see Materials and Methods).
DISCUSSION
In this study, we show that on a molar basis GagΔp6 is a better NA chaperone than NC, facilitating annealing at about 4-fold lower concentrations, albeit at a 10-fold reduced rate relative to NC. Inositol phosphates (IPs) stimulate GagΔp6-facilitated tRNA3Lys annealing to the PBS to a level that approaches that of NC. IP6-independent enhancement in annealing also occurs if the MA domain is deleted from Gag or if positively charged residues known to interact with IPs (e.g., K30 and K32) are neutralized. Furthermore, IPs compete very effectively for NA binding to MA but not NC, a finding in accordance with recent studies showing that MA bound preferentially to PIP2-containing liposomes in the presence of NA (4, 13). The HIV-1 MA domain alone shows little or no chaperone activity (Table 1), but MA binds NA with moderate affinity, and mutations affecting the basic character of MA modulate Gag's chaperone activity (i.e., GagΔp6-3 M and GagΔp6-K30,32N). Taken together, these findings suggest that Gag binds to NA with both NC and MA domains and that this binding mode is reflected in reduced chaperone activity.
The conformation of HIV-1 Gag in solution has been measured by hydrodynamic techniques and small-angle neutron scattering (17). Gag was found to be flexible, with a distribution of conformations, including conformers in which the MA and NC domains are closer than would be expected for a completely extended protein (17). Moreover, IPs have been shown to bind to Gag at both ends in solution, possibly stabilizing the folded conformation (19). In the immature virion, Gag is extended (23) and has thus been proposed to undergo a large conformational change during the process of assembly (4, 17, 19). Previously, it was observed that mixing RNA and GagΔp6 in vitro leads to the formation of small, aberrant VLPs 25 to 30 nm in diameter (9). Interestingly, adding IPs to this mixture results in a change in the morphology of VLPs assembled in vitro—i.e., the formation of 100- to 120-nm-diameter WT particles resembling immature HIV-1 virions (8). However, a GagΔp6 variant lacking residues 16 to 99 of MA, which includes the PIP2 binding site, does not require IPs for WT VLP formation. These findings are in agreement with the present study and other evidence (4, 17, 52) suggesting that Gag may adopt the MA/NC-bound state in the presence of NA.
An IP-induced conformational change within Gag could account for the enhancement in Gag's chaperone activity although additional experiments are needed to test this possibility. An alternative explanation is that IPs prevent MA-NA interactions without inducing a conformational change upon binding to MA. Future studies using biophysical techniques to directly observe Gag's conformation in the presence of IPs and NA are planned. In either case, the enhanced chaperone activity of Gag in the presence of IP6 may be due to more effective NA coating by the NC domain. Alternatively, the increase may be due to a subtler rearrangement of NC-NA interactions resulting in more efficient chaperone activity.
High GagΔp6 concentrations (3 μM; 1:1 nucleotide-to-Gag ratio) do not stimulate the tRNA annealing rate (Fig. 2). This was surprising since NC binds NA with a higher affinity than MA (Table 1), and one might expect that the NC domains of excess GagΔp6 might compete out the MA domain of bound GagΔp6 molecules, thereby favoring the NC-bound state. Whereas high Gag concentration evidently cannot drive this change in vitro, expression of large amounts of Gag in cells (i.e., in the presence of PIP2 or other factors) leads to an enrichment of Gag on membranes (52).
MA may interact with RNA for a variety of reasons, such as to prevent premature assembly in the cytoplasm or on inappropriate membranes (4, 13), to prevent premature myristoyl exposure (52, 60, 66), or to act along with NC in viral RNA selection and packaging (51, 72). CANC and GagΔp6-K30,32N show elevated tRNA annealing rates, which appear to be correlated with the fact that neither protein has an MA domain capable of interacting with NA as effectively as GagΔp6. Moreover, the GagΔp6-3 M variant binds with higher affinity to NA and is a less efficient chaperone than WT GagΔp6 (Table 1). It is important to note that all of the Gag variants studied here lack the p6 domain for technical reasons. Whether the acidic nature of p6 diminishes the affinity of the C terminus of Gag for NA remains to be established.
NC facilitates tRNA annealing ∼105-fold over the background rate (31), whereas the rate of GagΔp6-facilitated annealing is at least 10-fold lower. No differences in aggregation and duplex destabilization were observed for the Gag variants tested here (Fig. 4). Given the NA binding affinity of MA (Table 1), the difference in binding modes is likely responsible for Gag's weaker chaperone activity. Without NA binding, an NA chaperone cannot act on its substrate, but for Gag, additional binding contributed by its MA domain limits its chaperone activity. This finding is not without precedent as mutations that reduce the StpA NA chaperone protein's binding affinity increase its overall chaperone activity (46).
Recent structure-probing analysis of the HIV-1 genome has found that the RNA structure differs between NC-bound and unbound states (74). Although the structure of the genome in the presence of Gag is unknown, it is possible that a more reverse transcription-competent tRNA/genome binary complex is achieved by the NC protein only after proteolysis of the Gag precursor (1). In accordance with this idea, a recent report suggests that Gag-annealed tRNA is not as stable as NC-annealed tRNA (29). This finding may be explained by Gag's preference to bind NA with both its MA and NC domains, resulting in a less stably annealed tRNA structure relative to an NC-annealed complex.
Our findings suggest that prior to PM binding, MA-RNA interactions may prevent Gag from prematurely annealing tRNA3Lys. Recent single-virion fluorescence experiments support a model in which relatively few Gag molecules traffic with the RNA genome to the site of assembly on the PM (36). These Gag molecules are likely clustered around the ψ region of the genome (36). Regulation of annealing may benefit the virus by preventing premature reverse transcription (33, 67, 75) or cleavage by Dicer (76). Inhibition of Gag's chaperone activity during assembly may also serve to regulate RNA remodeling of the 5′ UTR (1) and modulate interactions with RNA binding proteins such as RNA helicase A (6). Studies are under way to test these possibilities.
In cells, PIP2-Gag interaction preferentially targets Gag to the PM rather than other cellular membranes, and it has been hypothesized that this interaction may even target Gag to lipid rafts (61). Our new findings suggest that PIP2 interaction may also activate Gag's chaperone activity. However, the interaction between HIV-1 MA and PIP2 is relatively weak in vitro, especially compared to more highly phosphorylated IPs such as inositol pentakisphosphate (IP5) and IP6 (8, 61). For example, in the present study, 150 times more PIP2 than IP6 was required to enhance Gag's chaperone activity. In cells, IP6 is present as a structural cofactor in ADAR2 (41) and as a nuclear mRNA export signal synthesized from PI(4,5)P2 (48). It is unclear what role, if any, IP5 or IP6 may play in HIV-1 biology, but our results suggest that MA-IP interactions in the cytosol could, in principle, modulate Gag-NA interactions prior to membrane binding.
Acknowledgments
We thank Robert J. Gorelick for providing HIV-1 NC and Meng Sun for preparation of HIV-1 MA protein used in these studies. We thank Judith G. Levin for helpful discussions.
This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by NIH grant GM065056 (to K.M.-F.). C.P.J. was supported by NIH Training Grant T32 GM008512.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1.Abbink, T. E., and B. Berkhout. 2008. HIV-1 reverse transcription initiation: a potential target for novel antivirals? Virus Res. 134:4-18. [DOI] [PubMed] [Google Scholar]
- 2.Ako-Adjei, D., M. C. Johnson, and V. M. Vogt. 2005. The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles. J. Virol. 79:13463-13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldovini, A., and R. A. Young. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 64:1920-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfadhli, A., R. L. Barklis, and E. Barklis. 2009. HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology 387:466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfadhli, A., A. Still, and E. Barklis. 2009. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 83:12196-12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolinger, C., and K. Boris-Lawrie. 2009. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology 6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess, R. R. 1991. Use of polyethyleneimine in purification of DNA-binding proteins. Methods Enzymol. 208:3-10. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, S., et al. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. U. S. A. 98:10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cen, S., et al. 2001. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J. Virol. 75:5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, R., et al. 2008. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 82:11228-11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chukkapalli, V., I. B. Hogue, V. Boyko, W. S. Hu, and A. Ono. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag membrane binding. J. Virol. 82:2405-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chukkapalli, V., S. J. Oh, and A. Ono. 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. U. S. A. 107:1600-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin, J. M., S. H. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 15.Cruceanu, M., et al. 2006. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 34:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darlix, J. L., M. Lapadat-Tapolsky, H. de Rocquigny, and B. P. Roques. 1995. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 254:523-537. [DOI] [PubMed] [Google Scholar]
- 17.Datta, S. A., et al. 2007. Conformation of the HIV-1 Gag protein in solution. J. Mol. Biol. 365:812-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datta, S. A., and A. Rein. 2009. Preparation of recombinant HIV-1 Gag protein and assembly of virus-like particles in vitro. Methods Mol. Biol. 485:197-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datta, S. A., et al. 2007. Interactions between HIV-1 Gag molecules in solution: an inositol phosphate-mediated switch. J. Mol. Biol. 365:799-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Guzman, R. N., et al. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 ψ-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 21.Feng, Y. X., et al. 1999. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 73:4251-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fossen, T., et al. 2005. Solution structure of the human immunodeficiency virus type 1 p6 protein. J. Biol. Chem. 280:42515-42527. [DOI] [PubMed] [Google Scholar]
- 23.Fuller, S. D., T. Wilk, B. E. Gowen, H. G. Krausslich, and V. M. Vogt. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729-738. [DOI] [PubMed] [Google Scholar]
- 24.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 25.Ganser-Pornillos, B. K., M. Yeager, and W. I. Sundquist. 2008. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18:203-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrus, J. E., et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 27.Gheysen, D., et al. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 28.Gorelick, R. J., et al. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo, F., J. Saadatmand, M. Niu, and L. Kleiman. 2009. Roles of Gag and NCp7 in facilitating tRNA(Lys)(3) Annealing to viral RNA in human immunodeficiency virus type 1. J. Virol. 83:8099-8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo, J., et al. 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 74:8980-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hargittai, M. R., R. J. Gorelick, I. Rouzina, and K. Musier-Forsyth. 2004. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 337:951-968. [DOI] [PubMed] [Google Scholar]
- 32.Hong, M. K., et al. 2003. Nucleic acid conformational changes essential for HIV-1 nucleocapsid protein-mediated inhibition of self-priming in minus-strand transfer. J. Mol. Biol. 325:1-10. [DOI] [PubMed] [Google Scholar]
- 33.Houzet, L., et al. 2008. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 36:2311-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, Y., et al. 1997. Primer tRNA3Lys on the viral genome exists in unextended and two-base extended forms within mature human immunodeficiency virus type 1. J. Virol. 71:726-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang, M., et al. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jouvenet, N., S. M. Simon, and P. D. Bieniasz. 2009. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. U. S. A. 106:19114-19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kafaie, J., R. Song, L. Abrahamyan, A. J. Mouland, and M. Laughrea. 2008. Mapping of nucleocapsid residues important for HIV-1 genomic RNA dimerization and packaging. Virology 375:592-610. [DOI] [PubMed] [Google Scholar]
- 38.Kleiman, L., C. P. Jones, and K. Musier-Forsyth. 2010. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 584:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levin, J. G., J. Guo, I. Rouzina, and K. Musier-Forsyth. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80:217-286. [DOI] [PubMed] [Google Scholar]
- 40.Lochrie, M. A., et al. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 Gag polyprotein. Nucleic Acids Res. 25:2902-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macbeth, M. R., et al. 2005. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309:1534-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marenchino, M., D. W. Armbruster, and M. Hennig. 2009. Rapid and efficient purification of RNA-binding proteins: application to HIV-1 Rev. Protein Expr. Purif. 63:112-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mascarenhas, A. P., and K. Musier-Forsyth. 2009. The capsid protein of human immunodeficiency virus: interactions of HIV-1 capsid with host protein factors. FEBS J. 276:6118-6127. [DOI] [PubMed] [Google Scholar]
- 44.Massiah, M. A., et al. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198-223. [DOI] [PubMed] [Google Scholar]
- 45.Massiah, M. A., et al. 1996. Comparison of the NMR and X-ray structures of the HIV-1 matrix protein: evidence for conformational changes during viral assembly. Protein Sci. 5:2391-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayer, O., L. Rajkowitsch, C. Lorenz, R. Konrat, and R. Schroeder. 2007. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 35:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Momany, C., et al. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 48.Odom, A. R., A. Stahlberg, S. R. Wente, and J. D. York. 2000. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287:2026-2029. [DOI] [PubMed] [Google Scholar]
- 49.Ono, A. 2010. HIV-1 assembly at the plasma membrane. Vaccine 28(Suppl. 2):B55-B59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono, A., S. D. Ablan, S. J. Lockett, K. Nagashima, and E. O. Freed. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 101:14889-14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ott, D. E., L. V. Coren, and T. D. Gagliardi. 2005. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J. Virol. 79:13839-13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perez-Caballero, D., T. Hatziioannou, J. Martin-Serrano, and P. D. Bieniasz. 2004. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on Gag precursor-membrane interactions. J. Virol. 78:9560-9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prats, A. C., et al. 1988. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 7:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purohit, P., S. Dupont, M. Stevenson, and M. R. Green. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7:576-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qualley, D. F., et al. 2010. C-terminal domain modulates the nucleic acid chaperone activity of human T-cell leukemia virus type 1 nucleocapsid protein via an electrostatic mechanism. J. Biol. Chem. 285:295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajkowitsch, L., et al. 2007. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 4:118-130. [DOI] [PubMed] [Google Scholar]
- 57.Ratner, L., et al. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 58.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 59.Roldan, A., O. U. Warren, R. S. Russell, C. Liang, and M. A. Wainberg. 2005. A HIV-1 minimal Gag protein is superior to nucleocapsid at in vitro tRNALys3 annealing and exhibits multimerization-induced inhibition of reverse transcription. J. Biol. Chem. 280:17488-17496. [DOI] [PubMed] [Google Scholar]
- 60.Saad, J. S., et al. 2007. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 366:574-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saad, J. S., et al. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. U. S. A. 103:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shiba, K., et al. 1997. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues Escherichia coli double-defective mutant. J. Biol. Chem. 272:22809-22816. [DOI] [PubMed] [Google Scholar]
- 63.Shkriabai, N., et al. 2006. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 45:4077-4083. [DOI] [PubMed] [Google Scholar]
- 64.Stewart-Maynard, K. M., et al. 2008. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 82:10129-10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Göttlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 66.Tang, C., et al. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. U. S. A. 101:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas, J. A., W. J. Bosche, T. L. Shatzer, D. G. Johnson, and R. J. Gorelick. 2008. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J. Virol. 82:9318-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas, J. A., and R. J. Gorelick. 2008. Nucleocapsid protein function in early infection processes. Virus Res. 134:39-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vo, M. N., G. Barany, I. Rouzina, and K. Musier-Forsyth. 2009. Effect of Mg2+ and Na+ on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: implications for reverse transcription. J. Mol. Biol. 386:773-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vo, M. N., G. Barany, I. Rouzina, and K. Musier-Forsyth. 2009. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop “kissing” to “zipper.” J. Mol. Biol. 386:789-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vo, M. N., G. Barany, I. Rouzina, and K. Musier-Forsyth. 2006. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J. Mol. Biol. 363:244-261. [DOI] [PubMed] [Google Scholar]
- 72.Wang, H., K. M. Norris, and L. M. Mansky. 2003. Involvement of the matrix and nucleocapsid domains of the bovine leukemia virus Gag polyprotein precursor in viral RNA packaging. J. Virol. 77:9431-9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilk, T., et al. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilkinson, K. A., et al. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu, T., et al. 2010. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: biological implications. Virology 405:556-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yeung, M. L., et al. 2009. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 37:6575-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yoo, S., et al. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780-795. [DOI] [PubMed] [Google Scholar]