Abstract
The hepatitis C virus (HCV) NS2 protein is essential for particle assembly, but its function in this process is unknown. We previously identified critical genetic interactions between NS2 and the viral E1-E2 glycoprotein and NS3-NS4A enzyme complexes. Based on these data, we hypothesized that interactions between these viral proteins are essential for HCV particle assembly. To identify interaction partners of NS2, we developed methods to site-specifically biotinylate NS2 in vivo and affinity capture NS2-containing protein complexes from virus-producing cells with streptavidin magnetic beads. By using these methods, we confirmed that NS2 physically interacts with E1, E2, and NS3 but did not stably interact with viral core or NS5A proteins. We further characterized these protein complexes by blue native polyacrylamide gel electrophoresis and identified ≈520-kDa and ≈680-kDa complexes containing E2, NS2, and NS3. The formation of NS2 protein complexes was dependent on coexpression of the viral p7 protein and enhanced by cotranslation of viral proteins as a polyprotein. Further characterization indicated that the glycoprotein complex interacts with NS2 via E2, and the pattern of N-linked glycosylation on E1 and E2 suggested that these interactions occur in the early secretory pathway. Importantly, several mutations that inhibited virus assembly were shown to inhibit NS2 protein complex formation, and NS2 was essential for mediating the interaction between E2 and NS3. These studies demonstrate that NS2 plays a central organizing role in HCV particle assembly by bringing together viral structural and nonstructural proteins.
Hepatitis C virus (HCV) is a member of the Flaviviridae family of enveloped, positive-strand RNA viruses (67). The HCV genome is 9.6 kb in length and encodes a single large open reading frame, which is translated as a large polyprotein (8, 39). This polyprotein is cleaved co- and posttranslationally by viral and host proteases into distinct protein products (Fig. 1 A).
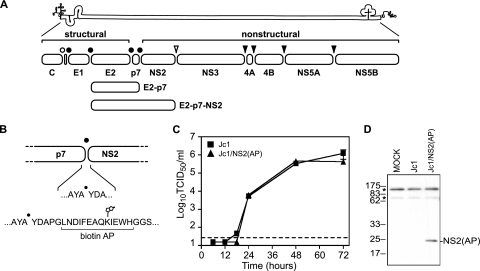
FIG. 1.
HCV genome and NS2 affinity purification system. (A) HCV genome and polyprotein. The open bullet represents a signal peptide peptidase cleavage site; closed bullets represent signal peptidase cleavage sites; the open arrowhead represents the NS2-3 cysteine autoprotease cleavage site; closed arrowheads represent NS3-4A serine protease cleavage sites. (B) Schematic of the AP inserted into the N terminus of NS2. The biotinylated lysine residue is indicated. (C) Time course of Jc1 and Jc1/NS2(AP) virus production after RNA electroporation. Viral infectivities are represented as tissue-culture infectious doses (50% endpoint) per ml (TCID50/ml). Values are averages of results from three independent transfections; errors represent standard deviations from the means. The dotted line represents the limit of detection of our titering assay. (D) HCV NS2(AP) is biotinylated in Huh-7.5(BirA) cells. Huh-7.5(BirA) cells were transfected with Jc1 and Jc1/NS2(AP) RNA and harvested 48 h posttransfection. Cells were lysed in SDS-PAGE buffer, and proteins were separated by SDS-PAGE and transferred to PVDF membranes. Biotinylated proteins were visualized by using the Vectastain ABC kit. Asterisks indicate two endogenously biotinylated mitochondrial proteins, most likely pyruvate carboxylase (upper band) and propionyl coenzyme A carboxylase (lower band) (29).
The N-terminal region of the polyprotein encodes three structural proteins: core, which presumably forms viral nucleocapsids, and two glycoproteins, E1 and E2, which mediate viral entry. E1 and E2 form heterodimers that are retained in the endoplasmic reticulum (ER), the likely site of viral budding (32). This heterodimerization involves charged residues within the C-terminal membrane anchors of E1 and E2 as well as regions in the glycoprotein ectodomains (9, 51, 78).
The C-terminal region of the polyprotein encodes seven nonstructural (NS) proteins: p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (8, 39). Numerous functions have been described for the HCV NS proteins. The small p7 protein has ion channel activity and is required for virus particle assembly (24, 54, 64). NS2 encodes the active sites for the NS2-3 cysteine autoprotease (19, 20, 43) and plays an essential but undefined role in virus particle assembly (23, 24). NS3 is a bifunctional protein that encodes an N-terminal serine protease domain and a C-terminal NTPase/RNA helicase domain. NS3 is important for RNA replication (27, 31), modulating innate antiviral defenses (33, 36, 42, 47), and has been implicated in virus particle assembly (45, 55). NS4A is a small protein that binds NS3, functions as a cofactor for NS3 serine protease and RNA helicase activities, and anchors the NS3-4A enzyme complex to cellular membranes (6, 13, 28, 73). NS4B is a multispanning membrane protein that is important for organizing the membrane-bound viral RNA replication machinery (reviewed in reference 17). NS5A is an RNA-binding phosphoprotein that plays essential roles in viral RNA replication (1, 2, 66) and virus particle assembly (3, 65). NS5B encodes the RNA-dependent RNA polymerase (74).
During translation and processing of the viral polyprotein, cleavage of core/E1, E1/E2, E2/p7, and p7/NS2 are mediated by host signal peptidase (4, 41). Inefficient or delayed cleavage by signal peptidase can lead to the accumulation of E2-p7 and E2-p7-NS2 intermediates (Fig. 1A) (34, 49, 56, 63). NS2/NS3 cleavage is mediated by the NS2-3 cysteine autoprotease (19, 20, 62). The NS3/4A, NS4A/4B, NS4B/5A, and NS5A/5B cleavages are mediated by the NS3-4A serine protease (4, 18, 20).
HCV NS2 contains an N-terminal membrane anchor that likely contains three transmembrane helices (23, 55) and a C-terminal domain that homodimerizes to form a cysteine protease that contains two composite active sites (43). The only known substrate of this protease is the NS2/3 cleavage site. While NS2 encodes the catalytic residues of the cysteine protease, formation of the active protease requires interactions between NS2 and the N-terminal region of NS3 prior to cleavage (19, 20, 62).
In addition to its role in viral gene expression, NS2 plays an essential but unknown role in virus particle assembly. Several cell culture-adaptive mutations that increase virus yields have been mapped to NS2 (16, 21, 25, 60, 76), and mutagenesis of NS2 identified several residues that are important for infectious virus production (10, 23, 55, 71, 77). We previously identified second-site mutations in E1 (A78T), E2 (I360T), NS3 (Q221L), and NS4A (E42G) that suppressed defects in virus particle assembly caused by specific NS2 mutations (55). These genetically defined interactions led us to hypothesize that NS2 protein can physically interact with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes and that these interactions are required for HCV particle assembly. To address this hypothesis, we established an efficient NS2-specific affinity purification system from virus-producing cells and identified essential physical interactions between NS2, E1, E2, and NS3 as well as a role for other viral proteins in NS2 complex assembly. Further characterization of NS2-containing protein complexes by blue native polyacrylamide gel electrophoresis (BN-PAGE) identified distinct membrane-associated high-molecular-mass NS2 protein complexes present during HCV infection. These complementary biochemical and genetic studies indicate the presence of dynamic protein-protein interactions required for HCV particle assembly and highlight the role of NS2 protein complexes in this process.
MATERIALS AND METHODS
Cell lines and culture conditions.
All cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (FCS) (HyClone, Logan, UT) and 1 mM nonessential amino acids (Invitrogen, Carlsbad, CA) at 37°C with 5% CO2.
Huh-7.5 cells expressing the Escherichia coli biotin ligase BirA were generated by lentivirus transduction. Briefly, the pLenti/BirA vector (described below) was packaged in HEK-239T cells by cotransfection with pLP1, pLP2, and pLP-VSV-G (Invitrogen) by using Fugene 6 (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. After 48 h, the conditioned cell culture medium was clarified by centrifugation (1,847 × g for 10 min) and filtration (pore size, 0.2 μm; Corning, Corning, NY). Huh-7.5 cells were transduced by spin inoculation (1,282 × g for 30 min at 25°C) with clarified, packaged vector-containing media and 8 μg/ml Polybrene (Millipore, Temecula, CA) and 20 mM HEPES buffer, pH 7.5 (Invitrogen). Three days postinfection, the medium was replaced with standard growth medium containing 100 μg/ml of zeocin (Invitrogen), and zeocin-resistant cell populations were selected over the course of several weeks. Zeocin-resistant Huh-7.5(BirA) colonies were isolated with glass cloning cylinders, expanded, and screened for BirA expression. The Huh-7.5(BirA) clone used here grew with kinetics similar to those of standard Huh-7.5 cells and stably maintained BirA expression either with or without further zeocin selection.
Plasmids.
All viral constructs were constructed by standard molecular biology techniques. A list of primers used in these studies is shown in Table 1. Site-directed mutagenesis (SDM) was performed by using the QuikChange method (Stratagene, La Jolla, CA). Plasmid sequences were validated by DNA sequencing at the W. M. Keck Foundation Biotechnology Research Center at Yale University.
TABLE 1.
Primers used in these studies
| Primer | Primer sequence (5′-3′) |
|---|---|
| YO-0315 | CGCGCCGGGCCTGAACGACATCTTCGAGGCCCAGAAGATCGAGTGGCACGGCGGCAGCGGCGGCAGCGA |
| YO-0316 | CGCGTCGCTGCCGCCGCTGCCGCCGTGCCACTCGATCTTCTGGGCCTCGAAGATGTCGTTCAGGCCCGG |
| YO-0361 | CGCCACCTGAACATACCTACC |
| YO-0521 | GCATCACCACGCCGGTGTCCGCTGCCGAAGTGAAGAATTCCACCGGCCTGTACCACGTGACTAACGACTGCACCAA |
| YO-0522 | GTTAGTCACGTGGTACAGGCCGGTGGAATTCTTCACTTCGGCAGCGGACACCGGCGTGGTGATGCAGGACAG |
| YO-0527 | CGGTCTCCGCTGCCGAAGTGCCTCAACAGGCTTATGCTTATGAC |
| YO-0528 | GTCATAAGCATAAGCCTGTTGAGGCACTTCGGCAGCGGAGACCG |
| YO-0603 | TATGCAGGGAGCGTGGGCGCAAGTCGTTGTCATCCT |
| YO-0604 | AGGATGACAACGACTTGCGCCCACGCTCCCTGCATA |
| YO-0605 | TCTTATTCCTGCTCTTAGCGTGGGCCAGGGTTTGCGCCTG |
| YO-0657 | CAGGCGCAAACCCTGGCCCACGCTAAGAGCAGGAATAAGA |
| YO-0743 | TGGATGCTCATCTTGTTGGGCCAGGCCGAAGCATAAGTTTAAACCCTCTCCCTC |
| YO-0744 | CCAGCAGGCCCAGGGCCAGCAGGGGCAGCATGCGCTCCATGGTATCATCGTGTTTTTCAA |
| YO-0745 | GCCCTGGGCCTGCTGGCCGCCGGCTTCTGCCCCGCCGTGGAGGCCGCACTAGAGAAGCTGGTCAT |
| YO-0746 | GCCTGCTGGCCGCCGGCTTCTGCCCCGCCGTGGAGGCCTATGACGCGCCGGGCCTGAACGACATC |
| YO-0747 | ACGCGTCGCTGCCGCCGCTGCCGCCGTGCCACTCGATCTTCTGGGCCTCGAAGATGTCGTTCAGGCCCGGCG |
| YO-0750 | GGTGGAAGCTCCTTTAAGTTTAAACGCTCCCATCACTGCTTAT |
| YO-0751 | GCAGTGATGGGAGCGTTTAAACTTAAAGGAGCTTCCACCCCTT |
| YO-0764 | TGGTTTTCCTTTGAAAAACACGATGATACCATGGCACTAGAGAAGCTGGTCATCTTGCAC |
| YO-0765 | AGATGACCAGCTTCTCTAGTGCCATGGTATCATCGTGTTTTTCAAAGGAAAACCACGTCC |
| YO-0766 | TACTTTGAAAAACACGATGATACCATGTGACGCGCCGGGCCTGAACGACATCTTCGAGGC |
| YO-0767 | CGTTCAGGCCCGGCGCGTCATACATGGTATCATCGTGTTTTTCAAAGGAAAACCACGTCC |
| RU-O-6823 | CGCGCCCGGGCAGCGGCAGATCTGGCGGCAGCGGCGGCAGCGGCGGCAGCC |
| RU-O-6824 | CGCGGGCTGCCGCCGCTGCCGCCGCTGCCGCCAGATCTGCCGCTGCCCGGG |
| RU-O-7234 | CCGGGCCTGAACGACATCTTCGAGGCCCAGAAGATCGAGTGGCACGAGG |
| RU-O-7235 | GATCCCTCGTGCCACTCGATCTTCTGGGCCTCGAAGATGTCGTTCAGGC |
pLenti/BirA was constructed in multiple steps. First, the E. coli BirA gene (GenBank accession no. M10123) was synthesized in human codon-optimized form by Codon Devices (Cambridge, MA). To efficiently translate and direct BirA to the secretory compartment, a consensus Kozak sequence and human codon-optimized signal sequence (amino acids 756 to 778 of yellow fever virus strain 17D; GenBank accession no. X03700) were incorporated upstream of BirA during gene synthesis. The synthetic BirA gene was subcloned into pLenti4 (Invitrogen) by using common BamHI and XhoI restriction sites.
The genotype 2a HCV construct, pJc1·MluI, was previously described (55). The amino-terminal, biotin acceptor peptide (AP)-tagged NS2 construct pJc1/NS2(AP) was constructed by inserting the annealed oligonucleotides YO-0315 and YO-0316 into the MluI restriction site of pJc1·MluI.
The pJc1/NS2(AP) Δcore construct was made by inserting a 1,103-bp EcoRI/BsiWI fragment from pJc1/Gluc2A Δcore (55) into common restriction sites of pJc1/NS2(AP). The E1-E2 deletion construct pJc1/NS2(AP) ΔE1-E2 was constructed from pJc1/NS2(AP) as previously described (24, 69). The naturally occurring E1-E2-p7 deletion, D88 (50), was constructed from pJc1·MluI by SDM with the primers YO-0527 and YO-0528. The biotin acceptor peptide sequence was then inserted into the MluI site of pJc1/D88 as described above to generate pJc1/NS2(AP) D88.
pJc1/E2(AP) was constructed via a multistep process. First, pBDL549 was created by subcloning a 1,142-bp Acc65I/SalI fragment from pFL-J6/JFH (38) into pSL1180 (GE Healthcare). pBDL549 was cut with BssHII and ligated to annealed oligonucleotides RU-O-6823 and RU-O-6824, yielding pBDL578. pBDL578 was cut with XmaI and BglII and ligated to annealed oligonucleotides RU-O-7234 and RU-O-7235. The resulting construct, pBDL578/bio, encoded the AP tag and a small flexible linker, (GGS)3, just upstream of E2. The 9,773-bp AvrII/Acc65II and 1,583-bp SphI/AvrII fragments of pFL-J6/JFH were then ligated to a 1,101-bp Acc65I/SphI fragment of pBDL578/bio, yielding pFL-J6/JFH/E2(AP). Finally, the 1,304-bp BsiWI/BsaBI fragment from pFL-J6/JFH/E2(AP) was subcloned into pJc1.
pJc1/NS2(AP) E2-IRES-p7 was constructed via a multistep process. First, the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) was amplified from pYSGR-JFH (55) by using Phusion DNA polymerase (New England Biolabs, Ipswich, MA) and oligonucleotides YO-0743 and YO-0744. This 657-bp amplicon (PCR1) encoded the C-terminal 11 amino acids of E2 (including a unique BsaBI site), a stop codon, a PmeI site, and the EMCV IRES driving expression of a synthetic signal sequence (SSS) derived from human alpha1 anti-trypsin. The IRES-SSS-p7 junction was created by amplification of pJc1/AP-NS2 by using oligonucleotides YO-0745 and YO-0361, yielding the 688-bp PCR2. PCR1 and PCR2 were cross-primed and amplified by using primers YO-0743 and YO-0361, yielding PCR3, which was TA cloned into pCR2.1-TOPO (Invitrogen) and sequenced. When it was subsequently determined that the SSS failed to drive proper p7-NS5B expression in a full-length construct (data not shown), the SSS was removed from the pCR2.1/PCR3 intermediate via SDM with oligonucleotides YO-0764 and YO-0765. The E2-IRES-p7 junction was then subcloned into pJc1/NS2(AP) by using common BsaBI and MluI sites.
pJc1/NS2(AP) NS2-IRES-NS3 was constructed via a multistep process. First, a stop codon and PmeI site were introduced downstream of the NS2 gene via SDM with oligonucleotides YO-0750 and YO-0751. The EMCV IRES-NS3 junction was then subcloned into this intermediate from pYSGR-JFH1 (55) by using common PmeI and SpeI sites.
pJc1/NS2(AP) Δp7 was constructed via a multistep process. First, the 5′ end of the NS2(AP) gene was fused to the EMCV IRES and SSS via cross-priming of oligonucleotides YO-0746 and YO-0747. The 119-bp product, PCR4, was cross-primed with PCR1 and amplified by using primers YO-0743 and YO-0747 to yield a 771-bp product, PCR5, which was then TA cloned into pCR2.1-TOPO and sequenced. As before, the SSS was subsequently removed via SDM with oligonucleotides YO-0766 and YO-0767. The final product encoded the 3′ end of the E2 gene, a stop codon, and the EMCV IRES driving NS2(AP) expression. This insert was then subcloned as a 753-bp fragment into pJc1/NS2(AP) by using common BsaBI and MluI sites.
The E1 A4 epitope was introduced into pJc1·MluI via SDM with primers YO-0521 and YO-0522 to generate pJc1/E1(A4). The biotin acceptor peptide was then inserted into the MluI site of pJc1/E1(A4) as described above to generate pJc1/E1(A4)+NS2(AP). pJc1/E1(A4)+NS2(AP) constructs containing individual NS2 point mutations (K27A, W35A, Y39A, E45K, K81A, and P89A) were constructed by inserting the 1,321-bp MluI/SpeI fragment of pJc1/Gluc2A NS2 mutant constructs (55) into pJc1/E1(A4) via common restriction sites.
To facilitate SDM of the glycoprotein transmembrane domains, truncated forms of pJc1/NS2(AP) and pJc1/E2(AP) were constructed by religating the 5.6-kb fragments generated from digestion with XbaI and SpeI. The individual E1 K179Q and E2 D349W heterodimerization mutations were generated via SDM with primers YO-0603 and YO-0604 and primers YO-0605 and YO-0657, respectively. The E1 K179Q+E2 D349W double mutant was constructed via SDM by using the E1 K179Q construct as a template and primers YO-0605 and YO-0657. The individual and double mutations were then inserted into full-length pJc1/E1(A4)+NS2(AP) and pJc1/E1(A4)+E2(AP) by inserting the 1,698-bp BsiWI/NotI digestion fragment of the truncated plasmids into the same sites of the full-length constructs.
RNA in vitro transcription and transfection.
HCV expression constructs were linearized overnight with XbaI, and the 5′ overhang was removed by digestion with mung bean nuclease (New England Biolabs). The reaction mixtures were further digested with proteinase K, purified by phenol-chloroform extraction and ethanol precipitation, and resuspended at a concentration of 1 μg/μl in TE buffer (10 mM Tris-HCl [pH 7.4], 0.1 mM EDTA). Linearized plasmids (2 μg) were added to in vitro transcription mixtures (20 mM Tris-HCl [pH 7.5], 5 mM NaCl, 9 mM MgCl2, 3 mM [each] ATP, CTP, GTP, and UTP, 1 mM dithiothreitol [DTT], 12 units SUPERasin [Ambion, Alamo, CA], and 20 units T7 RNA polymerase [Epicentre, Madison, WI]) and incubated at 37°C for 1.5 h. HCV RNAs were purified by using RNeasy minikits (Qiagen, Valencia, CA) by following the manufacturer's instructions and stored in RNA storage solution (2 mM sodium citrate, pH 6.4) at −80°C.
For RNA transfections, cells were washed once with phosphate-buffered saline (PBS), trypsinized, and harvested into fresh culture medium. Cells were then washed twice with ice-cold PBS, resuspended at a concentration of 2 × 107 cells/ml in ice-cold PBS, and mixed (0.4 ml of cells) with 1 μg in vitro-transcribed RNA. The cell-RNA mix was placed in a 2-mm-gap electroporation cuvette (BTX Genetronics, San Diego, CA) and electroporated with five pulses at 820 V for 99 μs with a pause of 1.1 s between each pulse. Cells were allowed to recover for 10 min, added to 10 ml of complete culture medium, and plated on a 10-cm culture dish.
Infectivity measurements.
Viral infectivity was measured by using a standard endpoint dilution assay (37). In brief, virus-containing supernatant was collected at the indicated time points, centrifuged at 3,000 × g for 10 min, and filtered through a 45-μm-pore-size filter. Viral supernatants were then serially diluted into growth media and incubated with naïve Huh-7.5 cells (8 × 104 cells/ml) in a 96-well plate format. After 3 days, the media were removed, and cells were fixed with ice-cold methanol and stained for NS5A by using the NS5A-specific monoclonal antibody 9E10 (38). The 50% endpoint was calculated by using the method of Reed and Muench (57).
Affinity precipitation and Western blotting.
At 48 h posttransfection, cells were washed once with PBS, trypsinized, and harvested into complete culture medium. Cells were pelleted by centrifugation at 335 × g for 5 min, washed twice with ice-cold PBS, resuspended in lysis buffer (Tris-buffered saline [TBS; 20 mM Tris-HCl, pH 7.4, 150 mM NaCl] containing 1 mM EDTA, 1% Triton X-100, and complete protease inhibitor cocktail [Roche, Indianapolis, IN]), and incubated for 1 h on ice. Lysates were clarified by centrifugation at 10,000) × g for 5 min, and the supernatant was incubated for 1 h with 50 μl of Dynabeads M-280 streptavidin beads (Invitrogen, Carlsbad, CA). Beads were washed in lysis buffer and resuspended in protein sample buffer (50 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate [SDS], 0.1% bromophenol blue, 10% glycerol, 100 mM DTT). Samples were heated at 95°C for 10 min, centrifuged at 10,000 × g for 5 min, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were blocked in 5% nonfat milk and incubated with primary monoclonal antibodies to core (C7-50; Affinity Bioreagents, Golden, CO), E1 (A4 [11]; a kind gift from Jean Dubuisson), E2 (3/11 [14]; a kind gift from Jane McKeating), NS2 (6/H6 [10]; a kind gift from Charles M. Rice), NS3 (9G2; Virogen, Watertown, MA), and NS5A (9E10 [38]). Membranes were washed extensively with TBS with 1% Tween 20, incubated with secondary IgG-horseradish peroxidase (HRP) antibodies, and developed using the SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL). Biotinylated proteins were detected using the Vectastain Elite kit (Vector Laboratories, Burlingame, CA) by following the manufacturer's instructions. To examine E2 glycosylation, protein samples were prepared in 1× glycoprotein denaturing buffer (New England Biolabs), split into three equal portions, and treated for 2.5 h at 37°C with 500 U peptide:N-glycosidase F (PNGase F) or endoglycosidase H (Endo H) as per the manufacturer's instructions (New England Biolabs), or left untreated. Untreated samples were incubated in parallel with the buffer conditions used for PNGase F. Samples were denatured in SDS sample buffer and analyzed by Western blotting as described above.
Preparation of microsomal membranes.
Huh-7.5 and Huh-7.5(BirA) cells were transfected as described above, washed once with PBS, trypsinized, and harvested into complete media. Cells were pelleted by centrifugation at 335 × g for 5 min, washed twice with ice-cold PBS, resuspended in five volumes of cell homogenization buffer (20 mM Tris-HCl [pH 7.4], 1 mM EDTA, and complete protease inhibitor cocktail), and incubated on ice for 30 min. Cells were placed into a 45-ml nitrogen cavitation bomb (Parr Instrument Company, Moline, IL) and equilibrated with 500 lb/in2 of nitrogen gas for 15 min. Cells were lysed through nitrogen decompression, and lysates were centrifuged at 500 × g for 5 min to remove debris. Mitochondria were removed by centrifugation at 12,000 × g for 10 min, and the corresponding microsome-containing supernatant was centrifuged at 100,000 × g for 2 h to obtain the microsome-enriched pellet. The microsomal pellet was resuspended in storage buffer (20 mM Tris-HCl [pH 7.4], 1 mM EDTA, 250 mM sucrose), and the total protein content was determined by bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL) by following the manufacturer's instructions. Microsomes were adjusted to a concentration of 10 μg total protein/μl in storage buffer and stored at −80°C. Microsomes were enriched in viral proteins and contained viral RNA-dependent RNA polymerase activity (T. Kazakov, K. A. Stapleford, and B. D. Lindenbach, unpublished data). Furthermore, E2 and NS3 could be cocaptured with NS2(AP) from microsomes prepared in parallel from Jc1/NS2(AP)-infected Huh-7.5 cells (data not shown), confirming that NS2 complexes were membrane associated.
Blue native polyacrylamide gel electrophoresis (BN-PAGE).
Microsomal fractions (100 μg total protein) were thawed, solubilized in 100 μl of BN-PAGE buffer (20 mM Tris-HCl [pH 7.4], 50 mM NaCl, 1 mM EDTA, 10% glycerol, protease inhibitor cocktail, and 0.5% digitonin [Invitrogen]) for 1 h on ice, and centrifuged at 16,000 × g for 10 min to remove unsolubilized material. Coomassie brilliant blue G-250 (Invitrogen) was added to the clarified supernatant (final detergent concentration, 0.25% [vol/vol]), and protein complexes were separated on precast 4 to 16% Bis-Tris native gels (Invitrogen) per the manufacturer's instructions. Following electrophoresis, gels were incubated in SDS-PAGE buffer for 20 min and transferred to PVDF at 25 V for 3 h at 4°C. PVDF membranes were washed several times with methanol, and Western blotting was performed as previously described. Pilot experiments were used to optimize BN-PAGE conditions. Parameters that were varied included protein-detergent ratio, choice of detergent, and detergent supplier: digitonin (Invitrogen versus Calbiochem), Triton X-100 (Invitrogen), n-dodecyl-β-d-maltoside (Invitrogen versus Anatrace [Maumee, OH]), n-decyl-β-d-maltopyranoside (Anatrace), CHAPS (Anatrace), Cymal-5 (Anatrace), fos-choline-12 (Anatrace), and n-octyl-β-d-gycopyranoside (Anatrace). Most conditions (including Triton X-100) yielded high-molecular-mass NS2 complexes. However, digitonin gave superior band resolution and was therefore used for subsequent studies.
For two-dimensional BN-PAGE/SDS-PAGE (2D BN/SDS-PAGE) analysis, solubilized membrane fractions were separated by BN-PAGE as described above, and lanes were cut out of the gel and incubated in 50 mM Tris-HCl (pH 6.8), 2% SDS, and 1% β-mercaptoethanol for 15 min. Gel slices were then placed on top of a 10% SDS-PAGE gel, and proteins were separated, transferred, and immunoblotted as previously described.
RESULTS
A system to affinity capture functional NS2 from virus-producing cells.
Based on our prior genetic analysis (55), we hypothesized that NS2 contributes to virus particle assembly through physical interactions with E1, E2, NS3, and NS4A. To address this hypothesis, we used an affinity purification strategy to capture NS2 and its associated proteins. We took advantage of the efficient and high-affinity interaction between biotin and streptavidin for protein isolation (5, 29, 46, 52), and engineered viral constructs containing a biotin acceptor peptide (AP), GLNDIFEAQKIEWH (5), inserted at the amino terminus of HCV NS2 (Fig. 1B). When expressed in the presence of E. coli biotin ligase BirA, the AP is biotinylated on its lysine residue (Fig. 1B) and thus can be captured by streptavidin beads. Importantly, the insertion of this peptide into NS2 did not affect viral replication or production of infectious virus particles compared to results for the untagged strain Jc1 (Fig. 1C).
To biotinylate AP-tagged NS2 in virus-producing cells, we generated a stable cell line, Huh-7.5(BirA), that expresses a codon-optimized, secreted form of BirA (46). No differences in virus replication or infectious virus production were detected between Huh-7.5 and Huh-7.5(BirA) cells (data not shown), indicating that expression of BirA had no detectable effect on virus production. Furthermore, the AP-tagged construct produced intracellular protein levels similar to those of the untagged strain Jc1, and upon expression of these constructs in Huh-7.5(BirA) cells, NS2(AP) was specifically biotinylated, whereas untagged NS2 was not (Fig. 1D).
NS2 physically associates with E2 and NS3 in virus-producing cells.
We used the BirA/AP-tag system to determine whether NS2 physically interacts with other HCV proteins in virus-producing cells. NS2(AP) was specifically captured on streptavidin magnetic beads from Huh-7.5(BirA) cell lysates (Fig. 2 A, lane 4). Unless otherwise noted, all AP capture experiments were conducted by using lysates made during the peak of virus production, 48 h postelectroporation or postinfection. The specificity of biotinylated NS2(AP) capture was confirmed by the lack of untagged NS2 capture from Huh-7.5(BirA) cells (Fig. 2A, lane 3) and the lack of NS2(AP) capture from Huh-7.5 cells that did not express BirA (not shown). Notably, E2 and NS3 were specifically cocaptured with NS2(AP) (Fig. 2A, lane 4) but not with untagged NS2 expressed by Jc1 (Fig. 2A, lane 3). Furthermore, we did not detect stable interactions between NS2(AP) and core or NS5A (Fig. 2A, lane 4) or between NS2(AP) and NS5B (not shown). These data demonstrate that NS2 physically interacts with E2 and NS3 in lysates from virus-producing cells.
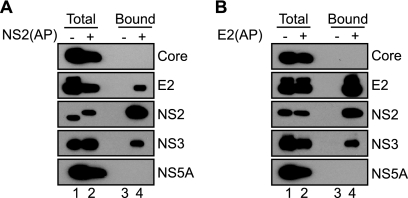
FIG. 2.
HCV NS2 interacts with E2 and NS3 in virus-producing cells. (A) Specific affinity purification of NS2(AP)-containing complexes from cells transfected with Jc1 or Jc1/NS2(AP), as indicated. Biotinylated proteins were captured on streptavidin beads and eluted into SDS-PAGE sample buffer, as described in Materials and Methods. Proteins present in lysates or eluates from streptavidin beads were separated by SDS-PAGE, transferred to PVDF membranes, and immunoblotted for the indicated viral proteins as described in Materials and Methods. “Total” indicates proteins in the clarified lysates; “Bound” indicates proteins captured by strepavidin beads; minus signs indicate untagged Jc1; plus signs Jc1/NS2(AP). (B) Specific affinity purification of E2(AP)-containing complexes from cells transfected with Jc1 or Jc1/E2(AP), as indicated. Samples were prepared and analyzed as in panel A.
To determine whether the interaction between NS2 and E2 was reciprocal and whether E2, NS2, and NS3 are in a large complex, we generated a Jc1 variant encoding the biotin AP at the amino terminus of E2. This construct replicated and produced infectious virus, but peak viral titers were ≈10-fold lower than for Jc1 (data not shown); given the position of the E2(AP) insertion, we hypothesize that this may be due to a defect in viral entry. Nevertheless, Jc1/E2(AP) expressed levels of viral proteins similar to those for Jc1 (Fig. 2B, lanes 1 and 2). Notably, NS2 and NS3 were specifically cocaptured with E2(AP) but not with untagged E2 (Fig. 2B, lanes 3 and 4). As seen for NS2(AP), stable interactions were not detected between E2(AP) and core or NS5A (Fig. 2B, lane 4). These data confirmed the interaction between E2, NS2, and NS3 and suggested that they form a large complex.
E2, NS2, and NS3 form discrete, membrane-associated high-molecular-mass complexes.
To independently confirm that NS2, E2, and NS3 form higher-order complexes and to estimate the sizes of these complexes, we used BN-PAGE. BN-PAGE is a useful method to separate native protein complexes based on their size and has been widely used to isolate enzymatically active mitochondrial electron transport proteins and other membrane protein complexes (48, 58, 59, 70, 72). First, BN-PAGE conditions were optimized as described in Materials and Methods by using microsomal membrane fractions isolated from Jc1-infected Huh-7.5 cells. Microsomal protein complexes were gently solubilized in digitonin, separated via BN-PAGE, transferred to PVDF membranes, and immunoblotted. Under these conditions, E2, NS2, and NS3 were solubilized and could be resolved by BN-PAGE, while core and NS5A were poorly solubilized and remained in the membrane pellet (not shown). E2, NS2, and NS3 gave reproducible patterns of multiple, discrete bands (Fig. 3 A). Some bands were specific for a given protein (Fig. 3A, open arrowheads). For instance, a small amount of NS2 migrated as a ≈40-kDa band, which is consistent with the native NS2 homodimer (43). However, E2, NS2, and NS3 were most abundantly present in two bands that comigrated at ≈680 kDa and ≈520 kDa (Fig. 3A, closed arrowheads). These data support the interaction of HCV E2, NS2, and NS3 proteins in virus-producing cells and suggest that they form at least two distinct high-molecular-mass complexes.
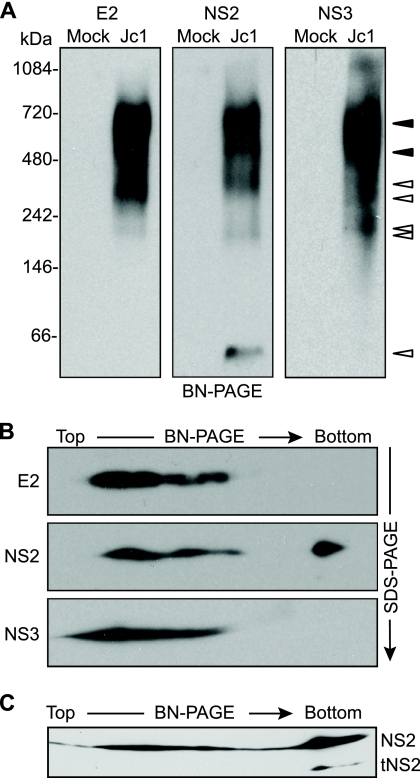
FIG. 3.
E2, NS2, and NS3 form discrete, membrane-associated high-molecular-mass complexes. (A) BN-PAGE separation of E2-, NS2-, and NS3-containing protein complexes. Microsomes were prepared at 48 h postelectroporation of Huh-7.5 cells with Jc1 RNA or cells transfected without RNA (Mock). Proteins were solubilized in 0.5% (vol/vol) digitonin, separated under native conditions on a 4 to 16% Bis-Tris polyacrylamide gel, and transferred to PVDF. The blot was cut into three strips and immunoblotted for E2, NS2, and NS3. Open arrowheads indicate bands that were specific for a given protein; closed arrowheads indicate bands in which E2, NS2, and NS3 comigrated (see Results). (B) 2D BN/SDS-PAGE analysis of E2-, NS2-, and NS3-containing protein complexes. Microsome-associated protein (20 μg) was solubilized in digitonin and separated in the first dimension by BN-PAGE and in the second dimension on a 10% SDS-PAGE gel. E2, NS2, and NS3 were detected by immunoblotting. (C) Microsome-associated protein (40 μg) was solubilized in digitonin and separated in the first dimension by BN-PAGE and in the second dimension on a 12% SDS-PAGE gel. NS2 was detected by immunoblotting.
To further confirm the identity of viral proteins identified by BN-PAGE and to determine whether processing intermediates or other forms of these proteins were present in large protein complexes, we used two-dimensional BN-PAGE/SDS-PAGE (2D BN/SDS-PAGE) (26, 61). To do this, BN-PAGE lanes were excised, rotated 90°, and electrophoresed by denaturing SDS-PAGE, followed by transfer to PVDF and immunoblotting. This approach confirmed that denatured E2, NS2, and NS3 migrated with apparent molecular masses of 70 kDa, 24 kDa, and 68 kDa, respectively (Fig. 3B). By loading more protein and by using an SDS-PAGE gel with a higher percentage of polyacrylamide in the second dimension, a 17-kDa form of NS2 was observed in the ≈40-kDa native band but not in the high-molecular-mass native bands (Fig. 3C). A similar, truncated form of NS2 was previously identified as tNS2 (23). This suggested that tNS2 may be excluded from high-molecular-mass E2-NS2-NS3 complexes.
Characterizing interactions between NS2 and the glycoprotein complex.
Since the HCV glycoproteins form a heterodimeric complex, NS2 likely also interacts with E1. However, we were unable to address this directly because the E1-specific antibodies available to us did not react with E1 protein from the J6 strain. We therefore introduced the E1 epitope A4 from HCV strain H77 (11) into Jc1, Jc1/NS2(AP), and Jc1/E2(AP). These constructs replicated and produced infectious virus, although peak viral titers were reduced ≈40-fold compared to those in unmodified Jc1 (data not shown). Nevertheless, E1 was expressed in RNA-transfected Huh-7.5(BirA) cells (Fig. 4 A, lanes 1 to 5) and specifically cocaptured with NS2(AP), E2, and NS3 (Fig. 4A, lanes 6 and 7). These data confirmed that E1 is part of the E2-NS2-NS3 complex.
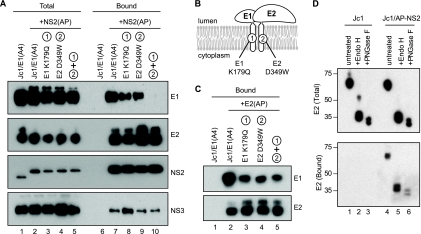
FIG. 4.
Characterization of the NS2-associated glycoproteins. (A) NS2 interacts with E1-E2 through E2. NS2(AP) was affinity captured from Huh-7.5(BirA) cell lysates at 48 h postelectroporation with the indicated constructs and analyzed as in Fig. 2A. All constructs encoded the A4 epitope in the E1 protein; the indicated constructs also encoded AP-tagged NS2, with or without mutations in E1 and/or E2 that blocked E1-E2 heterodimerization. (B) Schematic of glycoprotein heterodimerization mutations. (C) E1 K179Q and E2 D349W mutations inhibit E1-E2 heterodimerization. E2(AP) was affinity captured from Huh-7.5(BirA) cell lysates at 48 h postelectroporation with the indicated constructs and analyzed as in Fig. 2A. All constructs encoded the A4 epitope in the E1 protein; the indicated constructs also encoded AP-tagged E2; the indicated constructs also contained mutations in E1 and/or E2 that blocked E1-E2 heterodimerization. (D) NS2-interacting E2 contains high-mannose glycans. (Top) Cell lysates from Jc1- or Jc1/NS2(AP)-transfected Huh-7.5(BirA) cells were treated with Endo H, PNGase F, or left untreated. Proteins were separated by SDS-PAGE, transferred to a PVDF membrane, and immunoblotted for E2. (Bottom) NS2(AP)-associated E2 was cocaptured on streptavidin beads as in Fig. 2A, treated with Endo H, PNGase F, or left untreated, separated by SDS-PAGE, transferred to a PVDF membrane, and detected by immunoblotting.
We hypothesized that NS2 could interact with the glycoproteins via direct or indirect interactions with E1 or E2 or may be specific for heterodimeric E1-E2. To discern between these possibilities, we took advantage of mutations in the transmembrane domains of E1 and E2 (Fig. 4B) that inhibit the formation of functional E1-E2 heterodimers (9). These mutations were introduced individually or in combination in the context of Jc1/E1(A4)+NS2(AP). In addition, to confirm that these mutations inhibited E1-E2 heterodimer formation, we also introduced them into Jc1/E1(A4)+E2(AP). As shown in Fig. 4C, E1 was specifically cocaptured with E2(AP) but not with untagged E2 (lanes 1 and 2), consistent with the formation of E1-E2 heterodimers. The E1 K179Q and E2 D349W mutations, alone or in combination, decreased the amount of E1 that was cocaptured with E2(AP), confirming that these mutations inhibited heterodimer formation (Fig. 4C, lanes 3 to 5). In the context of Jc1/E1(A4)+NS2(AP), these mutations progressively reduced the amount of E1 that was cocaptured by NS2(AP) but had no effect on the amount of E2 or NS3 that was cocaptured by NS2(AP) (Fig. 4A, lanes 7 to 10). Taken together, these data suggest that the viral glycoproteins are incorporated into the NS2 complex via interactions with E2.
Next, we examined the glycosylation status of the viral glycoproteins in the NS2 complex. The HCV glycoproteins are retained within the ER and contain high-mannose N-linked glycans that are modified to complex glycans during transit of virus particles through the secretory pathway (11, 68). Therefore, the glycosylation status of the viral glycoproteins can be used as a marker for their localization within the cell. If the interactions between NS2 and the viral glycoproteins occur early in the secretory pathway, the NS2-associated form of E2 should be sensitive to treatment with either Endo H or PNGase F, whereas interactions that occur late in the secretory pathway should contain E2 that is resistant to Endo H but sensitive to PNGase F. As shown in the top panel of Fig. 4D, nearly all of the E2 present in Huh-7.5(BirA) cell lysates was sensitive to both Endo H and PNGase F. Endo H digestion produced a doublet of bands at around 34 kDa and 40 kDa, consistent with complete cleavage within the 11 N-acetylglucosamine disaccharide cores on E2 and E2-p7 (34, 49). Similarly, PNGase F digestion produced a slightly faster-migrating pair of bands, consistent with the complete removal of glycans from E2 and E2-p7. After affinity purifying NS2(AP) protein complexes from Huh-7.5(BirA) cell lysates and treating them with Endo H or PNGase F, we found that NS2-associated E2 was sensitive to both Endo H and PNGase F (Fig. 4D, bottom panel, lanes 5 and 6). Interestingly, both E2 and the putative E2-p7 were associated with NS2. Similarly, NS2-associated E1(A4) was also sensitive to Endo H- and PNGase F-digestion (data not shown). These data indicate that NS2 interacts with the glycoprotein complex in an early (i.e., pre-Golgi) compartment within the secretory pathway and that E2-p7 may be part of the NS2 complex.
NS2 complex formation is enhanced by cotranslation as a polyprotein.
Incomplete or delayed signal peptidase cleavage of the HCV polyprotein can lead to the formation of E2-p7 and E2-p7-NS2 (Fig. 1A) (34, 49, 56, 63). In addition, NS2 and NS3 must interact prior to NS2/3 cleavage to form the NS2-3 cysteine autoprotease (19, 20, 62). To determine whether these processing intermediates are required for NS2 complex assembly, we ablated their formation by inserting a stop codon and encephalomyocarditis virus (ECMV) internal ribosomal entry site (IRES) between E2-p7 and NS2-NS3 (Fig. 5 A). As previously described for similar bicistronic constructs (23, 24), these genomes replicated and produced normal levels of viral proteins (Fig. 5B, lanes 1 to 4) and viral RNA (Fig. 5C). Interestingly, the E2-IRES-p7 construct had reduced levels of NS2-E2 interaction but normal levels of NS2-NS3 interaction compared to those of Jc1/NS2(AP) (Fig. 5B, lanes 6 and 7). In contrast, the NS2-IRES-NS3 construct had reduced levels of NS2-NS3 interaction but normal levels of NS2-E2 interaction (Fig. 5B, lanes 6 and 8). Taken together, these data suggest that NS2 can interact with E2 and NS3 in trans but that formation of these interactions was enhanced by cotranslation within the viral polyprotein.
FIG. 5.
NS2 complex formation is enhanced by cotranslation as a polyprotein. (A) Schematic of bicistronic HCV constructs containing insertions of a stop codon and ECMV IRES element (indicated by E·I). Protein cleavage sites are labeled as in Fig. 1A; the AP tag is indicated by a black box. (B) Affinity purification of NS2(AP)-containing complexes. Huh-7.5(BirA) cells were transfected with the indicated constructs and samples were prepared and analyzed as in Fig. 2A. (C) Bicistronic HCV constructs replicate efficiently. Huh-7.5(BirA) cells were split at 48 h posttransfection with the indicated viral RNAs, reseeded, and allowed to grow for an additional 48 h. Total cellular RNAs were extracted, and the level of HCV RNA was quantified by quantitative reverse transcription (qRT)-PCR as previously described (37). Jc1 GNN was a replication-defective Jc1 genome containing inactivating mutations in the NS5B RNA polymerase active site. The y axis represents copies of HCV genome per 10 ng of total RNA. (D) Bicistronic HCVcc constructs produced reduced viral titers. Media were collected from Huh-7.5(BirA) cells at 48 h posttransfection with the indicated viral RNAs, and titers were determined on Huh-7.5 cells. Values represent averages of results from three independent experiments; error bars represent the standard deviations from the means.
Both E2-IRES-p7 and NS2-IRES-NS3 produced infectious virus particles, although viral titers were 50- to 100-fold lower than those of wild-type (WT) Jc1 or Jc1/NS2(AP) at 48 h posttransfection (Fig. 5D). The reduced viral titers were not attributable to reduced RNA replication (Fig. 5C) but did correlate with the decreased levels of NS2-E2 and NS2-NS3 interaction.
HCV p7 is essential for NS2 complex assembly.
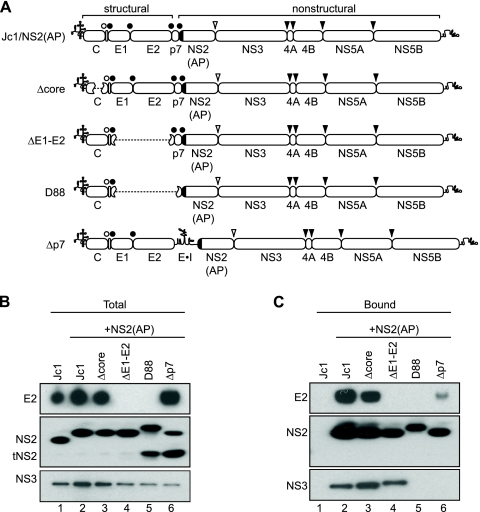
We next examined whether the HCV core, E1, E2, or p7 proteins are required for NS2 complex assembly by deleting them individually or in combination from Jc1/NS2(AP). The Δcore construct contained an in-frame deletion of Jc1 codons 62 to 150 (55), the ΔE1-E2 construct contained an in-frame deletion of E1-E2 (Jc1 codons 217 to 566 [24, 69]), and the D88 construct corresponded to a naturally occurring in-frame deletion of Jc1 codons 195 to 807 (50) (Fig. 6 A). In-frame deletion of p7 alone was previously shown to cause defects in polyprotein processing, yielding reduced levels of NS2 and accumulation of an E2-NS2 intermediate (7, 64). To avoid this, the Δp7 construct was created by inserting a stop codon and EMCV IRES between E2 and NS2(AP) (Fig. 6A). All of these deletion constructs replicated and expressed similar levels of viral proteins (Fig. 6B). Consistent with previous results (55, 64, 69), none of the deletion constructs produced infectious virus (data not shown). It was notable that D88 gave a slower-migrating form of NS2, likely due to the N-terminal extension of three residual amino acids from E1 and four amino acids from p7 (Fig. 6B, lane 5). Interestingly, both D88 and Δp7 gave rise to high levels of the 17-kDa truncated form of NS2, tNS2 (Fig. 6B, lanes 5 and 6).
FIG. 6.
HCV p7 is essential for NS2 complex assembly. (A) Schematic of deletion constructs. HCV genomes are annotated as in Fig. 5A. In-frame deletions are indicated by dashed lines. (B) Expression of E2, NS2, and NS3. Cellular lysates were prepared at 48 h posttransfection with the indicated constructs and analyzed by SDS-PAGE and immunoblotting with antibodies specific for E2, NS2, or NS3. (C) Affinity purification of NS2(AP)-associated proteins from deletion constructs. NS2(AP) was affinity captured from the lysates in panel A and analyzed as in Fig. 2A.
We affinity purified AP-containing protein complexes from the deletion mutants and probed for E2, NS2, and NS3 by Western blotting (Fig. 6C). Deletion of core had no discernible effect on the interaction of NS2 with E2 or NS3 (Fig. 6C, lane 3). Naturally, the ΔE1-E2 or D88 constructs lacked NS2-E2 interaction (Fig. 6C, lanes 4 and 5). While the ΔE1-E2 deletion had no effect on the interaction between NS2 and NS3 (Fig. 6C, lane 4), the D88 deletion completely disrupted the NS2-NS3 interaction (Fig. 6C, lane 5). These results suggested that p7 is necessary for NS2-NS3 interaction. Consistent with this, the Δp7 deletion disrupted NS2-NS3 interaction and inhibited NS2-E2 interaction (Fig. 6C, lane 6). The latter effect may in part be due to the bicistronic configuration (compare Fig. 5B, lane 7, and Fig. 6C, lane 6). In addition, tNS2 expressed in the D88 and Δp7 constructs was excluded from the NS2-containing complexes. These data indicate that the interaction between NS2 and NS3 is independent of core, E1, and E2 expression but dependent on p7 expression. Furthermore, p7 may contribute to the NS2-E2 interaction and help to stabilize NS2 and/or prevent its conversion into tNS2.
NS2 mutations that disrupt virus assembly interfere with NS2 complex assembly.
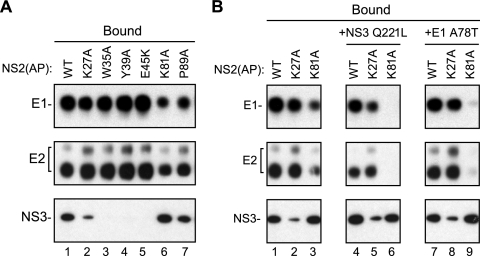
Our original hypothesis that NS2 interacts with the E1-E2 glycoprotein and NS3-4A enzyme complexes arose from our prior identification of second-site mutations in E1, E2, NS3, and NS4A that suppressed virus assembly defects caused by mutations in NS2 (55). We were therefore interested to know whether these NS2 mutations disrupted NS2 protein complexes and, if so, whether second-site suppressor mutations would restore these interactions. Six NS2 mutations that caused severe defects in virus assembly were introduced into Jc1/E1(A4)+NS2(AP), and the relative levels of NS2(AP)-associated E1, E2, and NS3 proteins were determined as above. As shown in Fig. 7 A, the NS2 K27A, W35A, Y39A, and E45K mutations disrupted interaction between NS2 and NS3 but had little effect on NS2-E1 or NS2-E2 interactions (Fig. 7A, lanes 1 to 5). In contrast, the NS2 K81A and P89A mutations partially inhibited NS2-E1 and NS2-E2 interactions but had no effect on NS2-NS3 interaction (Fig. 7A, lanes 6 and 7). These results indicate that specific NS2 mutations known to cause defects in virus assembly can preferentially inhibit interactions between NS2 and the E1-E2 glycoprotein or NS3-4A enzyme complexes.
FIG. 7.
Mutations in NS2 inhibit NS2 protein-protein interactions. (A) NS2(AP) was affinity captured from cell lysates at 48 h postelectroporation of Huh-7.5(BirA) cells with Jc1/NS2(AP), either with or without (WT) the indicated mutations in NS2. NS2(AP)-associated proteins were detected as in Fig. 2A. (B) Affinity purification of NS2(AP)-associated proteins with (K27A, K81A) or without (WT) mutations in NS2 and with (+NS3 Q221L, +E1 A78T) or without second-site suppressor mutations. NS2(AP)-associated proteins were detected as in Fig. 2A. This panel is a composite created from a single exposure; it is representative of additional experiments.
We previously showed that the defect in virus particle assembly of the NS2 K27A mutant can be suppressed by the second-site mutation NS3 Q221L, while the assembly defect of the NS2 K81A mutant can be suppressed by the E1 A78T second-site mutation and, curiously, is further inhibited by NS3 Q221L (55). We therefore tested whether these second-site mutations could restore (or inhibit) interactions within the NS2 complex. In the WT and NS2 K27A genetic background, the NS3 Q221L mutation slightly inhibited interaction of NS2 with the viral glycoproteins and slightly enhanced the NS2-NS3 interaction (Fig. 7B, compare lanes 1 and 2 to lanes 4 and 5). In the NS2 K81A genetic background, the NS3 Q221L mutation disrupted the NS2-glycoprotein interactions but had no effect on NS2-NS3 interaction (Fig. 7B, compare lanes 3 and 6). In contrast, the E1 A78T mutation had little effect on WT or K27A NS2 interactions but in the NS2 K81A genetic background further inhibited interactions between NS2 and the viral glycoproteins (Fig. 7B, compare lanes 1 to 3 to lanes 7 to 9). These data indicate that these second-site suppressor mutations do not restore NS2 interactions, and in some cases can inhibit them.
NS2 coordinates interaction between the E1-E2 glycoprotein and NS3-4A enzyme complexes.
The above data indicated that E1-E2 can interact with NS3-4A, possibly through interaction with NS2. To determine whether NS2 plays an essential role in mediating this interaction, we deleted NS2 by inserting a stop codon and EMCV IRES between p7 and NS3 (Fig. 8 A). This construct replicated and expressed normal levels of NS3 (Fig. 8B, lanes 1 to 3) and E2 (not shown). However, NS3 did not interact with E2(AP) in this context (Fig. 8B, lane 6), indicating that NS2 is essential for this interaction.
FIG. 8.
NS2 plays a central role in coordinating interaction between the E1-E2 glycoprotein and NS3-4A enzyme complexes. (A) Structure of Jc1/E2(AP) and ΔNS2. Constructs are labeled as in Fig. 5A. (B) NS2 is required for E2-NS3 interaction. Protein lysates were prepared from Huh-7.5(BirA) cells transfected with the indicated constructs at 48 h postelectroporation. NS3 present in lysates (Total) or eluates from streptavidin beads (Bound) was separated by SDS-PAGE, transferred to PVDF membranes, and detected by immunoblotting. (C) NS2 mutations inhibit E2-NS3 interaction. Huh-7.5(BirA) cells were transfected with Jc1/E2(AP) or Jc1/E2(AP) containing the indicated NS2 mutations, and lysates were prepared 48 h postelectroporation. E2(AP)-associated NS3 protein (Bound) was detected as in Fig. 2B and 8A.
Our earlier results indicated that bicistronic constructs had reduced protein-protein interactions (Fig. 5), which could have contributed to the lack of E2-NS3 interaction in the ΔNS2 construct. Therefore, to better clarify the role of NS2 in coordinating E1-E2 and NS3-4A interaction, we introduced NS2 mutations that inhibit NS2-E2 or NS2-NS3 interactions into Jc1/E1(A4)+E2(AP). Consistent with our analysis of NS2-NS3 interaction (Fig. 7), the NS2 K27A mutation partially inhibited the interaction between E2(AP) and NS3, while the NS2 W35A, Y39A, and E45K mutations completely blocked the interaction between E2(AP) and NS3 (Fig. 8C, lanes 2 to 5). Similarly, consistent with our analysis of E2-NS2 interaction (Fig. 7), the NS2 K81A and P89A mutations strongly inhibited the interaction between E2(AP) and NS3 (Fig. 8C, lanes 7 and 8). All NS2 mutants replicated efficiently and expressed normal levels of E2, NS2, and NS3 (data not shown). These data confirm that NS2 plays an essential role in coordinating the interaction between the E1-E2 glycoprotein and NS3-4A enzyme complexes.
DISCUSSION
The role of NS2 in HCV particle assembly is incompletely understood, in part due to a lack of biochemical systems to study the function of NS2 in the context of the viral life cycle. In this study we developed an efficient affinity purification system to specifically capture NS2 and its associated proteins from genotype 2a virus-producing cells. By using this system, we showed that NS2 physically interacts with the viral E1, E2, and NS3 proteins. In contrast, we did not detect stable interaction of NS2 with either core or NS5A proteins. Due to a lack of immunological detection reagents, we were unable to determine whether the viral p7 or NS4A proteins are part of the NS2 complex. Nevertheless, it appeared that E2-p7 was part of this complex (Fig. 4D), and p7 was essential for NS2 complex formation (Fig. 6). Consistent with this, Ma and colleagues recently showed that NS2 interacts with an epitope-tagged form of p7 during the replication of a genotype 1a/2a chimeric virus and that p7 is required for the interaction of NS2 with E1, E2, and NS3 (44). Moreover, these authors found that mutations in p7 affected the apparent topology of NS2 (44). Thus, p7 is likely to be part of the NS2 complex. In addition, we predict that NS4A is also part of the NS2 complex. NS4A heterodimerizes with NS3, contributes to the proper folding of the NS3 serine protease domain, and stabilizes NS3 (4, 13, 35, 73). Furthermore, we previously found that the NS4A E42G mutation partially suppressed the virus assembly defect caused by the NS2 D62A mutation (55).
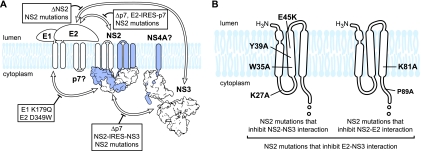
Based on the considerations described above, we propose a model of the NS2 protein complex containing E1, E2, p7, NS2, NS3, and NS4A (Fig. 9 A). This model takes into account the cocapture of E1, E2, and NS3 with biotinylated NS2(AP) and the reciprocal cocapture of E1, NS2, and NS3 with biotinylated E2(AP) (Fig. 2). Our model was also supported by BN-PAGE analysis, which showed that E2, NS2, and NS3 were all found in native, high-molecular-mass complexes that comigrated at ≈680 kDa and ≈520 kDa, as well as several protein-specific complexes (Fig. 3). These native, high-molecular mass complexes were not resolved when microsomes were prepared from Jc1/Δp7-transfected cells (data not shown), further indicating that their formation is dependent on p7 expression.
FIG. 9.
Model of HCV NS2 complex assembly. (A) NS2 complex. Proteins within the NS2 complex (E1, E2, NS2, and NS3) are indicated; proteins suspected to be in the NS2 complex (p7, NS4A) are shown with question marks. White arrows denote interactions identified in this work; boxes and thick black lines indicate inhibitors of these interactions. For contrast, NS4A and one monomer of NS2 are shown in blue. (B) NS2 mutations that inhibit NS2-NS3, NS2-E2, or E2-NS3 interaction. The locations of these mutations are mapped onto a model of the NS2 transmembrane domains (23, 55); the cysteine protease domain is not shown.
Importantly, the relevance of the NS2 complex was supported by several insertion, deletion, and point mutations that inhibited virus assembly and inhibited NS2 complex formation, as summarized in Fig. 9A. Mutations in the first cytoplasmic loop and second putative transmembrane domain of NS2 inhibited NS2-NS3 interaction, while mutations in the third putative transmembrane domain and cytoplasmic stem region of NS2 inhibited NS2-E2 interaction (Fig. 9B). These data indicate that discrete regions of NS2 are critical for NS3 and E2 interaction, although we do not yet know whether these are direct interactions or indirect interactions that involve intermediate protein(s). Importantly, mutations that inhibited the interaction of NS2 with either E2 or NS3 also inhibited the interaction of E2 with NS3 (Fig. 9A and B). Therefore, NS2 plays a central organizing role in bringing together the E1-E2 glycoprotein and NS3-4A enzyme complexes.
The stoichiometry of the NS2 complexes remains to be quantitatively characterized. Given that the NS2-associated proteins were concentrated by the capture process, it is clear that only a fraction of E1, E2, and NS3 were cocaptured with NS2 (Fig. 2). This was supported by our BN-PAGE analysis, which showed that E2, NS2, and NS3 were all found in complexes of different sizes as well as complexes of similar sizes (Fig. 3).
While several NS2 mutations coordinately inhibited NS2 interactions and virus assembly, it was surprising that the NS3 Q221L and E1 A78T mutations, which we previously showed suppressed specific NS2 defects in virus assembly (55), did not restore interactions with NS2 (Fig. 7). This finding has two important implications. First, these suppressor mutations do not simply restore protein-protein interaction surfaces on E1 or NS3. Second, the complete set of NS2 interactions is not absolutely required for virus assembly. One possible explanation for this is that NS2 may activate or alter the activity of the E1-E2 glycoprotein or NS3-4A enzyme complex, and these suppressor mutations may confer this change in activation status in an NS2-independent manner, effectively bypassing the need for a subset of NS2 interactions.
Our results largely agree with the work of Ma and colleagues, who recently showed that a yellow fluorescent protein (YFP)-tagged NS2 protein from an H77-JFH1 chimeric virus interacts with E1, E2, p7, NS3, and NS5A (44). Furthermore, both studies found that p7 is essential for NS2 complex assembly and that E1-E2 is dispensable for NS2-NS3 interaction. Thus, these interactions are likely conserved across different HCV genotypes.
Although we were able to identify interactions between NS2 and E1 and between E2 and NS3, we did not detect stable interactions between NS2 and core, NS5A, or NS5B (Fig. 2 and data not shown). This is particularly interesting given that core has been shown to interact with the E1 glycoprotein (40), a member of the NS2 protein complex. Thus, E1 may interact with NS2 prior to its interaction with core, perhaps upstream of virus particle assembly. Furthermore, Ma and colleagues found that NS5A did not coimmunoprecipitate with NS2-YFP, which was consistent with our findings, but that NS2 coimmunoprecipitated with YFP-NS5A (44). One possibility for this apparent discrepancy is that the N-terminal region of NS2 may be sterically obstructed and thereby inefficient at affinity capture when NS2 is interacting with NS5A. Alternatively, it could be that these interactions exist but are weak or unstable under our protein capture conditions.
Similarly to the previous work of Jirasko and colleagues, we observed a truncated form of NS2, tNS2 (23). In BN-PAGE, tNS2 associated only with the low-molecular-mass (≈40-kDa) native NS2 complex (Fig. 3), suggesting that it was excluded from the high-molecular-mass complexes. Consistent with this, tNS2 accumulation was significantly increased in the D88 and Δp7 constructs (Fig. 6B), yet tNS2 was not cocaptured with NS2(AP) (Fig. 6C). Thus, p7 may help to stabilize NS2 and prevent its conversion into tNS2. While the cleavage event(s) that generates tNS2 remains undefined, it is tempting to speculate that this cleavage may function early in the viral life cycle to prevent premature virus assembly or participate in the turnover or disassembly of the NS2 complex in later stages of virus assembly. It is notable that the yellow fever virus NS2A protein performs an essential role in virus assembly, and this function is mediated through NS2A cleavage by the viral NS2B-3 serine protease (30, 53). Thus, HCV NS2 and flavivirus NS2A proteins may perform similar but as-yet-undefined functions in virus particle assembly.
The HCV NS2 protein complex may also contain host proteins. Based on yeast two-hybrid analyses and pulldown of proteins overexpressed in HepG2 cells, NS2 reportedly interacts with cell death-inducing DFFA-like effector B (CIDE-B) (12). This is potentially interesting, as CIDE-B was recently shown to play a role in the lipidation of very-low-density lipoprotein particles (75) and could be related to the coordination of HCV and very-low-density lipoprotein particle assembly (15, 22). We therefore tested whether CIDE-B could be specifically cocaptured with NS2(AP), but we did not detect such an interaction (data not shown). Again, these negative results could be due to transient or weak interactions or to differences in experimental systems such as viral strains, cell types, etc. Nevertheless, our NS2 affinity capture system should be useful for the future identification of host proteins within the NS2 protein complex.
In summary, these studies demonstrate that HCV NS2 physically interacts with the viral E1-E2 glycoprotein and NS3-NS4A enzyme complexes during virus assembly. The relevance of these interactions was demonstrated by numerous mutations that coordinately disrupted the NS2 complex and virus particle assembly. Moreover, mutations that disrupted NS2 interactions also disrupted the E2-NS3 interaction. Thus, NS2 is responsible for bringing the glycoprotein and enzyme subcomplexes together, perhaps to initiate the early steps in virus assembly.
Acknowledgments
We thank J. Bloom and N. Counihan for helpful comments on the manuscript, C. Peters for technical support, and P. Flynn, Y. Jiang, and T. Drozdz for administrative support. We are grateful to S. Wagner for advice on BN-PAGE, J. Dubuisson for suggesting the use of the A4 epitope, H. Greenberg and J. Dubuisson for providing the A4 antibody, J. A. McKeating for providing the 3/11 antibody, and C. M. Rice for providing the 6H6 antibody and Huh-7.5 cells.
This work was funded through NIH grants AI087925 and AI076259 (both to B.D.L.).
Footnotes
Published ahead of print on 8 December 2010.
REFERENCES
- 1.Appel, N., U. Herian, and R. Bartenschlager. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79:896-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appel, N., T. Pietschmann, and R. Bartenschlager. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 79:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appel, N., et al. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckett, D., E. Kovaleva, and P. J. Schatz. 1999. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8:921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beran, R. K., B. D. Lindenbach, and A. M. Pyle. 2009. The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase. J. Virol. 83:3268-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brohm, C., et al. 2009. Characterization of determinants important for hepatitis C virus p7 function in morphogenesis by using trans-complementation. J. Virol. 83:11682-11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo, Q.-L., et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciczora, Y., N. Callens, F. Penin, E. I. Pecheur, and J. Dubuisson. 2007. Transmembrane domains of hepatitis C virus envelope glycoproteins: residues involved in E1E2 heterodimerization and involvement of these domains in virus entry. J. Virol. 81:2372-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dentzer, T. G., I. C. Lorenz, M. J. Evans, and C. M. Rice. 2009. Determinants of the hepatitis C virus nonstructural protein 2 protease domain required for production of infectious virus. J. Virol. 83:12702-12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubuisson, J., et al. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdtmann, L., et al. 2003. The hepatitis C virus NS2 protein is an inhibitor of CIDE-B-induced apoptosis. J. Biol. Chem. 278:18256-18264. [DOI] [PubMed] [Google Scholar]
- 13.Failla, C., L. Tomei, and R. De Francesco. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 68:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flint, M., et al. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gastaminza, P., et al. 2008. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 82:2120-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottwein, J. M., et al. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364-377. [DOI] [PubMed] [Google Scholar]
- 17.Gouttenoire, J., F. Penin, and D. Moradpour. 2010. Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory. Rev. Med. Virol. 20:117-129. [DOI] [PubMed] [Google Scholar]
- 18.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. U. S. A. 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hijikata, M., et al. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen, T. B., et al. 2008. Highly efficient JFH1-based cell-culture system for hepatitis C virus genotype 5a: failure of homologous neutralizing-antibody treatment to control infection. J. Infect. Dis. 198:1756-1765. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, J., and G. Luo. 2009. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 83:12680-12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jirasko, V., et al. 2008. Structural and functional characterization of non-structural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283:28546-28562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato, T., et al. 2008. Hepatitis C virus JFH-1 strain infection in chimpanzees is associated with low pathogenicity and emergence of an adaptive mutation. Hepatology 48:732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klepsch, M., et al. 2008. Immobilization of the first dimension in 2D blue native/SDS-PAGE allows the relative quantification of membrane proteomes. Methods 46:48-53. [DOI] [PubMed] [Google Scholar]
- 27.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuang, W. F., et al. 2004. Hepatitis C virus NS3 RNA helicase activity is modulated by the two domains of NS3 and NS4A. Biochem. Biophys. Res. Commun. 317:211-217. [DOI] [PubMed] [Google Scholar]
- 29.Kulman, J. D., M. Satake, and J. E. Harris. 2007. A versatile system for site-specific enzymatic biotinylation and regulated expression of proteins in cultured mammalian cells. Protein Expr. Purif. 52:320-328. [DOI] [PubMed] [Google Scholar]
- 30.Kümmerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lam, A. M., and D. N. Frick. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 80:404-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavie, M., A. Goffard, and J. Dubuisson. 2007. Assembly of a functional HCV glycoprotein heterodimer. Curr. Issues Mol. Biol. 9:71-86. [PubMed] [Google Scholar]
- 33.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 102:17717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, C., B. D. Lindenbach, B. M. Pragai, D. W. McCourt, and C. M. Rice. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, R., et al. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 80:6072-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindenbach, B. D. 2009. Measuring HCV infectivity produced in cell culture and in vivo. Methods Mol. Biol. 510:329-336. [DOI] [PubMed] [Google Scholar]
- 38.Lindenbach, B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 39.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe et al. (ed.), Fields virology, 5th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 40.Lo, S. Y., M. J. Selby, and J. H. Ou. 1996. Interaction between hepatitis C virus core protein and E1 envelope protein. J. Virol. 70:5177-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohmann, V., J. O. Koch, and R. Bartenschlager. 1996. Processing pathways of the hepatitis C virus proteins. J. Hepatol. 24:11-19. [PubMed] [Google Scholar]
- 42.Loo, Y. M., et al. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 103:6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorenz, I. C., J. Marcotrigiano, T. G. Dentzer, and C. M. Rice. 2006. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442:831-835. [DOI] [PubMed] [Google Scholar]
- 44.Ma, Y., et al. 2011. Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J. Virol. 85:86-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma, Y., J. Yates, Y. Liang, S. M. Lemon, and M. Yi. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624-7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mechold, U., C. Gilbert, and V. Ogryzko. 2005. Codon optimization of the BirA enzyme gene leads to higher expression and an improved efficiency of biotinylation of target proteins in mammalian cells. J. Biotechnol. 116:245-249. [DOI] [PubMed] [Google Scholar]
- 47.Meylan, E., et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 48.Mine, A., et al. 2010. Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of red clover necrotic mosaic virus. J. Virol. 84:6070-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizushima, H., et al. 1994. Two hepatitis C virus glycoprotein E2 products with different C termini. J. Virol. 68:6215-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noppornpanth, S., et al. 2007. Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients. J. Virol. 81:12496-12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Op De Beeck, A., et al. 2000. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J. Biol. Chem. 275:31428-31437. [DOI] [PubMed] [Google Scholar]
- 52.Parrott, M. B., and M. A. Barry. 2001. Metabolic biotinylation of secreted and cell surface proteins from mammalian cells. Biochem. Biophys. Res. Commun. 281:993-1000. [DOI] [PubMed] [Google Scholar]
- 53.Patkar, C. G., and R. J. Kuhn. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J. Virol. 82:3342-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pavlovic, D., et al. 2003. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. U. S. A. 100:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phan, T., R. K. Beran, C. Peters, I. C. Lorenz, and B. D. Lindenbach. 2009. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 83:8379-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pietschmann, T., et al. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 58.Schagger, H. 2001. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65:231-244. [DOI] [PubMed] [Google Scholar]
- 59.Schagger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 60.Scheel, T. K., et al. 2008. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc. Natl. Acad. Sci. U. S. A. 105:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schlegel, S., M. Klepsch, D. Wickstrom, S. Wagner, and J. W. de Gier. 2010. Comparative analysis of cytoplasmic membrane proteomes of Escherichia coli using 2D blue native/SDS-PAGE. Methods Mol. Biol. 619:257-269. [DOI] [PubMed] [Google Scholar]
- 62.Schregel, V., S. Jacobi, F. Penin, and N. Tautz. 2009. Hepatitis C virus NS2 is a protease stimulated by cofactor domains in NS3. Proc. Natl. Acad. Sci. U. S. A. 106:5342-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Selby, M. J., E. Glazer, F. Masiarz, and M. Houghton. 1994. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology 204:114-122. [DOI] [PubMed] [Google Scholar]
- 64.Steinmann, E., et al. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tellinghuisen, T. L., K. L. Foss, J. C. Treadaway, and C. M. Rice. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thiel, H.-J., et al. 2005. Family Flaviviridae, p. 979-996. In C. M. Fauquet et al. (ed.), Virus taxonomy. VIIIth Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- 68.Vieyres, G., et al. 2010. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J. Virol. 84:10159-10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wakita, T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, L., and B. Dobberstein. 1999. Oligomeric complexes involved in translocation of proteins across the membrane of the endoplasmic reticulum. FEBS Lett. 457:316-322. [DOI] [PubMed] [Google Scholar]
- 71.Welbourn, S., et al. 2009. Investigation of a role for lysine residues in non-structural proteins 2 and 2/3 of the hepatitis C virus for their degradation and virus assembly. J. Gen. Virol. 90:1071-1080. [DOI] [PubMed] [Google Scholar]
- 72.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue native PAGE. Nat. Protoc. 1:418-428. [DOI] [PubMed] [Google Scholar]
- 73.Wölk, B., et al. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamashita, T., et al. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 75.Ye, J., et al. 2009. Cideb, an ER- and lipid droplet-associated protein, mediates VLDL lipidation and maturation by interacting with apolipoprotein B. Cell Metab. 9:177-190. [DOI] [PubMed] [Google Scholar]
- 76.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2009. trans-Complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 5:e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yi, M., Y. Nakamoto, S. Kaneko, T. Yamashita, and S. Murakami. 1997. Delineation of regions important for heteromeric association of hepatitis C virus E1 and E2. Virology 231:119-129. [DOI] [PubMed] [Google Scholar]