Abstract
Background & objectives:
Hepatitis E is the main cause of enterically transmitted non-A, non-B hepatitis in developing countries. In the developed countries such as the USA, Japan and Taiwan, the viruses infecting humans and swine share the same genotype with a high sequence similarity. Genotype 1 circulates in humans whereas genotype 4 in pigs in India. The present study was designed to investigate the presence of anti-HEV antibodies and HEV-RNA in swine population from north India, to investigate the genotype prevalent in it, and to compare it with other swine and human HEV strains from India.
Methods:
A total of 67 serum samples were collected from pigs of age period (1-6 months) from Indian Veterinary Research Institute (IVRI), Izatnagar, Bareily and subjected to anti-HEV IgG and HEV RNA detection. A phylogenetic tree was constructed using the neighbor-joining method and evaluated using the interior branch test method with MEGA 4 software.
Results:
Anti-HEV IgG and HEV RNA was found in 38.8 and 4.5 per cent of swine samples studied respectively. The above samples were observed to be of genotype 4e. The three new sequences had nucleotide similarity with other swine sequences in genotype 4 ranging from 80-98 per cent.
Interpretation & conclusions:
The three sequences observed in the present study showed nucleotide similarity with other swine sequences from southern and western India. The present study suggests that genotype 4 ‘e’ is prevalent in the north India.
Keywords: Genotype, hepatitis E virus , phylogenetic analysis, swine
Hepatitis E is the main cause of enterically transmitted non-A, non-B hepatitis in developing countries. Hepatitis E virus (HEV) is a member of the genus Hepevirus. It is a non-enveloped, single-stranded RNA virus of approximately 7.2 kb in length1. Its genome is encoded by 3 separate but partially overlapping open reading frames (ORFs)2. ORF1 likely encodes non-structural viral proteins, ORF2 encodes the putative capsid protein and ORF3 encodes a cytoskeleton-associated phosphoprotein3–5. Four major genotypes of mammalian HEV have been identified on the basis of complete genome sequences6. Genotype 1 includes human isolates from Asia and North America, genotype 2 comprises human isolates from Mexico and some African countries, genotypes 3 and 4 include human and swine strains isolated in industrialized countries as well as developing areas.
HEV-RNA and antibodies to HEV have been found in a wide variety of animals, especially swine7–10. It was hypothesized that zoonosis was involved in the transmission of HEV, especially for the cases in non-endemic areas. Studies by Meng et al11–13 provided initial evidence for the possibility of such spread in US. Subsequently, circulation of swine HEV was documented in several countries such as Taiwan14, Japan15, The Netherlands16, Canada17 and India18.
In countries such as the USA, Japan and Taiwan, the viruses infecting humans and swine share the same genotype with a high sequence similarity14–16. However, studies from India reported that genotype 1 circulates in humans whereas genotype 4 in pigs18–20. The aim of the present study was to investigate the presence of anti-HEV antibodies and HEV-RNA in swine population from north India, to investigate the genotype prevalent in swine, and to compare it with other swine and human HEV strains from India and different areas of the world.
Material & Methods
Samples: A total of 67 serum samples were collected from pigs of age period (1-6 months) from Indian Veterinary Research Institute (IVRI), Bareily, India in July 2005. The serum samples were stored at -40°C until tested for anti-HEV IgG and HEV RNA.
ELISA for anti-HEV IgG: All serum samples were thawed at room temperature and tested with IgG anti-HEV ELISA kits (Genelabs Diagnostics, Singapore). This commercially available assay is based on the ORF2 and ORF3 recombinant proteins of the Burmese and Mexican strains of HEV. The ELISA was performed according to the protocols provided by the manufacturer. All the samples were assayed in duplicate.
RNA extraction and reverse transcription polymerase chain reaction: RNA was extracted from 100 µl of serum sample by using TRIZOL reagent (Invitrogen, USA) in accordance with the manufacturer’s protocol in PCR Hepatitis Lab, Department of Medicine, Maulana Azad Medical College, New Delhi. The viral RNA was finally dissolved in 20 µl Rnase-free water. The nested PCR was performed in all the samples using primers for genotypes 1 and 4, as these two genotypes have been reported from India18–21. The primers used for genotype 1 were external sense: 5'- CCG GAT CCA CAC ACA TCT GAG CTA CAT TCG TGA GCT- 3', external anti-sense: 5'- CCG AAT TCA AAG GCA TCC ATG GTG TTT GAG AAT GAC- 3', internal sense: 5'- GGA ATT CGA CTC CAC CCA GAA TTA CTT- 3', and internal anti-sense 5'- GGA ATT CAC AGC CGG CGA TCA GGA CAG- 3'. These two sets of primers were designed to produce 343 bp segment of ORF1 region21. The primers used for genotype 4 were external sense: 5'- AAT ACA CCT TAC ACT GGC GCC CT- 3', external anti-sense: 5'- TCA GCA AGA TTA AAT AAG GTC AGC GC- 3', internal sense: 5'- ACA CTG GCG CCC TCG GTC TGC T- 3', and internal anti-sense 5'- AGA TTA AAT AAG GTC AGC GCT ATA CCA C- 3'. These two sets of primers were designed to produce 246 bp segment of ORF2 region18.
The parameters for first-round PCR for both the sets of primers for genotypes 1 and 4 included initial denaturation 95°C for 5 min, followed by 35 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 50°C, extension for 1 min at 72°C and a final incubation at 72°C for 7 min. The parameters for the second-round PCR were similar.
Nucleotide sequencing and phylogenetic analysis: The nested PCR products were purified using QIA quick PCR purification Kit (QIAGEN, Inc. Germany). The purified DNA was subjected to direct sequencing. The 220-nt consensus sequences were aligned using CLUSTAL W programme and phylogenetic tree was constructed using the neighbor-joining method and evaluated using the interior branch test method with MEGA 4 software22. Prototype HEV strains used as references in the analysis and their Gen Bank accession numbers are shown in the Fig. The sequences determined in this study were deposited in Gen Bank database under the accession numbers EU003603, EU003604 and EU003605.
Fig.
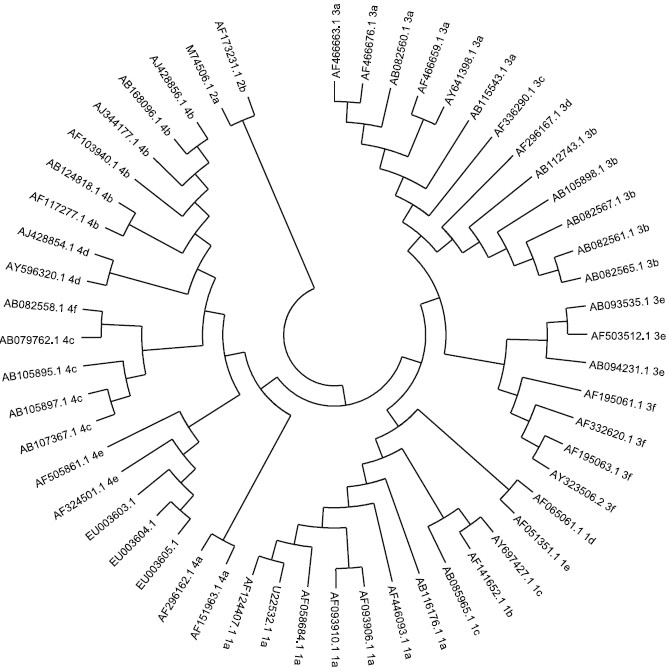
Phylogenetic relationship among swine and human strains of hepatitis E virus (HEV) representing the 4 major genotypes, based on 230 nucleotide fragment of ORF2 of the genome.
The study protocol was approved by the Institute Animal Ethical Committee of Maulana Azad Medical College, New Delhi.
Results
Of the 67 samples tested for IgG anti-HEV in the first round of ELISA, 26 samples were positive and 3 samples were in gray zone/cut-off index, however, when the test was repeated, these three samples showed negative results. Therefore, the overall presence of anti-HEV IgG in swine samples tested was found to be 38.8 per cent.
Using the primers for genotype 1, none of the 67 samples showed the presence of HEV RNA, which suggested absence of HEV genotype 1 infection among these swine samples. However, when the primers of genotype 4 were used, three samples (4.5%) showed the presence of HEV RNA. The occurrence of IgG anti-HEV and HEV RNA in the serum samples of swine at different months of age is shown in Table I. The serum samples positive for HEV RNA were also positive for anti-HEV IgG.
Table I.
Detection of IgG anti-HEV and HEV RNA in serum samples of pigs of various ages
| Age (months) | No. of samples | IgG anti-HEV N (%) | HEV RNA N (%) |
|---|---|---|---|
| 1-3 | 13 | 3 (23.1) | 2 (5.7) |
| 3-5 | 19 | 8 (42.1) | 1 (5.3) |
| >5 | 35 | 15 (42.8) | 0 (0) |
| Total | 67 | 26 (38.8) | 3 (4.5) |
The amplified PCR product was confirmed to be of HEV strain by direct sequence analysis using the BLAST programme. Partial sequences of 230 bp of HEV ORF2 were compared with others from the known genotypes and were found to cluster in genotype 4e group. The three sequences of the present study had nucleotide similarity (Table II) with other swine sequences in genotype 4 ranging from 80.6-97.8 per cent7,18,19. The closest relationship (87.3%) between these sequences and human strains in genotype 4 was between HEVS and Chinese and Japan human isolates. All the sequences formed single clustered of subgroup ‘e’ in the genotype 4.
Table II.
Nucleotide similarity of 220-base-pair fragment of open reading frame 2 of the HEV genome of swine and human strains of different genotypes, as compared with 3 strains isolated in this study
| Type | Isolate | Country | Host | Nucleotide identity per cent (%) |
||
|---|---|---|---|---|---|---|
| HEVS | HEVS1 | HEVS2 | ||||
| 4e* | EU003605 | India (North) | Pig | 97 | 97 | |
| 4e* | EU003604 | India (North) | Pig | 97 | - | 98.3 |
| 4e* | EU003603 | India (North) | Pig | 97 | 98.3 | - |
| 1a | AF124407 | India | Human | 72.8 | 72.6 | 71.6 |
| 1a | AF446093 | India | Human | 74.6 | 74.3 | 73.3 |
| 1a | AF058684 | Spain | Sewage | 73.2 | 73 | 72 |
| 1a | AF093906 | India | Human | 72.8 | 72.6 | 71.6 |
| 1a | U22532 | India | Human | 74.1 | 73.9 | 72.8 |
| 1a | AB116176 | Nepal | Human | 72.4 | 72.2 | 71.1 |
| 1b | AF141652 | China | Human | 74.6 | 74.3 | 73.3 |
| 1c | AB085965 | Nepal | Human | 73.7 | 74.8 | 73.7 |
| 1c | AY697427 | Kyrgyzstan | Human | 74.6 | 74.8 | 73.7 |
| 1d | AF065061 | Morocco | Human | 73.2 | 73 | 72 |
| 1e | AF051351 | Egypt | Human | 72.8 | 71.7 | 70.7 |
| 2a | M74506 | Mexico | Human | 72.0 | 71.8 | 70.8 |
| 2b | AF173231 | Nigeria | Human | 44.5 | 46 | 45.1 |
| 3a | AF466676 | US | Pig | 67.2 | 66.8 | 67.2 |
| 3a | AF466663 | US | Pig | 75.7 | 73.7 | 73.5 |
| 3a | AB082560 | Japan | Human | 77.2 | 75.2 | 75 |
| 3a | AB115543 | Japan | Human | 68.9 | 68 | 68.5 |
| 3a | AY641398 | Korea | Human | 75 | 73.9 | 72.8 |
| 3a | AF466667 | US | Pig | 77 | 74.6 | 74.8 |
| 3b | AB082567 | Japan | Human | 73.7 | 72.6 | 71.6 |
| 3b | AB112743 | Japan | Human | 68 | 67.2 | 67.6 |
| 3b | AB105898 | Japan | Pig | 74.3 | 72.8 | 71.8 |
| 3c | AF336290 | Netherlands | Pig | 67.9 | 67.9 | 68.7 |
| 3d | AF296167 | Taiwan | Pig | 72.2 | 71.1 | 70.1 |
| 3e | AF503512 | UK | Pig | 69.3 | 67.5 | 68.9 |
| 3e | AB093535 | Japan | Human | 67.6 | 66.7 | 67.2 |
| 3e | AB094231 | Japan | Pig | 67.6 | 66.7 | 67.2 |
| 3f | AF332620 | Netherlands | Pig | 68.9 | 67.6 | 68.5 |
| 3f | AF195061 | Spain | Human | 68.9 | 68.9 | 68.5 |
| 3f | AF195063 | Spain | Human | 69.7 | 68.7 | 69.3 |
| 3f | AY323506 | Spain | Pig | 68.9 | 68.5 | 68.9 |
| 4a | AF151963 | China | Human | 83.8 | 83 | 82.3 |
| 4a | AF296162 | Taiwan | Human | 81.6 | 80.9 | 80.2 |
| 4b | AJ344177 | China | Human | 87.3 | 86.5 | 84.9 |
| 4b | AB168096 | Japan | Human | 87.3 | 85.7 | 84.9 |
| 4b | AF103940 | China | Human | 87.3 | 85.7 | 84.9 |
| 4b | AB124818 | Indonesia | Pig | 85.1 | 84.3 | 82.8 |
| 4b | AJ428856 | China | Pig | 86.4 | 84.8 | 84.1 |
| 4b | AF117277 | Taiwan | Human | 86 | 84.3 | 83.6 |
| 4c | AB105895 | Japan | Human | 84.2 | 83.5 | 82.8 |
| 4c | AB107367 | Japan | Human | 84.2 | 83.5 | 82.8 |
| 4c | AB079762 | Japan | Human | 82.9 | 81.3 | 80.6 |
| 4c | AB105897 | Japan | Human | 84.6 | 83.9 | 83.2 |
| 4d | AY596320 | China | Pig | 81.1 | 82.2 | 80.6 |
| 4d | AJ428854 | China | Pig | 83.3 | 84.3 | 82.8 |
| 4e | AF324501 | India (West) | Pig | 97.4 | 97.8 | 96.1 |
| 4e | AF505861 | India (South) | Pig | 93.9 | 91.7 | 90.9 |
| 4f | AB082558 | Japan | Human | 83.3 | 81.7 | 81 |
Present study.
Source: The nucleotide sequences of above HEV isolates were retrieved from Genbank
Discussion
Anti-HEV antibodies have been shown among pigs and other animals in several HEV-endemic and non-endemic countries, including India7,8,10. Pigs stand out as being an animal group with the highest rate of anti-HEV seropositivity. In the present study, the anti-HEV IgG positivity (38.8%) among pigs was somewhat lower than 43-74.4 per cent (western & south India) and 97.5 per cent (Lucknow) reported previously among Indian pigs10,18,19. This may be due to the difference in age of pigs at which the samples have been drawn. In the present study, it appears that anti-HEV IgG positivity increased with increasing age of pigs.
Serum samples from pigs older than 5 months were tested negative, similar to the study from Lucknow10, which showed presence of HEV RNA in only one of the 200 serum samples collected from adult pigs. However, infection with HEV is associated with a short time of detectable HEV RNA in serum, which is followed by development of anti-HEV antibodies that may last for a long time and pre-existing anti-HEV IgG can prevent HEV viraemia23. Thus, detection of HEV RNA is less likely in older pigs than in the younger24,25.
The strongest evidence in favour of animal-to-human transmission of a pathogen is provided by an identity or close resemblance of isolates from these sources. Swine-to-human transmission hypothesis for HEV was supported by accumulated evidences26–29. The most direct evidence of animal-to-human transmission of HEV came from Japan, where four human cases of hepatitis E were linked to the consumption of uncooked deer meat, based on 99.7-100 per cent nucleotide sequence homology between the virus recovered from patients and the left-over meat15. In the present study, the three sequences were phylogenetically related to the genotype 4 and shared 71.6-74.6 per cent homology with human isolates of India, based on the partial ORF2 sequences. The sequences showed least nucleotide homology with genotype 2 ‘b’ which ranged from 44.5-46 per cent.
In the present study, the strains phylogenetically clustered into genotype 4 and formed single subgroup ‘e’, sharing 90.9-97.8 per cent homology with inter-subgroup and 80.2-83.8, 82.8-87.3, 80.6-84.6, 80.6-84.3, and 81-83.3 per cent intra-subgroup identity homology with subgroup ‘a’, ‘b’, ‘c’, ‘d’ and ‘f’ respectively. Moreover, the present sequences showed 80.2-81.9 per cent homology with the only swine sequence reported from Lucknow19. This suggests that in north India, different subgroups may be present; such high difference between the nucleotide identities of swine sequences is not observed in west and south India.
In conclusion, the study confirms the circulation of genotype 4 ‘e’ in swine from north India similar to southern and western India suggesting genotype 4 ‘e’ to be predominant in Indian pigs. Other subgroups may also be present which can only be identified by sequencing more samples from north India.
Acknowledgments
The authors are grateful to staff of Indian Veterinary Research Institute, Izatngar, Bariely for providing the swine serum samples.
References
- 1.Reyes GR, Purdy MA, Kim JP, Luk KC, Young LM, Fry KE, et al. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247:1335–9. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 2.Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, et al. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–31. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jameel S, Zafrullah M, Ozdener MH, Panda SK. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J Virol. 1996;70:207–16. doi: 10.1128/jvi.70.1.207-216.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li TC, Yamakawa Y, Suzuki K, Tatsumi M, Razak MA, Uchida T, et al. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J Virol. 1997;71:7207–13. doi: 10.1128/jvi.71.10.7207-7213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zafrullah M, Ozdener MH, Panda SK, Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol. 1997;71:9045–53. doi: 10.1128/jvi.71.12.9045-9053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purcell RH, Emerson SU. Hepatitis E virus. In: Knipe DM, Howley PM, editors. Field virology. 4th ed. Philadelphia, PA, USA: Lippincott Williams & Wilkins; 2003. pp. 3051–62. [Google Scholar]
- 7.Wang YC, Zhang HY, Xia NS, Peng G, Lan HY, Zhuang H, et al. Prevalence, isolation, and partial sequence analysis of hepatitis E virus from domestic animals in China. J Med Virol. 2002;67:516–21. doi: 10.1002/jmv.10131. [DOI] [PubMed] [Google Scholar]
- 8.Goens SD, Perdue ML. Hepatitis E viruses in humans and animals. Anim Health Res Rev. 2004;5:145–56. doi: 10.1079/ahr200495. [DOI] [PubMed] [Google Scholar]
- 9.Saad MD, Hussein HA, Bashandy MM, Kamel HH, Earhart KC, Fryauff DJ, et al. Hepatitis E virus infection in work horses in Egypt. Infect Genet Evol. 2007;7:368–73. doi: 10.1016/j.meegid.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Shukla P, Chauhan UK, Naik S, Anderson D, Aggarwal R. Hepatitis E virus infection among animals in northern India: an unlikely source of human disease. J Viral Hepat. 2007;14:310–7. doi: 10.1111/j.1365-2893.2006.00815.x. [DOI] [PubMed] [Google Scholar]
- 11.Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, et al. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–5. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng XJ, Halbur PG, Shapiro MS, Govindarajan S, Bruna JD, Mushahwar IK, et al. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J Virol. 1998;72:9714–21. doi: 10.1128/jvi.72.12.9714-9721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng XJ, Dea S, Engle RE, Friendship R, Lyoo YS, Sirinarumitr T, et al. Prevalence of antibodies to the hepatitis E virus in pigs from countries where hepatitis E is common or is rare in the human population. J Med Virol. 1999;59:297–302. [PubMed] [Google Scholar]
- 14.Hsieh SY, Meng XJ, Wu YH, Liu ST, Tam AW, Lin DY, et al. Identity of a novel swine hepatitis E virus in Taiwan forming a monophyletic group with Taiwan isolates of human hepatitis E virus. J Clin Microbiol. 1999;37:3828–34. doi: 10.1128/jcm.37.12.3828-3834.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishizawa T, Takahashi M, Mizuo H, Miyajima H, Gotanda Y, Okamoto H. Characterization of Japanese swine and human hepatitis E virus isolates of genotype IV with 99 % identity over the entire genome. J Gen Virol. 2003;84:1245–51. doi: 10.1099/vir.0.19052-0. [DOI] [PubMed] [Google Scholar]
- 16.van der Poel WH, Verschoor F, van der Heide R, Herrera MI, Vivo A, Kooreman M, et al. Hepatitis E virus sequences in swine related to sequences in humans, The Netherlands. Emerg Infect Dis. 2001;7:970–6. doi: 10.3201/eid0706.010608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei Y, Yoo D. Genetic characterization and sequence heterogeneity of a canadian isolate of Swine hepatitis E virus. J Clin Microbiol. 2002;40:4021–9. doi: 10.1128/JCM.40.11.4021-4029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arankalle VA, Chobe LP, Joshi MV, Chadha MS, Kundu B, Walimbe AM. Human and swine hepatitis E viruses from Western India belong to different genotypes. J Hepatol. 2002;36:417–25. doi: 10.1016/s0168-8278(01)00297-5. [DOI] [PubMed] [Google Scholar]
- 19.Arankalle VA, Chobe LP, Walimbe AM, Yergolkar PN, Jacob GP. Swine HEV infection in south India and phylogenetic analysis (1985-1999) J Med Virol. 2003;69:391–6. doi: 10.1002/jmv.10301. [DOI] [PubMed] [Google Scholar]
- 20.Chobe LP, Lole KS, Arankalle VA. Full genome sequence and analysis of Indian swine hepatitis E virus isolate of genotype 4. Vet Microbiol. 2006;114:240–51. doi: 10.1016/j.vetmic.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Madan K, Gopalkrishna V, Kar P, Sharma JK, Das UP, Das BC. Detection of hepatitis C and E virus genomes in sera of patients with acute viral hepatitis and fulminant hepatitis by their simultaneous amplification in PCR. J Gastroenterol Hepatol. 1998;13:125–30. doi: 10.1111/j.1440-1746.1998.tb00626.x. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 23.Wu JC, Chen CM, Chiang TY, Tsai WH, Jeng WJ, Sheen IJ, et al. Spread of hepatitis E virus among different-aged pigs: two-year survey in Taiwan. J Med Virol. 2002;66:488–92. doi: 10.1002/jmv.2170. [DOI] [PubMed] [Google Scholar]
- 24.Fernández-Barredo S, Galiana C, García A, Gómez-Muñoz MT, Vega S, Rodríguez-Iglesias MA, et al. Prevalence and genetic characterization of hepatitis E virus in paired samples of feces and serum from naturally infected pigs. Can J Vet Res. 2007;71:236–40. [PMC free article] [PubMed] [Google Scholar]
- 25.Leblanc D, Ward P, Gagné MJ, Poitras E, Müller P, Trottier YL, et al. Presence of hepatitis E virus in a naturally infected swine herd from nursery to slaughter. Int J Food Microbiol. 2007;117:160–6. doi: 10.1016/j.ijfoodmicro.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Pina S, Buti M, Cotrina M, Piella J, Girones R. HEV identified in serum from humans with acute hepatitis and in sewage of animal origin in Spain. J Hepatol. 2000;33:826–33. doi: 10.1016/s0168-8278(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y, Ge S, Zhang J, Guo Q, Ng MH, Wang F, et al. Swine as a principal reservoir of hepatitis E virus that infects humans in eastern China. J Infect Dis. 2006;193:1643–9. doi: 10.1086/504293. [DOI] [PubMed] [Google Scholar]
- 28.Melenhorst WB, Gu YL, Jaspers WJ, Verhage AH. Locally acquired hepatitis E in the Netherlands: associated with the consumption of raw pig meat? Scand J Infect Dis. 2007;39:454–6. doi: 10.1080/00365540601087590. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Gracia MT, Mateos ML, Galiana C, Fernández-Barredo S, García A, Gómez MT, et al. Autochthonous hepatitis E infection in a slaughterhouse worker. Am J Trop Med Hyg. 2007;77:893–6. [PubMed] [Google Scholar]


