Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) encodes ORF57, which promotes the accumulation of specific KSHV mRNA targets, including ORF59 mRNA. We report that the cellular export NXF1 cofactors RBM15 and OTT3 participate in ORF57-enhanced expression of KSHV ORF59. We also found that ectopic expression of RBM15 or OTT3 augments ORF59 production in the absence of ORF57. While RBM15 promotes the accumulation of ORF59 RNA predominantly in the nucleus compared to the levels in the cytoplasm, we found that ORF57 shifted the nucleocytoplasmic balance by increasing ORF59 RNA accumulation in the cytoplasm more than in the nucleus. By promoting the accumulation of cytoplasmic ORF59 RNA, ORF57 offsets the nuclear RNA accumulation mediated by RBM15 by preventing nuclear ORF59 RNA from hyperpolyadenylation. ORF57 interacts directly with the RBM15 C-terminal portion containing the SPOC domain to reduce RBM15 binding to ORF59 RNA. Although ORF57 homologs Epstein-Barr virus (EBV) EB2, herpes simplex virus (HSV) ICP27, varicella-zoster virus (VZV) IE4/ORF4, and cytomegalovirus (CMV) UL69 also interact with RBM15 and OTT3, EBV EB2, which also promotes ORF59 expression, does not function like KSHV ORF57 to efficiently prevent RBM15-mediated nuclear accumulation of ORF59 RNA and RBM15's association with polyadenylated RNAs. Collectively, our data provide novel insight elucidating a molecular mechanism by which ORF57 promotes the expression of viral intronless genes.
Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 is a viral early protein. Like its homologues herpes simplex virus type 1 (HSV-1) ICP27 (38), Epstein-Barr virus (EBV) EB2 (40), human cytomegalovirus (HCMV) UL69 (47), and varicella-zoster virus (VZV) IE4/ORF4 (32), KSHV ORF57 regulates virus gene expression at the posttranscriptional level (23). The KSHV genome with a disrupted ORF57 gene cannot efficiently express a subset of viral lytic genes (20). ORF57 functions as a viral splicing factor and promotes viral RNA splicing (19, 21, 23). ORF57 also promotes the expression of viral intronless genes, such as ORF59, a viral DNA polymerase processivity factor. The specific ORF57 interactions with its targeted RNAs are facilitated by cellular proteins (23). It has been proposed that ORF57 stimulates RNA export via its interaction with Aly/REF, a cellular RNA-binding protein serving as an adaptor for the nuclear RNA export receptor NXF1/TAP (24). However, recent reports indicate that Aly/REF-ORF57 interaction does not play an important role in the ORF57-mediated enhancement of ORF59 expression, since Aly/REF knockdown in host cells did not affect the function of ORF57 (22, 29). In contrast, a recent study indicates that ORF57 appears to recruit the entire TREX through its interaction with Aly/REF to facilitate the export of a viral late transcript, ORF47 (2). Since ORF57 specifically binds to ORF59 RNA only in the presence of cellular proteins, it is thought that the cellular adaptors may act as cofactors. In support of this, the posttranscriptional regulator EB2 of EBV, a member of the gammaherpesvirus subfamily, was reported to interact with OTT3 (also referred to as RNA binding motif protein 15B [RBM15B]) and OTT3 was shown to have posttranscriptional regulatory function (6).
OTT3 (6, 26), RNA binding motif protein 15 (RBM15 [OTT1]) (16, 26), and SHARP (SMRT/HDAC1-associated repressor protein) (30, 41) are members of the SPEN (split ends) protein family. These nuclear proteins share a domain structure comprising three highly conserved N-terminal RNA-binding motifs (RNA recognition motif [RRM]) and a highly conserved C-terminal SPOC (Spen paralogue and orthologue C-terminal) domain, with 52%, 68%, and 82% sequence identity for RRM1, RRM2, and RRM3, respectively, and 58% amino acid (aa) sequence identity in their SPOC domain (6, 16, 37, 50). SPEN homologs are conserved in eukaryotes and are found from Caenorhabditis elegans to humans but show distinct functional properties (6, 13, 49). While SHARP (3,664 aa residues) acts at the transcriptional level (3, 10, 12, 30, 31, 39, 41) and has also been found in association with spliceosomes (51), both of the two smaller proteins, RBM15 (957 to 977 aa residues for the different isoforms) and OTT3 (890 aa residues), act at the posttranscriptional level (6, 13, 49). We and others have reported that the SPOC-containing C-terminal region of RBM15 and OTT3 mediates the RNA export function (6, 13, 49), while SHARP lacks this function, supporting divergent functions of RBM15 and OTT3. Ectopically expressed OTT3 was reported to act as a splicing regulator (6). RBM15 was determined to bind directly to the RNA transport element (RTE) (13), which is exported via the NXF1 pathway (42). RTE is present in murine intracisternal A-particle retroelements (28, 43) and is essential for the activity of the retrotransposon (52). RBM15 and OTT3 were shown to promote the export and expression of RTE-containing transcripts (13, 49). Both proteins localize to the nucleus and associate with the splicing factor compartment (SC35-containing speckles) and the nuclear envelope but do not shuttle to the cytoplasm (6, 13, 49). Both proteins can also interact with each other (49). RBM15 and OTT3 act as cofactors to the nuclear export receptor NXF1 and interact directly with NXF1 via the C-terminal region (13, 49). We recently reported that RBM15 also provides a direct molecular link between NXF1 and DEAD family RNA helicase DBP5 (53), supporting the model in which RBM15 acts locally at the nuclear pore complex, facilitating the recognition of NXF1-mRNP complexes by DBP5 during translocation and thereby contributing to efficient mRNA export. Genetic knockout of RBM15 is embryonic lethal in homozygous mice (35, 36, 49), indicating that OTT3 cannot compensate for the loss of RBM15. These findings support the model that, in addition to shared functions, these factors have distinct biological roles in animal development. In the study presented herein, we demonstrated that both RBM15 and OTT3, like KSHV ORF57, promote KSHV ORF59 expression. We identified the direct interaction of KSHV ORF57 with RBM15 and OTT3 and showed that the interaction with RBM15 and OTT3 is conserved among ORF57 homologs of four other herpesvirus family members.
MATERIALS AND METHODS
Cell cultures and cotransfection.
HEK293 and HeLa cell lines from ATCC (Manassas, VA) were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum as described before (22). The cells, at 2.5 × 105 per ml in a 60-cm dish, were cotransfected with plasmid DNA at 20, 50, or 100 ng for the ORF57 expression vector, at 20 or 100 ng for the RBM15 expression vector, at 100 ng for the OTT3 expression vector, and at 300 ng for the ORF59 expression vector in the presence of Lipofectamine 2000 (Invitrogen, Carlsbad, CA) or Fugene HD (Roche Applied Science, Indianapolis, IN).
Plasmids.
Vector pEGFP-N1 was obtained from Clontech (Mountain View, CA). All other plasmids used in this study were previously described, as follows: pVM18 (FLAG-tagged KSHV ORF59), pVM68 (3×FLAG-tagged KSHV ORF57), pVM90 (3×FLAG-tagged KSHV ORF57 mutant mtNLS2 + 3), pVM8 (green fluorescent protein [GFP]-tagged wild-type [wt] ORF57) and its mutant pVM36 (mtNLS2 + 3) (22), FLAG-tagged RBM15 and OTT3 or hemagglutinin (HA)-tagged OTT3, as well as their truncation mutants (13, 49), or HA-tagged Ref1-II (53). Enhanced GFP (EGFP)-tagged ICP27 (a gift from R. Sandri-Goldin), Myc-tagged EB2 (a gift from S. Swaminathan), VZV IE4-V5 (a gift from C. Sadzot), and FLAG-tagged UL69 (a gift from T. Stamminger) were used.
RNA interference (RNAi).
HeLa cells were transfected with 40 nM ON-TARGETplus SMARTpool small interfering RNA (siRNA) (Thermo Scientific, Waltham, MA) targeting endogenous RBM15 protein or a nonspecific siRNA. At 24 h after siRNA transfection, the cells were transfected with 500 ng of ORF59 expression vector (pVM18) and incubated for an additional 24 h. Total protein and RNA were isolated as described previously (22) for Western and Northern blot analysis, respectively.
Western blot analysis.
Protein extracts were prepared by direct cell lysis in SDS protein sample buffer and analyzed in Western blots as described previously (22). The following monoclonal antibodies were used in the study: anti-GFP (BD Biosciences, Mountain View, CA), anti-FLAG (Sigma, St. Louis, MO), anti-myc (Sigma), anti-HA (Sigma), anti-tubulin (Sigma), anti-RBM15 (Protein Tech Group, Chicago, IL), anti-V5 horseradish peroxidase (HRP) (Invitrogen), and anti-ORF59 (Advanced Biotechnologies, Columbia, MD). Rabbit anti-ORF57 was previously described (20).
Isolation of nuclear and cytoplasmic RNA.
Cytoplasmic and nuclear total RNAs were fractionated as described previously (22). Briefly, after transfection, the cells were trypsinized, washed with phosphate-buffered saline (PBS), and resuspended in cold buffer A (50 mM Tris, pH 8.0, 140 mM NaCl, 1.5 MgCl2, 0.2% NP-40, 1 mM dithiothreitol [DTT], and 200 U RNasin). After 5 min of incubation on ice, the nuclei were pelleted by spinning for 2 min at 3,000 × g at 4°C. Both the supernatant for the cytoplasmic fraction and the pellet for the nuclear fraction were collected for RNA isolation by using TRIzol reagent (Invitrogen).
Northern blot analysis.
RNA (∼5 μg) was separated in agarose gel and analyzed in Northern blots as described previously (22). The following oligonucleotide probes labeled with γ-32P were used in this study: oVM73 (5′-GTCCACCCTGACCCCATAGT-3′) for ORF59, oZMZ270 (5′-TGAGTCCTTCCACGATACCAAA-3′) for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), oVM11 (5′-CTCGTCTTCCAGTGTCGGTG-3′) for ORF57, and oST197 (5′-AAAATATGGAACGCTTCACGA-3′) for U6 snRNA. The hybridization signal was captured using a Molecular Dynamics PhosphorImager Storm 860 and analyzed with ImageQuant software.
Indirect immunofluorescence staining.
For indirect immunofluorescence staining (IFA), cells growing on coverslips were fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized for 10 min at room temperature with 0.5% Triton X-100, and blocked with 2% bovine serum albumin (BSA) in PBS with the addition of 0.02% Tween 20 (blocking buffer) for 1 h at 37°C. Primary antibodies were diluted in blocking buffer and incubated with samples for 1 h at 37°C, followed by 3 washes with PBS. Secondary antibodies conjugated with Alexa Fluor 546 dye (Molecular Probes, Eugene, OR) were diluted 1:500 in blocking buffer and incubated for 1 h at 37°C. After a final 3 washes with PBS, the cell nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole) and all samples were mounted in ProLong Gold mounting medium (Invitrogen). Confocal fluorescence images were collected using a Zeiss LSM510 META (Carl Zeiss MicroImaging, Inc., Thornwood, NY) laser scanning microscope, and colocalization analysis was performed using the colocalization module of Imaris version 6.0 software (Bitplane) as described previously (21).
Immunoprecipitation.
For coimmunoprecipitation (co-IP) assays, HEK293 cells were transfected with the indicated expression constructs and harvested 2 days later. The cells were extracted under stringent conditions (400 mM NaCl, 50 mM KCl, 0.2% Triton X-100, 15 mM HEPES, pH 7.9, 10% glycerol, protease inhibitors) (13). The extracts were treated with RNase A (0.5 mg/ml) for 10 min at room temperature, incubated with anti-FLAG M2 or anti-HA antibody beads for 1 h, and subsequently washed with lysis buffer containing 2 M urea, as described previously (49). Co-IPs under physiological conditions were performed in the same buffer (0.2% Triton X-100, 15 mM HEPES, pH 7.9, 10% glycerol, protease inhibitors) containing 150 mM NaCl, using a wash buffer which did not contain urea. The protein complexes were eluted by boiling the beads in SDS-PAGE loading buffer. The complexes were subjected to Western blot analysis, and the blots were probed with anti-GFP, anti-FLAG, anti-HA, anti-V5, or anti-myc antibodies. In vitro protein binding assays were performed (48) using Escherichia coli-expressed purified glutathione S-transferase (GST)-tagged RBM15 (aa 530 to 977), GST-tagged OTT3 (aa 488 to 890), or GST only and metabolically labeled, reticulocyte-produced ORF57, UAP56, or firefly luciferase protein, respectively.
RNase H digestion.
The length of the mRNA poly(A) tail was determined by RNase H digestion as described by Murray and Schoenberg (27), with small modifications. Briefly, 10 μg of RNA in a total volume of 7 μl was denatured for 5 min at 85°C. Two microliters of 5 nmol oligo(dT16) (Applied Biosystems, Carlsbad, CA) was added to RNA and annealed for 10 min at 42°C. Each RNA sample was supplemented with 1 μl of 10× RNase H buffer (200 mM Tris, pH 8.0, 500 mM KCl, 100 mM MgCl2, 10 mM DTT) and split into two tubes (5 μl each). One tube of the RNA was digested with 10 U of RNase H (Fermentas, Glen Burnie, MD) for 30 min at 30°C, and the other tube of the RNA served as an undigested RNA control. After digestion, 15 μl of formaldehyde loading dye was added directly to each sample and the samples were incubated for 15 min at 75°C and then analyzed by Northern blotting as described above.
UV cross-linking and isolation of polyadenylated RNA.
The detection of proteins associated with polyadenylated RNAs was performed as described previously (33). Briefly, transfected cells washed with PBS were exposed to UV light (4,800 mJ/cm2) and resuspended in PBS. One third of the cell suspension was spun, and the cell pellet was lysed directly in SDS protein sample buffer to serve as an input control. The rest of the cells were used for the isolation of polyadenylated RNAs using an Illustra QuickPrep micro mRNA purification kit (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions. The eluted mRNA-protein complexes were treated with RNase T1 (5 U/sample) at 37°C for 15 min and analyzed by Western blotting.
UV cross-linking and immunoprecipitation (CLIP).
HEK293 cells (5 × 105 cells/well in a 6-well plate) were cotransfected with RBM15-FLAG and ORF59-FLAG, 500 ng each, in the presence or absence of ORF57-GFP. At 24 h after cotransfection, the cells were washed with PBS and UV irradiated (4,800 mJ/cm2). Total cell extracts after UV cross-linking were prepared by lysis directly in radioimmunoprecipitation (RIPA) buffer with protease inhibitors added. Protein-RNA complexes were immunoprecipitated as previously described (22) using anti-FLAG M2 mouse monoclonal antibody. Nonspecific mouse IgG was used as a negative control. Immunoprecipitated RNA digested by proteinase K was amplified by RT-PCR with ORF59-specific oligonucleotides (5′ primer oST179, 5′-TAATACGACTCACTATAGG/GTCACCTGATTGGCAGCC-3′, and 3′ primer oST180, 5′-AACGCATCGAG/CGGTGACTGTGTCTGTCAGC-3′; slashes separate the nonspecific sequences from the specific viral sequences) or with GAPDH-specific oligonucleotides (21) and analyzed in agarose gel.
Quantitative RT-PCR and RNA stability analysis.
RNA samples obtained from CLIP assays were treated with Turbo DNase I (Ambion, Austin, TX) according to the manufacturer's instructions and subjected to TaqMan quantitative reverse transcription-PCR (qRT-PCR) using TaqMan reverse transcription reagents and TaqMan universal master mix (Applied Biosystems). KSHV ORF59 transcripts were amplified and detected by using the following TaqMan primers: ORF59 probe (5′ 56-FAM-AAACCGATCTGTGTCTGCCGAGG-3IABkFG 3′), ORF59 primer 1 (5′ TTAGAAGTGGAAGGTGTGCC 3′), and ORF59 primer 2 (5′ TCCTGGAGTCCGGTATAGAATC 3′). After background subtraction, the cycle threshold (CT) values of the qRT-PCR data from 2 repeats, each in triplicate, were analyzed by the 2−ΔΔCT or 2−ΔCT method (14) and are presented as bar graphs with means ± standard deviations.
For RNA stability assays, HEK293 cells were transfected with ORF59 expression vector pVM18 plus FLAG-tagged ORF57 expression vector pVM7, FLAG-tagged RBM15 or empty pFLAG-CMV-5.1 vector control. Since the expression level of ORF59 is very low in the absence of ORF57 or RBM15, HEK293 cells (1 × 106) in 60-mm dishes were cotransfected with 2 μg of ORF59 expression plasmid plus the appropriate amount of pFLAG-CMV-5.1 vector to keep the ratio as 5:1 or 15:1 as a control for the ORF57 or RBM15 expression vectors, using FuGene HD (Roche). Eighteen hours after transfection, actinomycin D (ActD) was added to the cells at a final concentration of 10 μg/ml, and total cell RNA was extracted at the designated time points by using an RNeasy kit (Qiagen), followed by poly(A) mRNA enrichment with an Oligotex mini mRNA kit (Qiagen). Approximately 100 to 200 ng of poly(A) mRNA was reverse transcribed with a poly(T) oligonucleotide. Real-time PCR for ORF59 RNA and GAPDH RNA was carried out on the cDNA in duplicates by using TaqMan Universal Master mix (Applied Biosystems). The GAPDH RNA level in each reaction mixture, determined by using TaqMan GAPDH control probes (catalog number 402869; Applied Biosystems), served as a loading control and was used to quantify the ORF59 RNA. The relative level of expression of ORF59 was determined using the CT method as described above and converted to a percentage, with the amount at the moment of the addition of ActD (time zero), set as 100%, representing the percentage of remaining mRNA. A nonlinear regression analysis on the raw data (fold percent) was performed, choosing an exponential decay model {[fold % = α·exp(β·t)], where α is the intercept at time zero β is the decay rate, and t is time}. The α was constrained to be either less than or equal to 100% (α ≤ 100%) or equal to 100% (α = 100%), and the β to be less than or equal to zero (β ≤ 0). Weighted linear regression results were graphed, and a regression curve was employed for half-life calculation. Comparison of parameter estimates (β only if α equals 100%, and α and β if α is less than or equal to 100%) between the treatments was performed by using a model with a dummy variable.
RESULTS
RBM15 promotes ORF59 protein expression in the absence or presence of ORF57.
As ORF57 promoted intronless ORF59 expression in our previous studies (19, 20, 22), we tested whether RBM15 and OTT3 would potentiate ORF57 to enhance ORF59 expression upon cotransfection of HEK293 cells. Unexpectedly, we found that cotransfection of the ORF59 plasmid with RBM15 alone led to increased levels of both ORF59 protein and RNA in the absence of ORF57 (Fig. 1 A and B, compare lanes 1 and 2 as well as the corresponding bar graphs of RNA levels). The enhanced expression of ORF59 protein and RNA was observed in cells transfected with 20 ng of the RBM15 expression vector. Comparing the exogenous to the endogenous RBM15 levels in transfected HEK293 cells showed that the level of exogenous RBM15 with the 20-ng transfection was about 30% less than its endogenous level in our experimental conditions (see Fig. S1 in the supplemental material). Our data indicate that ectopic RBM15 near its endogenous level was enough to further augment ORF59 expression in the absence of ORF57.
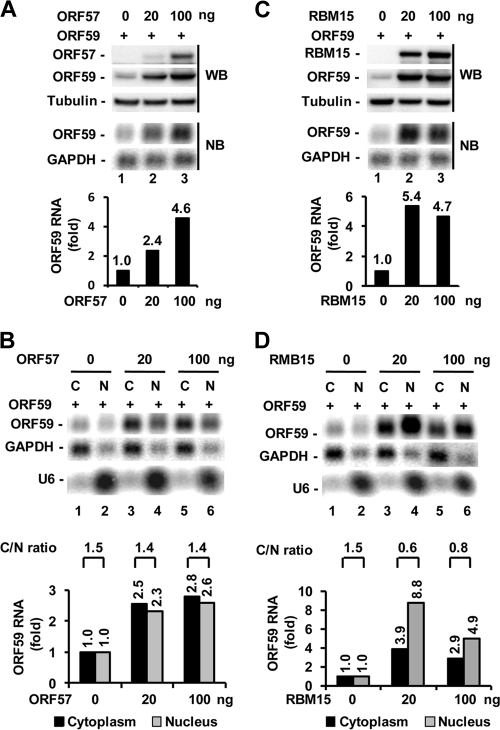
FIG. 1.
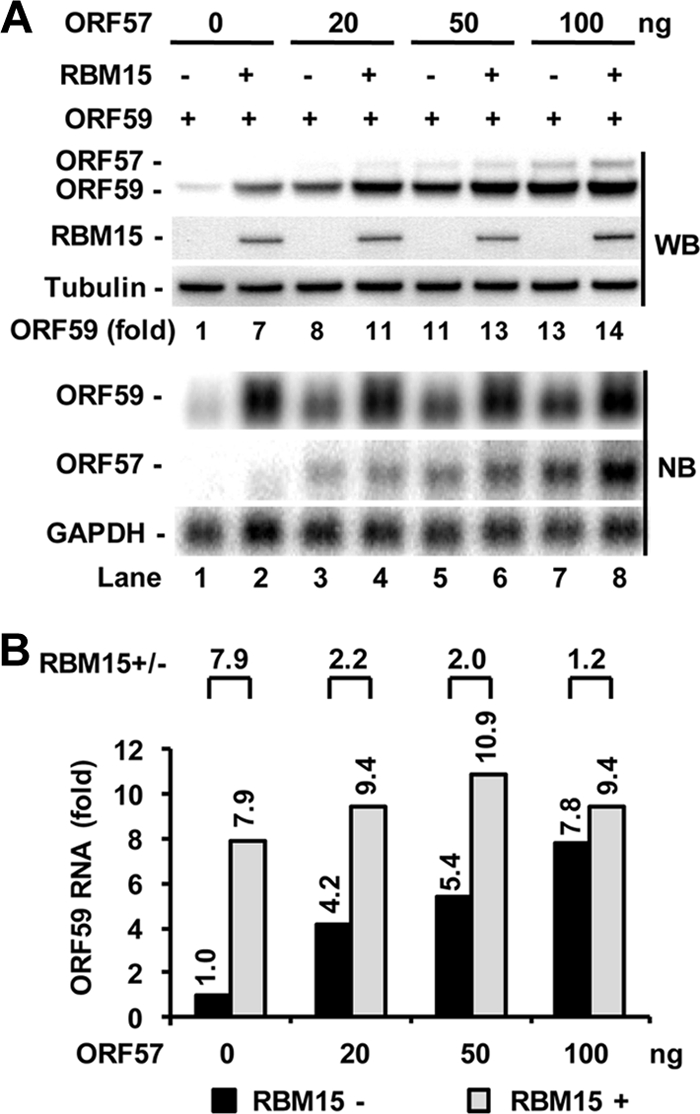
Ectopic RBM15 promotes and cooperates with ORF57 in ORF59 expression. HEK293 cells at 2.5 × 105 per ml were cotransfected with 20 ng of RBM15-FLAG expression vector in combination with 300 ng of ORF59-FLAG expression vector in the absence or presence of an increased amount of ORF57-FLAG expression vector as indicated. Protein and RNA samples were prepared 24 h after cotransfection and analyzed, respectively, by Western blot (WB) and Northern blot (NB) assays. Tubulin served as a protein loading control for Western blotting, and GAPDH RNA served as an RNA loading control for Northern blotting. (A) RBM15 regulation of ORF59 expression in the absence or presence of ORF57. Relative ORF59 protein levels (fold) were quantified based on each band's density after normalization to that of tubulin as a sample loading control, with the ORF59 protein level in lane 1 being set as 1. (B) Bar graphs show relative level of ORF59 RNA in each sample from Northern blot analysis in panel A after being normalized to the level of the corresponding GAPDH RNA for each sample loaded. Shown above the bar graph is the relative ratio of ORF59 RNA in the presence versus the absence of RBM15 for each pair of samples.
We next tested whether RBM15 at this low dose (20 ng) could synergize with ORF57 to further augment ORF59 expression in HEK293 cells by cotransfection. As expected, ORF57 alone was found to promote the expression of both ORF59 protein and RNA (Fig. 1A and B, compare lane 1 with lanes 3, 5, and 7 and the corresponding bar graphs of RNA levels) in a dose-dependent manner. ORF57 at 20 ng was better than RBM15 in promoting ORF59 protein expression, but the opposite was true for RBM15 in the increase of ORF59 RNA (Fig. 1A, compare lane 3 to lane 2). These data indicate that a proportion of RBM15-enhanced ORF59 RNA was not translated. Although RBM15 at the tested dose was found to synergize with ORF57 in promoting ORF59 expression, this synergy at both the protein and the RNA level was only moderate and became diminished with increasing ORF57 levels (Fig. 1A and B, compare lane 2 with lanes 4, 6, and 8 and the corresponding protein levels and bar graphs of RNA levels). Interestingly, this synergy may reflect a modulation by RBM15 of ORF57 expression, as expected (18). Under our experimental conditions, we found that RBM15 could slightly increase the expression of ORF57 protein and RNA in each ORF57 dose tested (Fig. 1A and B, compare lanes 3, 5, and 7 with lanes 4, 6, and 8).
Distinct effects of ORF57 and RBM15 in the promotion of ORF59 RNA accumulation.
To examine whether ORF57 and RBM15 act on ORF59 expression using a similar mechanism, we compared a dose response to ORF57 and RBM15 for ORF59 expression by Western and Northern blot analyses. Ectopic ORF57 and RBM15 were found to increase the levels of ORF59 protein and mRNA in a dose-dependent manner for ORF57 but not RBM15 (Fig. 2 A and C), which is likely due to saturating levels reached above the endogenous RBM15 levels (see Fig. S1 in the supplemental material). Analysis of the fractionated RNAs revealed two distinct profiles for ORF57 and RBM15 in the accumulation of ORF59 RNA. ORF57 promoted more prominent cytoplasmic than nuclear accumulation of ORF59 RNA in two tested concentrations (Fig. 2B), whereas RBM15 enhanced the accumulation of ORF59 RNA more prominently in the nucleus than in the cytoplasm (Fig. 2D). When the relative cytoplasmic versus nuclear (C/N) ratio of ORF59 RNA was taken into account, we did not see a noticeable effect of ORF57 on ORF59 RNA export at any dose (Fig. 2B). However, we found that ectopic RBM15 led to a greater increase of nuclear ORF59 RNA than of cytoplasmic ORF59 RNA (Fig. 2D). Although both RBM15 and ORF57 act by increasing the nuclear and cytoplasmic ORF59 mRNA levels, the presence of exogenous RBM15 led to preferential nuclear accumulation of ORF59 RNA. RNA decay analyses after actinomycin D suppression of polymerase II transcription indicate that ORF57 was capable of stabilizing ORF59 RNA and RBM15 was not. In the presence of ORF57, the ORF59 RNA half-life increased from 1.7 h to 4.1 h (P value, <0.0001; see Fig. S2 in the supplemental material).
FIG. 2.
Distinct effects of ORF57 and RBM15 in promotion of ORF59 expression. HEK293 cells were cotransfected with an ORF59-FLAG expression vector (300 ng) together with the indicated amount of ORF57-FLAG or RBM15-FLAG vector. Total protein and RNA, as well as fractionated RNA, were prepared and analyzed, respectively, by Western blot (WB) and Northern blot (NB) assays. (A and C) Dose-dependent effect of ORF57 but not RBM15 on the expression of ORF59 protein and RNA. Bar graphs show relative level of ORF59 RNA in each sample from Northern blot analysis after being normalized to the level of GAPDH RNA for sample loading. (B and D) ORF57 promotes ORF59 RNA accumulation both in the cytoplasm and in the nucleus, but RBM15 preferentially promotes ORF59 RNA accumulation in the nucleus. Bar graphs below each Northern blot show the relative level of ORF59 RNA in each sample after normalization to the level of the corresponding GAPDH for sample loading. U6 results indicate the nuclear fractionation efficiency. The cytoplasmic-to-nuclear RNA ratio (C/N ratio) of ORF59 RNA in each pair of samples from the Northern blot assay was calculated after normalization to the level of the corresponding GAPDH loading control and is shown above the bar graph.
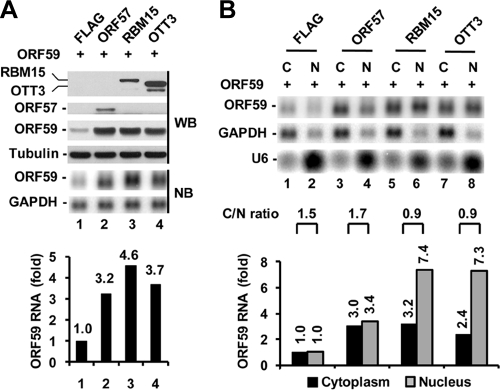
OTT3, a close member to RBM15 in the family, when used for cotransfection with ORF59, displays a function similar to that of RBM15 in the promotion of ORF59 expression. We found that OTT3 upon cotransfection noticeably enhanced the expression of both ORF59 protein and RNA (Fig. 3 A). RNA fractionation analyses revealed that OTT3-enhanced ORF59 RNA, consistent with the observations when using RBM15, preferentially accumulated in the nuclear fraction (Fig. 3B, compare lanes 7 and 8 with lanes 5 and 6 and the corresponding bar graphs), whereas the majority of ORF57-increased ORF59 RNA was found in the cytoplasmic fraction (Fig. 3B, lanes 3 and 4 and the corresponding bar graphs). Analysis of the relative C/N ratio of ORF59 indicated an efficient RNA export for ORF57-enhanced ORF59 RNA but a considerable export inefficiency for RBM15- or OTT3-increased nuclear ORF59 RNA (Fig. 3B).
FIG. 3.
OTT3 functions like RBM15 in promotion of ORF59 expression. (A) ORF59 expression in HEK293 cells 24 h after cotransfection with 300 ng of ORF59 and 100 ng of ORF57, RBM15, or OTT3. Bar graphs show relative level of ORF59 RNA in each sample from Northern blot analysis after being normalized to the level of GAPDH RNA for sample loading. (B) OTT3, which functions like RBM15, preferentially promotes ORF59 RNA accumulation in the nucleus. Bar graphs below each Northern blot show relative level of ORF59 RNA in each sample after normalization to the level of the corresponding GAPDH for sample loading. U6 results indicate nuclear fractionation efficiency. The C/N ratio of ORF59 RNA in each sample set calculated from the Northern blot is shown above the bar graph.
Endogenous RBM15 and intact RBM15 are important for ORF59 expression.
To further confirm the dependence of ORF59 on RBM15, we investigated whether endogenous RBM15 at its physiological level could affect ORF59 expression. RNAi was conducted to knock down endogenous RBM15 expression from the cells transfected with an ORF59 expression vector. Protein and RNA analyses of the cell samples demonstrated that the cells with RBM15 knockdown expressed lower levels of ORF59 protein and RNA than the cells receiving a nonspecific siRNA (Fig. 4 A). This appeared not to be an off-target effect, as the siRNA knockdown of RBM15 did not affect the expression of tubulin and GAPDH. Together, these data indicate that endogenous RBM15 plays an important role in ORF59 expression and that endogenous OTT3 cannot fully substitute for RBM15. Due to the lack of an anti-OTT3 antibody, we were unable to perform the same experiment for OTT3.
FIG. 4.
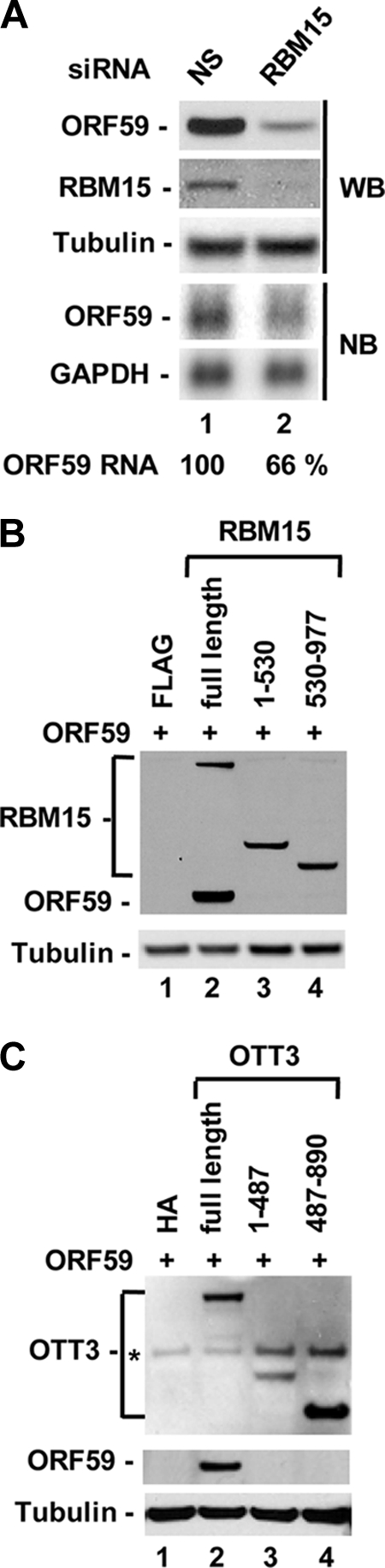
Endogenous RBM15 and full-length RBM15 or OTT3 are essential to promote ORF59 expression. (A) Endogenous RBM15 is required for ORF59 expression. HeLa cells, with or without knockdown of RBM15 expression by RNAi, were transfected with 500 ng of ORF59 expression vectors for 24 h and then examined for ORF59 and RBM15 expression by Western blot (WB) or Northern blot (NB) assays. The numbers below the Northern blots show the relative level of ORF59 RNA in each sample after normalization to the level of the corresponding GAPDH for sample loading. NS, nonspecific. (B and C) Full-length RBM15 and OTT3 are essential for ORF59 expression. HEK293 cells were cotransfected with 300 ng of ORF59-FLAG and 100 ng of full-length, N-terminal, or C-terminal RBM15-FLAG (B) or OTT3-HA (C). Protein samples were prepared 24 h after transfection and blotted with anti-FLAG or anti-HA antibodies. *, nonspecific. Tubulin served as sample loading control.
The study of deletion mutants showed that only the full-length forms of RBM15 and OTT3 were able to enhance ORF59 expression (Fig. 4B and C). Neither the N-terminal half of RBM15 or OTT3 which contains three RNA binding motifs nor their C-terminal half which interacts with proteins had any enhancement activity for ORF59 expression.
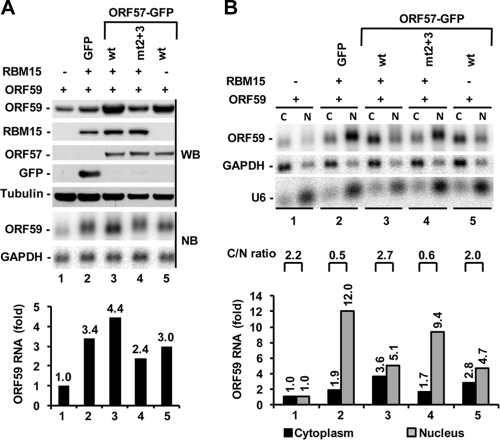
Wild-type ORF57 but not its mutant reduces RBM15 accumulation of nuclear ORF59 RNA.
Given the fact that both ORF57 and RBM15 increase the levels of ORF59 RNA (Fig. 1) with cell compartment preference (Fig. 2) and display some functional synergy in the promotion of ORF59 expression (Fig. 1), we compared wt ORF57 to its inactive mutant with point mutations in nuclear localization signals 2 and 3 (mtNLS2 + 3) in the regulation of RBM15 activity. As shown by the results in Fig. 5 A, RBM15 (lane 2) and ORF57 (lane 5) alone promoted the expression of ORF59 protein and RNA. When cotransfected, wt ORF57 (lane 3) but not its inactive mutant mtNLS2 + 3 (lane 4), which remains as a nuclear protein (22), exhibited a weak additive effect on the RBM15-enhanced expression of both ORF59 protein and RNA. The RNA fractionation analysis shown in Fig. 5B exhibited a similar correlation of the cytoplasmic ORF59 RNA level with the observed ORF59 protein and total RNA expression shown in Fig. 5A in response to RBM15 and/or ORF57. Despite the RBM15-mediated accumulation of nuclear ORF59 RNA, as expected (Fig. 5B, lane 2), the cells cotransfected with RBM15 and ORF57 showed that a large proportion of ORF59 RNA was accumulated in the cytoplasm in the presence of wt ORF57 but not in the presence of its inactive mutant mtNLS2 + 3 (Fig. 5B, compare lane 3 to lane 4 and the corresponding bar graphs), which does not interact with RBM15 and OTT3 (see Fig. 8 below). Analysis of the relative ORF59 RNA C/N ratio indicates that ectopic RBM15 specifically led to more nuclear ORF59 RNA (Fig. 5B), as shown in Fig. 2D, but the presence of ORF57 reversed the C/N ratio back to normal. These data clearly indicate that one of KSHV ORF57's functions is to reduce ectopic-RBM15-mediated nuclear accumulation of ORF59 RNA.
FIG. 5.
ORF57 offsets the nuclear RNA accumulation activity of RBM15 in promoting ORF59 expression. (A) ORF57 coordinates with RBM15 to additively promote ORF59 expression. (B) ORF57 promotes ORF59 RNA accumulation in the cytoplasm by offsetting the nuclear RNA accumulation mediated by ectopic RBM15. HEK293 cells were cotransfected with 300 ng of ORF59-FLAG in combination with or without 20 ng of RBM15-FLAG in the presence or absence of the indicated ORF57-GFP expression vector (100 ng). mtNLS2 + 3 (mt2 + 3), an inactive ORF57 mutant with point mutations in its nuclear localization signals 2 and 3, served as a control. A GFP-only plasmid was used as a vector control. Protein samples, total RNA, and fractionated RNA were prepared 24 h after cotransfection. Protein samples were examined by Western blot (WB) assay with anti-FLAG and anti-GFP antibodies (A, upper panel). Tubulin served as sample loading control. All RNA samples were analyzed by Northern blot (NB) assay (A, lower panel; B, upper panel). Bar graphs below each Northern blot in panels A and B show the relative level of ORF59 RNA in each sample after normalization to the level of the corresponding GAPDH for sample loading. U6 results show nuclear fractionation efficiency. Relative C/N ratio of ORF59 RNA in each sample set calculated from the Northern blot in panel B is shown above the bar graph.
ORF57 prevents RBM15-induced accumulation of hyperpolyadenylated nuclear ORF59 RNA.
Investigating how ectopic expression of RBM15 could lead to nuclear accumulation of ORF59 RNA, we noticed in our Northern blot analyses that the RBM15- or OTT3-induced nuclear ORF59 RNA migrated more slowly than its counterpart in the cytoplasm in electrophoresis (Fig. 3B, lanes 6 and 8, and Fig. 5B, lanes 2 and 4), indicating that the accumulated nuclear ORF59 RNA is of a larger size. Interestingly, ORF57 was found not only to reduce the RBM15-mediated nuclear accumulation of ORF59 RNA but also to eliminate the heterogeneity of ORF59 RNAs, thereby leading them to migrate as its normal size in electrophoresis (Fig. 5B, compare lane 3 to lane 5). We hypothesized that the longer nuclear ORF59 RNAs that are accumulated in the presence of RBM15 might be hyperpolyadenylated RNAs. To determine whether hyperpolyadenylation was a cause of the increase in length of nuclear ORF59 RNA, we extracted total or fractionated ORF59 RNAs from HEK293 cells 24 h after cotransfection with ORF59 and RBM15 or ORF57 and conducted RNase H digestion analyses after RNA hybridization with an oligo(dT16). As revealed by their slow migration in electrophoresis, shown in Fig. 6, the RBM15-accumulated nuclear ORF59 RNAs in the total RNA preparation (Fig. 6B, lane 5, top blot) and the nuclear fractionated RNA preparation (Fig. 6B lane 5, middle blot) had a larger size than ORF57-enhanced ORF59 RNAs (compare lanes 5 with lanes 3). However, all ORF59 RNAs analyzed by RNase H digestion, including total RNA and the fractionated nuclear ORF59 RNA, migrated by electrophoresis at the same position after the RNA poly(A) tail removal mediated by oligo(dT16) and RNase H digestion, indicating that the more slowly migrating nuclear ORF59 RNA was indeed hyperpolyadenylated in the presence of ectopic RBM15. RBM15's hyperpolyadenylation effect on ORF59 RNA in the nucleus also occurred with the inactive ORF57 mtNLS2 + 3, whose expression could be increased by ectopic RBM15 (see Fig. S3 in the supplemental material), but this effect did not occur with GAPDH RNA (Fig. 6B, compare lane 5 with lane 3 for total ORF59 and GAPDH RNA).
FIG. 6.
Determination of the RBM15-mediated hyperpolyadenylation of nuclear ORF59 RNA. (A) Strategy to determine RNA poly(A) tail length by RNase H digestion assay. Oligo(dT16) was annealed to purified RNAs, and poly(A) tails of RNAs were removed by incubation with RNase H, specifically digesting the RNA within the double-stranded DNA/RNA region. The digested RNA was separated in agarose gel and analyzed by Northern blotting. Undigested RNA served as a control. Bold lines represent mRNA transcripts with an m7G RNA cap (filled circle) on the 5′ end and a poly(A) tail (AAAAAAA) on the 3′ end. (B) Northern blot analyses of RNase H-digested total and fractionated nuclear RNA (5 μg/lane) extracted from HEK293 cells 24 h after cotransfection with 300 ng of ORF59-FLAG vector and 100 ng of ORF57, RBM15, or an empty vector. ORF59 transcripts were detected with a 32P-labeled oligonucleotide probe. Total GAPDH RNA served as a control RNA in the digestion. An, transcript with poly(A) tail; A0, transcript without poly(A) tail.
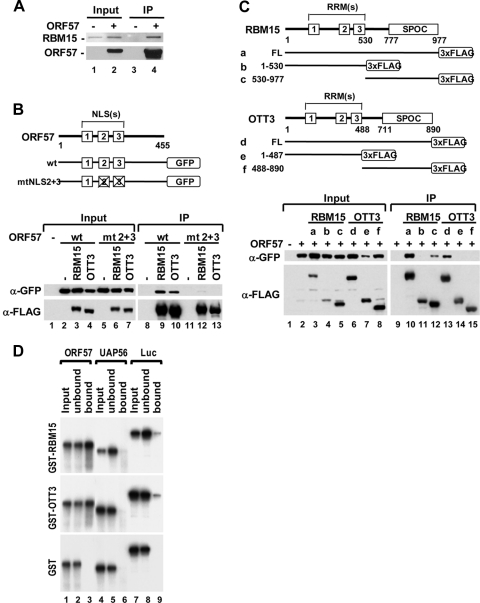
ORF57 colocalizes and interacts with RBM15 and OTT3.
Since ORF57, RBM15, and OTT3 are nuclear proteins (7, 22, 49), we visualized their expression and localization in cotransfected HeLa cells. Consistent with other reports (6, 7, 13, 49), we show that the majority of RBM15 and OTT3 localized to the nucleus. In support of the findings described above, we found that ORF57 colocalized with RBM15 and OTT3 in distinct, speckle-like nuclear structures (Fig. 7 A). The inactive ORF57 mutant (mtNLS2 + 3) lacking the ability to promote ORF59 expression (22) exhibited a diffuse distribution in the nucleus and did not display much colocalization with RBM15 (Fig. 7B). Thus, the cellular colocalization results suggest the presence of biochemical interaction of functional ORF57 with RBM15 and OTT3.
FIG. 7.
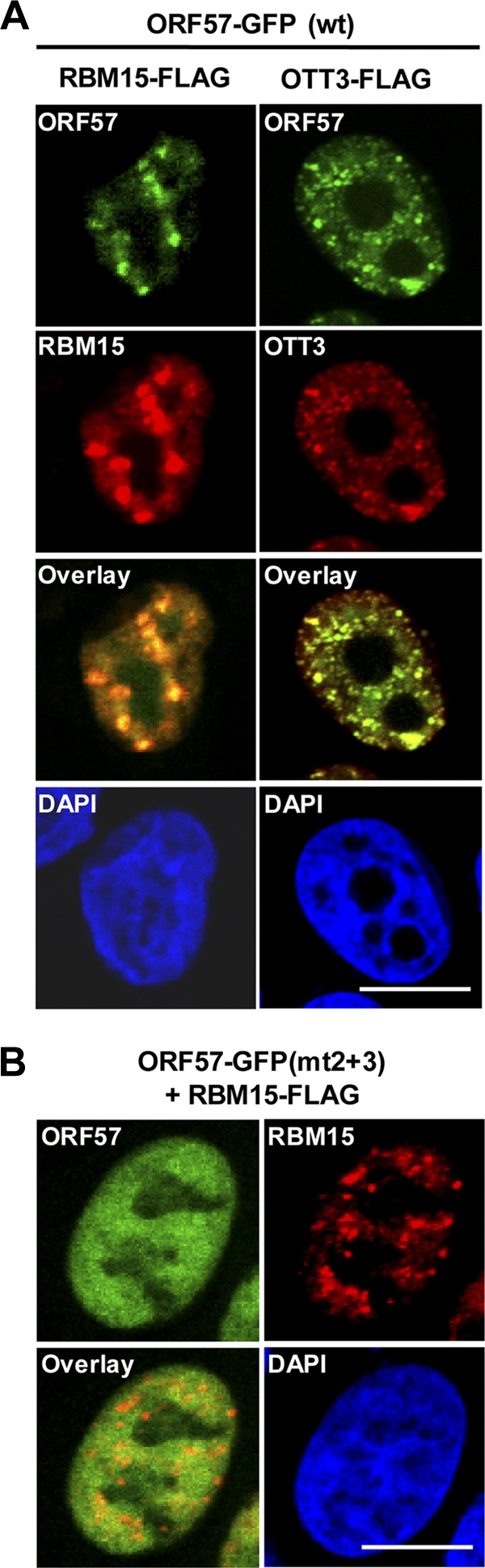
ORF57 colocalizes with RBM15 and OTT3 in vivo. (A and B) The confocal images were taken from HeLa cells 24 h after cotransfection with an equal amount (500 ng each) of wt ORF57-GFP (A) or its inactive mutant mtNLS2 + 3 (mt2 + 3) (B) and FLAG-tagged RBM15 or OTT3 constructs. Cells were stained with anti-FLAG antibody in immunofluorescent staining to visualize FLAG-tagged proteins. Cell nuclei were counterstained by DAPI. Scale bar, 10 μM.
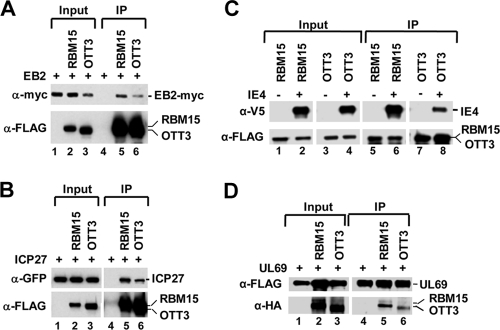
Next, we examined the possible interaction of ORF57 protein with RBM15 and OTT3. We first immunoprecipitated tagged ORF57 protein from extracts of transfected HEK293 cells. The extracts were digested with RNase A prior to IP to exclude the possibility of RNA-mediated protein-protein interaction in the IP pull-down assays. We found that, in HEK293 cells under physiological conditions (150 mM salt), ORF57 was associated with endogenous RBM15 (Fig. 8 A) but not with endogenous UAP56 (data not shown), as reported previously (2). Further studies by cotransfection of HEK293 cells showed that ORF57 was capable of interacting with both RBM15 and OTT3 tagged proteins (Fig. 8B, lanes 9 and 10). Next, we used previously characterized ORF57 mutants with point mutations in one of three nuclear localization signal (NLS) motifs (active mutants), as well as the inactive ORF57 mutant with a mutation in both NLS2 and NLS3 (mtNLS2 + 3) (22). As shown in Fig. 8B, the inactive ORF57 mutant (mtNLS2 + 3), which expresses as a nuclear protein, failed to interact with RBM15 and OTT3 (compare lanes 9 and 10 to lanes 12 and 13), confirming a correlation between ORF57 activity and its RBM15 binding capability. In contrast, the active ORF57 mutants (22) with single mutations in any one of the three individual NLS motifs also interacted with RBM15 or OTT3 (data not shown). These interactions appear to be specific to ORF57. Cellular RNA export factor Ref1-II, similar to UAP56 (13) and Y14 and 9G8 (53), was unable to coimmunoprecipitate with RBM15 or OTT3 (see Fig. S4 in the supplemental material). Mapping studies of RBM15 and OTT3 further revealed the interaction of both full-length proteins with ORF57 under high-stringency conditions (400 mM salt). We noted a relatively weaker binding of OTT3 to ORF57 (Fig. 8C, compare lane 13 to lane 10). The domain binding to ORF57 was mapped to the C-terminal region of RBM15 (Fig. 8C, lane 12). Similarly, interaction of OTT3 with ORF57 via its C-terminal region was also found when analyzed under physiological ionic strength (150 mM salt) (data not shown).
FIG. 8.
ORF57 interacts with RBM15 and OTT3. (A) ORF57 interacts with endogenous RBM15 in vivo. ORF57-FLAG expressed in HEK293 cells at 24 h after transfection was immunoprecipitated (IP) with anti-FLAG antibody. The proteins in the IP pulldowns were blotted with a polyclonal anti-RBM15 antibody or anti-ORF57. (B) RBM15 and OTT3 interact with the N-terminal nuclear localization signals (NLS) 2 and 3 of ORF57. HEK293 cell lysates were prepared 2 days after transfection and immunoprecipitated using anti-FLAG antibody. The IP complexes were blotted with anti-GFP for ORF57 or anti-FLAG antibody for RBM15 and OTT3. Shown at the top are diagrams of ORF57 proteins without (wt) or with (mtNLS2 + 3) mutation of NLS2 and NLS3. Lower panel shows interaction of wt ORF57 but not the inactive mutant mtNLS2 + 3 (mt2 + 3) with RBM15 and OTT3 in cotransfected HEK293 cells by IP-Western blotting. (C) Mapping of ORF57 interaction domains of RBM15 and OTT3. Shown at the top are the structures of wt RBM15 and OTT3 or their deletion mutant proteins. Both RBM15 and OTT3 contain an N-terminal portion with three RRMs (RNA recognition motifs) and a C-terminal SPOC domain. Shown for each protein, at a, b, and c for RBM15 and d, e, and f for OTT3, are diagrams of full-length (FL; a, d), N-terminal (b, e), and C-terminal (c, f) mutants that were cloned into a 3×FLAG vector. Numbers represent amino acid positions. Lower panel shows that RBM15 and OTT3 interact with ORF57. HEK293 cells were cotransfected with an ORF57-GFP expression vector in combination with an RBM15- or OTT3-FLAG expression vector. See more details for IP-Western blot assay in the description for panel B. (D) In vitro direct binding of ORF57 to the SPOC domain of RBM15 and OTT3. ORF57 and control proteins (UAP56 and firefly luciferase) that were expressed and metabolically labeled in reticulocyte extracts were tested for binding to E. coli-produced, purified GST-RBM15 (aa 530 to 977), GST-OTT3 (aa 488 to 890), or GST only. The 35S-radiolabeled proteins in the bound (GST pull-down) fractions and 1% aliquots of the respective input (load) and unbound fractions were analyzed by SDS-PAGE and autoradiography.
The direct interaction between RBM15 and ORF57 was further verified by in vitro GST pulldown assays using reticulocyte-produced ORF57, UAP56, or luciferase protein and E. coli-expressed, purified GST-tagged RBM15 (aa 530 to 977), GST-tagged OTT3 (aa 488 to 890), or GST alone. We confirmed that ORF57, like NXF1 (49), interacted specifically with RBM15 and OTT3, whereas UAP56 or luciferase protein did not (Fig. 8D). Collectively, these data indicate that ORF57 acts by promoting ORF59 mRNA expression through direct protein-protein interactions with RBM15 and OTT3.
The ORF57-RBM15 interaction interferes with RBM15 RNA binding.
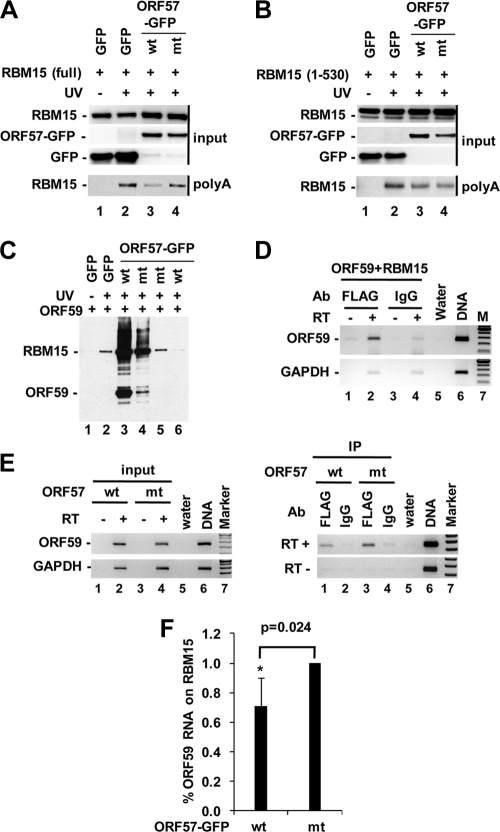
To determine whether the ORF57 colocalization and interaction with RBM15 would affect RBM15 binding to ORF59 RNA and thereby prevent ORF59 RNA from hyperpolyadenylation in the nucleus, we took two different approaches. First, we examined whether ORF57 could affect RBM15's association with polyadenylated mRNAs in HEK293 cells. As shown in Fig. 9 A, FLAG-tagged RBM15 could be UV cross-linked to polyadenylated mRNAs in the cells (compare lane 1 to lane 2), but the RBM15 cross-linking to the polyadenylated mRNAs appeared reduced in the presence of wt ORF57 over the presence of mutant ORF57 (compare lane 3 to lane 4). When a truncated RBM15 (aa 1 to 530), lacking a C-terminal SPOC domain and deficient in ORF57 interaction (Fig. 8C) but bearing all three RRMs, was used in the assay, this mutant RBM15 had the same RNA binding activity whether in the presence of wt or mutant ORF57 (Fig. 9B, compare lane 3 to lane 4). These data indicate that the ORF57 reduction of RBM15-mRNA interaction requires ORF57-RBM15 interaction. This reduction appeared to be unrelated to ORF59 protein. We found that only RBM15, and not ORF59, could be pulled down along with polyadenylated mRNAs (Fig. 9C). The associated RBM15 level on polyadenylated mRNA was reduced in the presence of wt ORF57 compared to the level in the presence of the mutant ORF57 that was incapable of interacting with RBM15 (Fig. 9C, compare lane 5 to lane 6).
FIG. 9.
ORF57 affects RBM15-RNA interactions in vivo. (A and B) ORF57-RBM15 SPOC domain interaction interferes with RBM15's association with polyadenylated RNA. HEK293 cells (5 × 105/well in a 6-well plate) were cotransfected with the following expression vectors (500 ng each): FLAG-tagged ORF59, FLAG-tagged full-length RBM15 (A) or SPOC domain deletion mutant (aa 1 to 530) (B) and GFP-tagged wild-type ORF57 (wt) or NLS2 + 3 mutant (mt). Empty vector pEGFP-N1 served as an ORF57 negative control. At 24 h after transfection, the cells were UV irradiated, and the polyadenylated [poly(A)] RNA transcripts were isolated. After RNase digestion, the proteins associated with poly(A) RNAs were detected by Western blotting by using anti-FLAG antibody for RBM15. Total cell lysates prior to poly(A) RNA selection served as input for each sample in Western blotting. (C to F) ORF57 interaction with RBM15 affects the binding of RBM15 to ORF59 RNA. HEK293 cells 24 h after cotransfection with RBM15 and ORF59 in the absence or presence of wt or mutant ORF57 were examined by poly(A) selection (C) or CLIP assays (D to F) followed by Western blot (C) or RT-PCR (D to F) analysis. (C) RBM15 protein but not ORF59 associates with poly(A) RNAs, which is preventable by wt ORF57. Lanes 3 and 4 contain cell lysate input prior to poly(A) mRNA selection, showing ectopic expression of FLAG-tagged RBM15 and ORF59. (D) RBM15 protein binds ORF59 RNA in the absence of ORF57. The RBM15-ORF59 RNA complex in the CLIP assay, after proteinase K digestion, was analyzed by RT-PCR analysis for ORF59 RNA or GAPDH RNA as a nonspecific RNA control. Preimmune rabbit serum IgG served as an IP control. RT, reverse transcriptase; M, 100-bp DNA ladder; DNA, ORF59 plasmid DNA for size control. (E and F) Coexpression of wt ORF57 with RBM15 reduces RBM15 binding to ORF59 RNA. See other details in the panel D legend. (E, left) Results show the same amount of ORF59 RNA from the input cell extracts as used for CLIP. GAPDH RNA in the extracts served as a loading control. (E, right) Results show that wt ORF57 but not ORF57 mtNLS2 + 3 prevents RBM15 association with ORF59 RNA in CLIP assays. (F) Bar graphs with means ± standard deviations show the ORF59 RNA level from each anti-FLAG CLIP, quantified by qRT-PCR and analyzed by two-tailed Student's t test. n = 2, each in triplicate. Ab, antibody.
Next, we performed CLIP assays to examine the specific effect of ORF57 on RBM15-ORF59 RNA interactions. HEK293 cells were cotransfected with FLAG-tagged RBM15 and FLAG-tagged ORF59 in the absence or presence of GFP-tagged wt or mutant ORF57 and UV irradiated. The protein-RNA complexes from the cells were coimmunoprecipitated by anti-FLAG antibody and digested with proteinase K. The RNAs in the digested complex were extracted and amplified by RT-PCR for ORF59 RNA as described previously (22). Using this strategy, we first confirmed that RBM15 binds ORF59 RNA in the cells in the absence of ORF57 (Fig. 9D, lane 2). Further analysis demonstrated that RBM15 bound ORF59 RNA in the presence of mutant ORF57, but this protein-RNA interaction was reduced in the presence of wt ORF57 (Fig. 9E, right panel, compare lane 3 to lane 1). Quantitative analysis of the ORF59 RNA levels from these two samples by qRT-PCR indicated a significant (P value, 0.024) reduction of ORF59 RNA on RBM15 protein in the presence of wt ORF57 (Fig. 9F). Collectively, these data provide compelling evidence that wt ORF57's interaction with RBM15 affects the association of RBM15 with ORF59 RNA.
The ORF57 homologs EB2, ICP27, UL69, and IE4 also interact with RBM15 and OTT3.
As EBV EB2 was reported to interact with OTT3 and RBM15 (6), we examined whether the interaction of KSHV ORF57 with RBM15 is a conserved function among other homologs in the herpesvirus family. We transfected HEK293 cells with RBM15 or OTT3 expression vectors in combination with EBV EB2 (6), HSV-1 ICP27 (38), VZV IE4 (32), or CMV UL69 (47) and analyzed the total cell extracts by co-IP. We found that EB2 (Fig. 10 A, lanes 5 and 6), ICP27 (Fig. 10B, lanes 5 and 6), IE4 (Fig. 10C, lanes 6 and 8), and UL69 (Fig. 10D, lanes 5 and 6), consistent with the results for ORF57 (Fig. 8C, lanes 10 and 13), all interact with RBM15 and OTT3. As described for ORF57 (Fig. 8C) and consistent with the report by Hiriart et al. (6), we found that EB2 interacts with the C-terminal portion of RBM15 (data not shown). These findings indicate that the interaction of RBM15 or OTT3 with ORF57 is indeed a highly conserved function among ORF57 homologs in the herpesvirus family.
FIG. 10.
The interaction with RBM15 and OTT3 proteins is conserved among ORF57 homologs. (A to C) Myc-tagged EBV EB2 (A), GFP-tagged HSV-1 ICP27 (B), and V5-tagged VZV IE4 (C) were expressed in HEK293 cells alone or together with FLAG-tagged RBM15 or OTT3, and the complexes were coimmunoprecipitated with anti-FLAG antibody under stringent salt conditions (400 mM). The IP complexes were blotted with anti-myc for EB2 (A), with anti-GFP for ICP27 (B), with anti-V5 to detect IE4 (C), and with anti-FLAG for RBM15 and OTT3 (A, B, and C, lower panels). (D) To determine the interaction of CMV UL69 with RBM15 and OTT3 in vivo, HEK293 cells were cotransfected with FLAG-tagged UL69 and HA-tagged RBM15 or HA-tagged OTT3. The protein complexes were coimmunoprecipitated with anti-FLAG antibody (UL69) under stringent salt conditions (400 mM) and blotted with anti-HA to detect RBM15 and OTT3 and with anti-FLAG to detect UL69.
EBV EB2 does not function like KSHV ORF57 to prevent RBM15-mediated nuclear accumulation of ORF59.
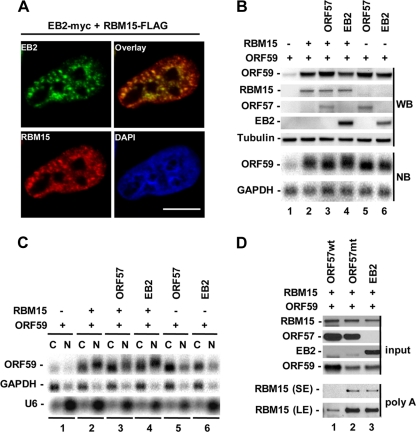
As EBV EB2 is the closest homologue to KSHV ORF57, we examined whether EB2 functions mechanistically like ORF57. We first determined EB2 colocalization with RBM15 by transient transfection of HeLa cells (Fig. 11 A) and then compared EB2 with ORF57 and RBM15 in promoting ORF59 expression. We found that EB2 protein in HEK293 cells exhibited a function similar to that of ORF57 and RBM15 in promoting ORF59 expression at both the protein and RNA level (Fig. 11B), as described previously (22), despite the fact that EB2 does not complement ORF57 function in an ORF57-null KSHV genome (4), nor does it complement ectopic RBM15 in Bac36Δ57 (our unpublished data). Further analyses of fractionated nuclear versus cytoplasmic RNA revealed that EB2, unlike ORF57, was unable to efficiently prevent RBM15-mediated accumulation of nuclear ORF59 RNA by cotransfection (Fig. 11C, compare lane 4 to lanes 2 and 3). Furthermore, EB2 exhibited no effect on RBM15's association with polyadenylated RNAs (Fig. 11D). Together, these data indicate that EB2, which interacts with RBM15 (Fig. 10A), may function differently from ORF57 in the promotion of ORF59 expression.
FIG. 11.
EBV EB2 does not prevent RBM15-mediated nuclear accumulation of ORF59 RNA and does not block RBM15-RNA interactions. (A) Colocalization of EB2-myc and RBM15-FLAG in HeLa cells by cotransfection. Scale bar, 10 μm. (B and C) EB2 promotes ORF59 expression but does not prevent RBM15-mediated nuclear accumulation of ORF59 RNA. HEK293 cells (2.5 × 105 cells/ml) were transfected with 300 ng of ORF59-FLAG with or without 20 ng RBM15-FLAG in addition to 100 ng empty vector, ORF57-FLAG, or EB2-myc. Twenty-four hours after transfection, the cells were collected for total protein and RNA analyses by Western blot (WB) and Northern blot (NB) assays, respectively (B). Fractionated cytoplasmic and nuclear RNA was also analyzed in Northern blot assays by using a 32P-labeled oligonucleotide probe specific for ORF59 RNA (C). (D) EB2 does not prevent RBM15's association with polyadenylated RNAs. HEK293 cells (5 × 105) were transfected with 500 ng each of ORF59-FLAG and RBM15-FLAG vectors together with 500 ng of an empty FLAG vector, ORF57-FLAG, or EB2-myc. Twenty-four hours after transfection, the cells were exposed to UV light and polyadenylated RNAs were isolated. RBM15 proteins bound onto polyadenylated RNAs were analyzed by Western blotting. SE, short exposure; LE, long exposure.
DISCUSSION
RNA transport from the nucleus to the cytoplasm is an essential step in the expression of mammalian and viral genes and requires several nuclear export proteins, including UAP56, URH49, Aly/REF, SRp20, 9G8, SF2/ASF, RBM15, OTT3, and the RNA export receptor NXF1/TAP (5, 8, 9, 11, 34, 46). These RNA binding and export factors, which function as RNA export adaptors, have been identified in two nuclear protein complexes, including the spliceosome and TREX complexes, to interact with the NXF1/p15 heterodimer, a major RNA exporter. Together, these complexes deliver the bound, export-ready RNA to the nuclear pore complex and, therefore, are responsible for the tightly coupled events of RNA transcription, splicing, and export in eukaryotic gene expression (15, 25, 45, 51). In this study, KSHV ORF57 was found to interact with the cellular RNA export cofactors RBM15 and OTT3. This protein-protein interaction affects the RNA binding activity of RBM15 in the nucleus and prevents RBM15-mediated nuclear accumulation of hyperpolyadenylated, intronless viral ORF59 RNA.
We have found that RBM15 and OTT3 themselves can function similarly to KSHV ORF57 to promote the expression of KSHV ORF59. The observed ability of RBM15 to promote the accumulation of ORF59 RNA in both cytoplasmic and nuclear fractions in the absence of ORF57 was unexpected since RBM15, and the related OTT3, were reported to participate in RNA export (6, 13, 49, 53). In contrast to ORF57, which promotes ORF59 expression in a dose-dependent manner, the maximum effect of ectopic RBM15 on ORF59 expression was found at a tested low dose (20 ng) that produced about 70% more RBM15 than the endogenous RBM15 level in HEK293 cells. Because RBM15 at the tested low dose also showed a moderate effect on ORF57 expression, the synergistic effect of ORF57 with RBM15 on ORF59 expression observed in this study could be the result of RBM15's effect on ORF57 expression, consistent with our previous observation that ORF57 expression depends on endogenous RBM15 and OTT3 (18). However, the function of ORF57 in ORF59 RNA expression differs from that of RBM15. The former accumulates more ORF59 RNA in the cytoplasm, but the latter accumulates the RNA more in the nucleus than in the cytoplasm. Therefore, ORF57 has a greater effect than RBM15 on the net outcome of ORF59 protein production when cotransfected at 20 ng.
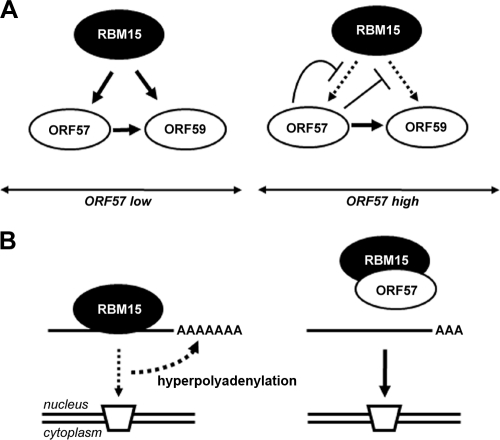
As the increase of ORF59 RNA expression by RBM15 is at the posttranscriptional level and RBM15 was unable to stabilize ORF59 RNA in this study, the cytoplasmic accumulation of ORF59 RNA by RBM15 could result from RBM15's promotion of RNA export (13, 18, 49). However, when ectopically expressed beyond its physiological level in the absence of ORF57, RBM15 adversely affects the RNA export machinery, leading to nuclear accumulation of ORF59 RNA, while a small proportion of the RNA remains exportable (Fig. 12). The effect of overexpressed RBM15 on ORF59 RNA hyperpolyadenylation also occurred with the ORF57 mtNLS2 + 3, which is a functionally inactive nuclear protein and lacks interaction with RBM15 and OTT3 (Fig. 8B). Other studies show that excess UAP56, another RNA export factor, inhibits the recruitment of Aly/REF to the spliced mRNP (15) and is toxic in C. elegans (17) and yeast (44). The mechanism by which RBM15 promotes the nuclear accumulation of ORF59 is now partially understood due to the findings in this report. We observed that the nuclear ORF59 RNA in the presence of ectopic RBM15 migrated slightly more slowly in electrophoresis due to its hyperpolyadenylation. Although it remains to be investigated whether this modification of ORF59 RNA is mediated directly by RBM15 or is the consequence of RBM15-mediated nuclear accumulation of ORF59 RNA, it is discernible that ORF57 can prevent ORF59 RNA from hyperpolyadenylation in the presence of ectopic RBM15 and reduce the RNA accumulation in the nucleus (Fig. 5B). Given the facts that RBM15 and OTT3 are RNA binding proteins (13, 49) and that the endogenous level of RBM15 promotes only limited expression of ORF59 protein in the absence of ORF57, our data indicate that RBM15 and OTT3 act as molecular links for ORF57, consistent with our previous reports of ORF57 binding specifically to its target RNAs only in the presence of cellular factors (22, 23) (Fig. 12). The inherent advantage of studying the KSHV ORF59 mRNA regulation rather than the previously described RTE-containing transcript (13, 49) lies in the fact that the ORF59 mRNA requires both cellular (RBM15 and OTT3) and viral (ORF57) factors, whereas RTE requires only cellular factors (RBM15, OTT3, and NXF1), in order to efficiently export from the nucleus. The ability to dissect the specific contributions of both the viral and cellular factors in ORF59 regulation contributes to our understanding of RNA export.
FIG. 12.
Proposed model for the effects of RBM15 and ORF57 on ORF59 expression. (A) RBM15 at physiological level promotes the expression of ORF59 at the early stage of KSHV lytic replication when ORF57 is at low concentration. Although ORF57 promotes ORF59 expression, ORF57 expression is also dependent on RBM15 (18). At a later stage of infection when ORF57 is high, its interaction with RBM15 prevents RBM15's association with ORF59 RNA and, consequently, reduces the function of RBM15 to promote ORF59 RNA export. ORF57 accumulates ORF59 RNA by increasing ORF59 RNA stability by an unknown mechanism. (B) Overexpression of RBM15 preferentially increases the nuclear over the cytoplasmic level of ORF59 RNA and stimulates hyperpolyadenylation of nuclear ORF59 RNA. In the presence of ORF57, ORF57 interacts with RBM15 and prevents the RBM15-mediated nuclear accumulation of ORF59 RNA.
ORF57 protein interacts with RBM15 and OTT3. The N-terminal NLS2 and NLS3 regions of ORF57 (22) are important for the high-affinity interaction with full-length RBM15 and OTT3. ORF57 interacts with the C-terminal regions of RBM15 and OTT3, which were previously shown to mediate interaction with the nuclear RNA export receptor NXF1 (13, 49). Interestingly, in addition to the interaction of the RBM15 or OTT3 protein with KSHV ORF57 in this study and the previously reported interaction with EBV EB2 (6), we found that posttranscriptional regulators of the other herpesvirus family members, such as the HSV-1 ICP27, the VZV IE4, and the CMV UL69, interact with the RBM15 or OTT3 proteins. The conservation of these interactions further supports their biological relevance. In vivo, we found ORF57 to colocalize with RBM15 and OTT3, and several reports have shown that these factors, when exogenously expressed, also colocalize with SC35 (1, 21, 49). Our previous studies demonstrated that ORF57 mainly associates with cellular protein-RNA complexes (23). Recently, we showed that ORF57-RBM15 interaction is required for ORF57 expression and functions in KSHV-infected cells (18). However, RBM15 is incapable of complementing ORF57's function in stable Bac36Δ57 cells containing an ORF57-null KSHV genome (our unpublished data). In this report, the biological relevance of the ORF57 interaction with RBM15 or OTT3 was further supported by the observation that ORF57-RBM15 interactions interfered with RBM15 binding to polyadenylated mRNAs and ORF59 RNA and, therefore, prevented the RBM15-/OTT3-mediated nuclear accumulation of ORF59 RNA. Interestingly, we found that EBV EB2, although it interacts with RBM15 and OTT3, does not function like KSHV ORF57 to prevent RBM15-RNA interaction and, thereby, RBM15-mediated nuclear accumulation of ORF59. Despite the fact that ORF57 interacts with RNA export factors Aly/REF (2, 22, 24) and CBP80 (2) and other cellular factors (23), the ORF57-Aly/REF interaction was found not to be important for ORF57-mediated ORF59 expression (22, 29). Thus, the finding that ORF57 interacts with RNA export cofactors RBM15 and OTT3 and offsets the RBM15- and OTT3-mediated nuclear accumulation of ORF59 RNA provides further insight into how ORF57 promotes the expression of intronless viral genes.
Supplementary Material
Acknowledgments
We thank R. Sandri-Goldin, S. Swaminathan, C. Sadzot, and T. Stamminger for plasmids and A. S. Zolotukhin for discussions. We also thank David Liewehr and Seth Sternberg for statistical analyses.
This study was supported by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print on 24 November 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bello, L. J., et al. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80:3207-3215. [DOI] [PubMed] [Google Scholar]
- 2.Boyne, J. R., K. J. Colgan, and A. Whitehouse. 2008. Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 4:e1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doroquez, D. B., T. L. Orr-Weaver, and I. Rebay. 2007. Split ends antagonizes the Notch and potentiates the EGFR signaling pathways during Drosophila eye development. Mech. Dev. 124:792-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han, Z., and S. Swaminathan. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 80:5251-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hautbergue, G. M., M. L. Hung, A. P. Golovanov, L. Y. Lian, and S. A. Wilson. 2008. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. U. S. A. 105:5154-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiriart, E., et al. 2005. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J. Biol. Chem. 280:36935-36945. [DOI] [PubMed] [Google Scholar]
- 7.Juillard, F., et al. 2009. Epstein-Barr virus protein EB2 contains an N-terminal transferable nuclear export signal that promotes nucleocytoplasmic export by directly binding TAP/NXF1. J. Virol. 83:12759-12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapadia, F., A. Pryor, T. H. Chang, and L. F. Johnson. 2006. Nuclear localization of poly(A)+ mRNA following siRNA reduction of expression of the mammalian RNA helicases UAP56 and URH49. Gene 384:37-44. [DOI] [PubMed] [Google Scholar]
- 9.Kota, K. P., S. R. Wagner, E. Huerta, J. M. Underwood, and J. A. Nickerson. 2008. Binding of ATP to UAP56 is necessary for mRNA export. J. Cell Sci. 121:1526-1537. [DOI] [PubMed] [Google Scholar]
- 10.Kuroda, K., et al. 2003. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18:301-312. [DOI] [PubMed] [Google Scholar]
- 11.Lai, M. C., Y. H. Lee, and W. Y. Tarn. 2008. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell 19:3847-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, H. V., et al. 2003. Splits ends is a tissue/promoter specific regulator of Wingless signaling. Development 130:3125-3135. [DOI] [PubMed] [Google Scholar]
- 13.Lindtner, S., et al. 2006. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J. Biol. Chem. 281:36915-36928. [DOI] [PubMed] [Google Scholar]
- 14.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 15.Luo, M. L., et al. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413:644-647. [DOI] [PubMed] [Google Scholar]
- 16.Ma, Z., et al. 2001. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 28:220-221. [DOI] [PubMed] [Google Scholar]
- 17.MacMorris, M., C. Brocker, and T. Blumenthal. 2003. UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA 9:847-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majerciak, V., M. Deng, and Z. M. Zheng. 2010. Requirement of UAP56, URH49, RBM15, and OTT3 in the expression of Kaposi sarcoma-associated herpesvirus ORF57. Virology 407:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majerciak, V., M. Kruhlak, P. K. Dagur, J. P. McCoy, Jr., and Z. M. Zheng. 2010. Caspase-7 cleavage of Kaposi sarcoma-associated herpesvirus ORF57 confers a cellular function against viral lytic gene expression. J. Biol. Chem. 285:11297-11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majerciak, V., N. Pripuzova, J. P. McCoy, S. J. Gao, and Z. M. Zheng. 2007. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J. Virol. 81:1062-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majerciak, V., et al. 2008. Kaposi's sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J. Virol. 82:2792-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majerciak, V., K. Yamanegi, S. H. Nie, and Z. M. Zheng. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 281:28365-28378. [DOI] [PubMed] [Google Scholar]
- 23.Majerciak, V., and Z. M. Zheng. 2009. Kaposi's sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front. Biosci. 14:1516-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik, P., D. J. Blackbourn, and J. B. Clements. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279:33001-33011. [DOI] [PubMed] [Google Scholar]
- 25.Masuda, S., et al. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19:1512-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mercher, T., et al. 2001. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 98:5776-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray, E. L., and D. R. Schoenberg. 2008. Assays for determining poly(A) tail length and the polarity of mRNA decay in mammalian cells. Methods Enzymol. 448:483-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nappi, F., et al. 2001. Identification of a novel posttranscriptional regulatory element by using a rev- and RRE-mutated human immunodeficiency virus type 1 DNA proviral clone as a molecular trap. J. Virol. 75:4558-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nekorchuk, M., Z. Han, T. T. Hsieh, and S. Swaminathan. 2007. Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J. Virol. 81:9990-9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oswald, F., et al. 2002. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 21:5417-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oswald, F., et al. 2005. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell Biol. 25:10379-10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ote, I., et al. 2009. Varicella-zoster virus IE4 protein interacts with SR proteins and exports mRNAs through the TAP/NXF1 pathway. PLoS ONE. 4:e7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinol-Roma, S., S. A. Adam, Y. D. Choi, and G. Dreyfuss. 1989. Ultraviolet-induced cross-linking of RNA to proteins in vivo. Methods Enzymol. 180:410-418. [DOI] [PubMed] [Google Scholar]
- 34.Pryor, A., et al. 2004. Growth-regulated expression and G0-specific turnover of the mRNA that encodes URH49, a mammalian DExH/D box protein that is highly related to the mRNA export protein UAP56. Nucleic Acids Res. 32:1857-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raffel, G. D., et al. 2009. Ott1 (Rbm15) is essential for placental vascular branching morphogenesis and embryonic development of the heart and spleen. Mol. Cell Biol. 29:333-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raffel, G. D., et al. 2007. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc. Natl. Acad. Sci. U. S. A. 104:6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rebay, I., et al. 2000. A genetic screen for novel components of the Ras/mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154:695-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandri-Goldin, R. M. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13:5241-5256. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber, S. L., A. Preiss, A. C. Nagel, I. Wech, and D. Maier. 2002. Genetic screen for modifiers of the rough eye phenotype resulting from overexpression of the Notch antagonist hairless in Drosophila. Genesis 33:141-152. [DOI] [PubMed] [Google Scholar]
- 40.Sergeant, A., H. Gruffat, and E. Manet. 2008. The Epstein-Barr virus (EBV) protein EB is an mRNA export factor essential for virus production. Front. Biosci. 13:3798-3813. [DOI] [PubMed] [Google Scholar]
- 41.Shi, Y., et al. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 15:1140-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smulevitch, S., et al. 2006. RTE and CTE mRNA export elements synergistically increase expression of unstable, Rev-dependent HIV and SIV mRNAs. Retrovirology 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smulevitch, S., et al. 2005. Structural and functional analysis of the RNA transport element, a member of an extensive family present in the mouse genome. J. Virol. 79:2356-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413:648-652. [DOI] [PubMed] [Google Scholar]
- 45.Strasser, K., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 46.Tintaru, A. M., et al. 2007. Structural and functional analysis of RNA and TAP binding to SF2/ASF. EMBO Rep. 8:756-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toth, Z., and T. Stamminger. 2008. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front. Biosci. 13:2939-2949. [DOI] [PubMed] [Google Scholar]
- 48.Tretyakova, I., et al. 2005. Nuclear export factor family protein participates in cytoplasmic mRNA trafficking. J. Biol. Chem. 280:31981-31990. [DOI] [PubMed] [Google Scholar]
- 49.Uranishi, H., et al. 2009. The RNA binding motif protein 15B (RBM15B/OTT3) acts as co-factor of the nuclear export receptor NXF1. J. Biol. Chem. 284:26106-26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiellette, E. L., et al. 1999. spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development 126:5373-5385. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]
- 52.Zolotukhin, A. S., et al. 2008. The RNA transport element RTE is essential for IAP LTR-retrotransposon mobility. Virology 377:88-99. [DOI] [PubMed] [Google Scholar]
- 53.Zolotukhin, A. S., et al. 2009. Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res. 37:7151-7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.