Abstract
Aims
Ubiquitin performs essential roles in a myriad of signalling pathways required for cellular function and survival. Recently, we reported that disruption of the stress-inducible ubiquitin-encoding gene Ubb reduces ubiquitin content in the hypothalamus and leads to adult-onset obesity coupled with a loss of arcuate nucleus neurones and disrupted energy homeostasis in mice. Neuropeptides expressed in the hypothalamus control both metabolic and sleep behaviours. In order to demonstrate that the loss of Ubb results in broad hypothalamic abnormalities, we attempted to determine whether metabolic and sleep behaviours were altered in Ubb knockout mice.
Methods
Metabolic rate and energy expenditure were measured in a metabolic chamber, and sleep stage was monitored via electroencephalographic/electromyographic recording. The presence of neurodegeneration and increased reactive gliosis in the hypothalamus were also evaluated.
Results
We found that Ubb disruption leads to early-onset reduced activity and metabolic rate. Additionally, we have demonstrated that sleep behaviour is altered and sleep homeostasis is disrupted in Ubb knockout mice. These early metabolic and sleep abnormalities are accompanied by persistent reactive gliosis and the loss of arcuate nucleus neurones, but are independent of neurodegeneration in the lateral hypothalamus.
Conclusions
Ubb knockout mice exhibit phenotypes consistent with hypothalamic dysfunction. Our data also indicate that Ubb is essential for the maintenance of the ubiquitin levels required for proper regulation of metabolic and sleep behaviours in mice.
Keywords: glial fibrillary acidic protein, hypothalamus, knockout mice, metabolism, polyubiquitin gene, sleep
Introduction
Ubiquitin (Ub) is a highly conserved eukaryotic protein, which functions as a signal in a broad range of essential biological processes including regulated protein degradation, signal transduction, transcription and DNA repair [1,2]. Within the cell, Ub exists in a dynamic equilibrium between free and conjugated pools that are maintained principally by the opposing actions of the conjugation and deconjugation machinery. Moreover, it has been recently suggested that Ub homeostasis is regulated by a balance between deubiquitinating enzyme and its inhibitor Rfu1 (regulator of free ubiquitin chains 1) in yeast [3]. Ub is also subject to degradation by the proteasome, as well as to nonproteasomal processes, such as autophagy [4–6]. Cellular Ub levels must ultimately therefore be maintained by de novo synthesis from transcripts of the four ubiquitously expressed nuclear ubiquitin genes [7–12].
The critical role played by Ub in neuronal function and survival has been underscored by several previous biochemical and genetic findings, including the observation that the neurone-specific deubiquitinating enzyme, UCHL1, is among the most abundant proteins in the brain, by the genetic linkage of deletions in the UchL1 gene to gracile axonal dystrophy in mice [13] and by the observation that late-onset familial Parkinson’s disease in humans is associated with UchL1 mutations, albeit rarely [14]. Likewise, mutations in the Ataxia gene encoding for the deubiquitinating enzyme USP14 also result in neuronal dysfunction in Ataxia (axJ) mice [15]. Recently collected evidence suggests that the ataxia phenotype is linked to the depletion of cellular Ub or disequilibrium among Ub pools within cerebellar Purkinje cells [16,17], which dramatically underscores the need to maintain cellular Ub pool dynamics for neuronal function and survival.
We reported previously that the loss of Ubb, one of two polyubiquitin genes in mice, leads to sterility and gonadal degeneration [18], as well as an unusual metabolic syndrome characterized by hypothalamic neurodegeneration and adult-onset obesity [19]. The latter phenotypes were linked to progressive defects in the hypothalamic circuitry controlling energy balance. The loss of neurones in the arcuate nucleus is accompanied by attenuated transcriptional responses of genes encoding for orexigenic (NPY and AgRP) and anorexigenic (POMC) neuropeptides in response to food deprivation and severely reduced levels of basal AgRP mRNA. Neither the metabolic phenotype nor the transcriptional dysregulation of hypothalamic neuropeptides in Ubb−/− mice conforms to any previously described genetic model of hypothalamic dysfunction, but rather shares some features with rodents exposed neonatally to monosodium glutamate, an excitotoxin that preferentially damages neurones within the arcuate nucleus [20,21]. Similarly to monosodium glutamate-treated mice, Ubb−/− mice exhibit increased fat content but are not hyperphagic, thus suggesting disturbances in energy partitioning. Here, we report that the disruption of Ubb leads to early-onset hypo-activity accompanied by reduced metabolic rates with increased reactive gliosis prior to detectable neurodegeneration. We also show that Ubb is required for the maintenance of body temperature and sleep homeostasis. These data indicate that Ubb is crucial in the proper regulation of hypothalamic function.
Materials and methods
Mouse studies
Ubb−/− mice were generated as previously described [18]. All mice were kept in plastic cages with ad libitum access to food (regular chow) and water, with 12-h light cycle. As previously reported [19], under normal conditions, food intake of Ubb−/− mice, when normalized to their body weights, was comparable to those of wild-type littermates. All procedures followed the NIH (National Institutes of Health) guidelines with the approval of Stanford University Administrative Panel on Laboratory Animal Care. Fasting/refeeding study was carried out as previously described [19].
Metabolic monitoring
Wild-type and Ubb−/− mice (4- and 15-week-old males, n = 6 per genotype per age) were individually housed in Oxymax Comprehensive Laboratory Animal Monitoring System (CLAMS; Columbus Instruments, Columbus, OH, USA) sealed chambers, each of which was equipped with an O2 electrochemical sensor, a CO2 infrared sensor and infrared beam activity sensors. The airflow rate was 0.5 l/min per cage. Mice were placed in the chamber 1 day prior to the start of measurements to allow for acclimation to the new environment.The metabolic data collected include the volume of O2 consumed (VO2; ml/kg/h), volume of CO2 generated (VCO2; ml/kg/h), respiratory exchange ratio (RER) (RER = VCO2/VO2) and heat produced (indirectly calculated from gas exchange data; kcal/h). O2 consumption and CO2 production were measured over a 2-min period, which was repeated every 10 min. VO2 and VCO2 values were normalized to the body weights of the mice (ml/kg/h) and corrected for the effective mass factor (0.75). The infrared beam interruptions in both horizontal (X) and vertical (Z) directions were used to quantify the motor activity of mice. Any horizontal beam breakage was recorded as total activity count (Xtot), and two or more consecutive horizontal beam breakages were recorded as ambulatory activity count (Xamb). Any vertical beam breakage was recorded as total activity count (Ztot). Because Xamb and Ztot data were tightly correlated with the Xtot data in both wild-type and Ubb−/− mice, only the Xtot data were plotted. During the recording, the mice were food-deprived with free access to water only. Similar results were obtained with ad libitum access to both food and water (data not shown).
Mouse surgery
Under 1–3% isoflurane inflation with medical grade oxygen gas, 4- to 6-month-old wild-type (n = 10) and Ubb−/− (n = 7) mice were anesthetized and implanted with four screw electrodes for electroencephalography (EEG) and two wire electrodes for electromyography (EMG) for polysomnogram recording. The EEG electrodes were screwed into the brain skull 1.5 mm lateral and 1.5 mm anterior to the bregma, and 3 mm lateral and 3.5 mm posterior to the bregma. Two multistranded stainless steel EMG wire electrodes were inserted into the neck extensor muscle. These electrodes were secured with pin-connectors onto the skull with dental cement. A telemetry transmitter (G2 E-mitter; Mini Mitter, Bend, OR, USA) was also implanted into the abdomen to measure the core body temperature and locomotor activity using a computer-based recording system (Vitalview Series 4000 System; Mini Mitter, Bend, OR, USA).
Sleep stage analysis
After a 1-week surgical recovery period, the mice were moved to specially modified Nalgene micro-isolator cages, each of which was equipped with a low-torque slip-ring commutator (Biella Engineering, Irvine, CA, USA), and the cages were placed in the recording chamber. The head connector of the animal was connected to a slip-ring commutator through a 15–20 cm length of light-weight 6-strand shielded signal cable (NMUF6/30-4046SJ; Cooner Wire, Chatsworth, CA, USA). The commutator output was connected to the amplifier. The mice were permitted full freedom of movement in the recording cages under a 12-h light and 12-h dark cycle with ad libitum feeding throughout the experiment. After a 1-week acclimation period in the recording cage, 24 h of polygraph recording was conducted for the baseline measurement, followed by another 24 h of recording with sleep deprivation for 6 h either during the first or second half of the light cycle to assess sleep homeostasis. As a separate recording session, the mice were maintained under 24-h dark cycle (constant dark) conditions for 2 weeks for measurement of the intrinsic circadian rhythms of the mice. The polysomnogram signals were acquired using a Grass Instruments model 12 amplifier (Grass Technologies, West Warwick, RI, USA) and digitized at 128 Hz using a computer-based recording system (Vital Recorder; Kissei Comtec, Matsumoto, Japan). Among the four EEG screw electrodes, a combination of one frontal and one parietal electrode was used. The remaining screw electrodes were used as anchors of the head-stage and/or as spares. Sleep stage was manually scored on the basis of the EEG and EMG results in 10-s epochs using sleep analysis software (SleepSign; Kissei Comtec, Matsumoto, Japan). We applied 30 Hz of high-cut digital filter for the EEG and 20–50 Hz of band-pass filter for the EMG. Wakefulness was defined as low-amplitude and mixed-frequency (>4 Hz) EEG with continuous large fluctuations in EMG; slow-wave or non-rapid eye movement (NREM) sleep was defined as high-amplitude and low-frequency (0.25–4 Hz) EEG with no fluctuation in EMG, and rapid eye movement (REM) sleep was defined as low-amplitude and high-frequency EEG (similar to wake stage, but with rhythmic α waves at 7–9 Hz) with no fluctuation in EMG. DREM (direct transitions from wakefulness to REM sleep) was scored in cases in which there were REM epoch(s) after four continued Wake epochs (40 s). For power spectral analysis, the delta (0.25–4 Hz) and theta (4–9 Hz) EEG power of each 10-s epoch were analysed via fast Fourier transformation. To assess the effects of sleep deprivation on EEG power, the total power in the delta range during NREM sleep and total power in the theta range during REM sleep were computed using SleepSign.
Confocal microscopy
Generation of free-floating sections, direct visualization of green fluorescent protein (GFP) fluorescence from GFP-puror fusion protein knocked in to the Ubb locus to monitor Ubb transcriptional activity, immunofluorescence and DNA visualization using TO-PRO-3 iodide were conducted as previously described [19]. For orexin (OreA), melanin-concentrating hormone (MCH), α-melanocyte-stimulating hormone (α-MSH), and glial fibrillary acidic protein (GFAP) immunofluorescence, anti-OreA (1:200; #H-003-03; Phoenix Pharmaceuticals, Burlingame, CA, USA), anti-MCH (1:1000; #H-070-47; Phoenix Pharmaceuticals), anti-α-MSH (1:200; #20074; Immunostar, Hudson, WI, USA) and anti-GFAP (1:1500; Z0334; Dako, Carpinteria, CA, USA) polyclonal antibodies and Alexa Fluor 555-conjugated goat anti-rabbit IgG (1:200; #A21428; Molecular Probes, Eugene, OR, USA) were utilized. For better visualization of GFP-positive neurones and to stain the α-MSH-containing neuronal cell bodies, the mice (except for the ones used to study GFAP immunoreactivity) were pretreated with i.c.v. colchicine injections (C9754; Sigma, Saint Louis, MO, USA) 48 h before euthanasia, as previously described [19]. Confocal images were collected with a Leica TCS SP2 laser scanning system with sequential image recording and tile scan to cover the large regions of the brain sections from 4-month-old Ubb+/− mice (n = 3; for colocalization of OreA and MCH-positive neurones to Ubb-expressing neurones), 5-month-old wild-type and Ubb−/− mice (n = 3 per genotype; for α-MSH, OreA and MCH-positive neurone counting), and 1- and 4-month-old wild-type and Ubb−/− mice (n = 3 per genotype; for GFAP immunoreactivity). Pseudo-colour images were generated with Leica confocal software, Ver. 2.61, and merged images were obtained using Adobe Photoshop CS3. For quantification of the number of neurones, a series of free-floating sections (25 μm thick) from −1.55 to −2.05 mm posterior to the bregma were used. Cell counts were obtained bilaterally from three coronal sections (200 μm interval) within the entire arcuate nucleus (for α-MSH) or within the entire hypothalamus (for OreA/MCH), and collections were anatomically matched among the different mice. Although α-MSH-expressing neurones were found to be evenly distributed among the three selected sections (−1.55, −1.75 and −1.95 mm posterior to bregma), OreA/MCH-expressing neurones were not evenly distributed (−1.6/−1.65, −1.8/−1.85 and −2.0/−2.05 mm posterior to bregma) and the number of neurones varied depending on the location of the sections from the bregma (data not shown). The number of neurones from each section was combined and compared between the two genotypes.
Immunohistochemistry
Brain sections were processed for NeuN immunofluorescence as previously described [22]. Briefly, sections were first removed from cryoprotectant and blocked in a solution containing 0.3% Triton X-100 and 3% normal goat serum in PBS (phosphate buffered saline) for 1 h, and then incubated with mouse antineuronal nuclei (NeuN; 1:500; Chemicon, Temecula, CA, USA) monoclonal anti-body. After washing in PBS, sections were incubated with Alexa Fluor 546-conjugated goat anti-mouse IgG (1:400). Sections were then washed and mounted as previously described. Images were visualized on an Olympus BX51 microscope equipped with 2.5× and 4× objectives.
Statistical analysis
Two-tailed unpaired Student’s t-test with equal or unequal variance, which was determined by F-test, was used to compare the data between the two groups. P < 0.05 was considered statistically significant.
Results
Early-onset reduction in metabolic rates in Ubb−/− mice
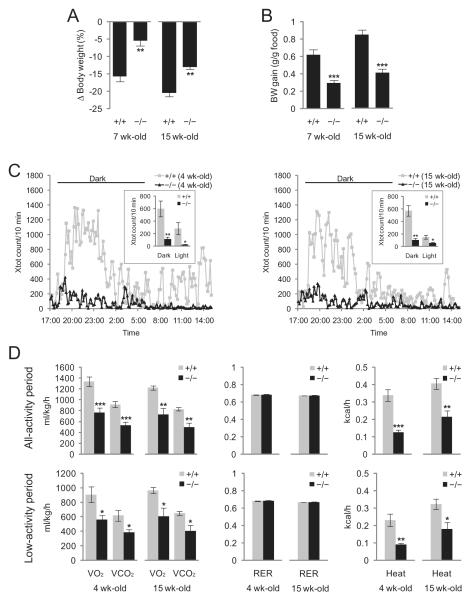
In a previous study, we reported that adult (15-week-old), but not young (7-week-old), mice lacking Ubb exhibit modest defects in compensatory hyperphagic responses to food deprivation, as well as a corresponding recovery of body weight upon refeeding, which correlated temporally with overt neuronal loss within the arcuate nucleus [19]. By way of contrast, both young and adult Ubb−/− mice lost significantly less body weight when fasting (Figure 1A), and gained significantly less weight per gram of food consumed during refeeding (Figure 1B), thereby suggesting that reduced metabolic activity may be an early consequence of Ubb disruption. Indeed, we found that Ubb−/− mice are markedly less active than their wild-type litter-mates, particularly during the dark phase, and that this dramatic difference is fully manifested at 4 weeks of age, nearly 3 months prior to the onset of frank obesity or detectable neuronal loss from the arcuate nucleus (Figure 1C). Energy expenditure, measured as O2 consumption and CO2 production, was reduced by nearly 40% in both young and old Ubb−/− mice (Figure 1D, left panel, top). This reduced energy expenditure in Ubb−/− mice, however, was not a simple consequence of reduced locomotor activity, as the difference persisted when determined at matching low levels of activity (Figure 1D, left panel, bottom). Heat production, an indicator of metabolic rate, was also reduced significantly in both young and old Ubb−/− mice (Figure 1D, right panel). Mice of both genotypes exhibited RER of approximately 0.7, thereby indicating that the loss of Ubb did not alter fuel preference (Figure 1D, middle panel). Thus, Ubb−/− mice exhibited dramatic reductions in metabolic rate and locomotor activity, thereby suggesting that Ubb is required for a broader range of neural circuits than those that directly control leptin-responsive signalling. Moreover, these defects are apparent considerably prior to the onset of obesity phenotype or neurodegeneration within the arcuate nucleus [19].
Figure 1.
Early-onset reduced metabolic rate and activity in Ubb−/− mice. (A) Loss of body weight after 48 h of fasting in 7- and 15-week-old wild-type (+/+) and Ubb−/− (−/−) mice (+/+, n = 9 or 11; −/−, n = 11). (B) Early-onset reduced feed efficiency in Ubb−/− mice after fasting and refeeding. Mice were fasted for 48 h and refed for 48 h, and feed efficiency was calculated as the gain in body weight (BW) per gram of food intake (+/+, n = 9 or 11; −/−, n = 11). (C) Early-onset reduced locomotor activity in male Ubb−/− mice (+/+, n = 6; −/−, n = 6). Movements of 4-week-old (left panel) and 15-week-old (right panel) mice were monitored in the metabolic chamber and expressed as the number of horizontal photobeam breaks per 10 min. (D) Early-onset reduced energy expenditure in male Ubb−/− mice (+/+, n = 6; −/−, n = 6). O2 consumption (VO2) and CO2 production (VCO2) were measured under normal (all-activity period) or resting (low-activity period) conditions (Xtot ≤ 20, in which Xtot is total number of horizontal movement with any beam breaks) (left panel) and heat production was determined by CV (caloric value) × VO2, in which CV = 3.815 + 1.232 × respiratory exchange ratio (RER) (right panel). RERs (VCO2/VO2) were normal in Ubb−/− mice (middle panel). The data were normalized to body weight and shown for a 24-h period. Similar results were obtained when the data were plotted for 12-h light or 12-h dark phase (data not shown). All data are expressed as the means ± SEM from the indicated number of individually housed mice. *P < 0.05; **P < 0.01; ***P < 0.001 vs. wild-type (+/+) mice.
Altered sleep behaviour in adult Ubb−/− mice
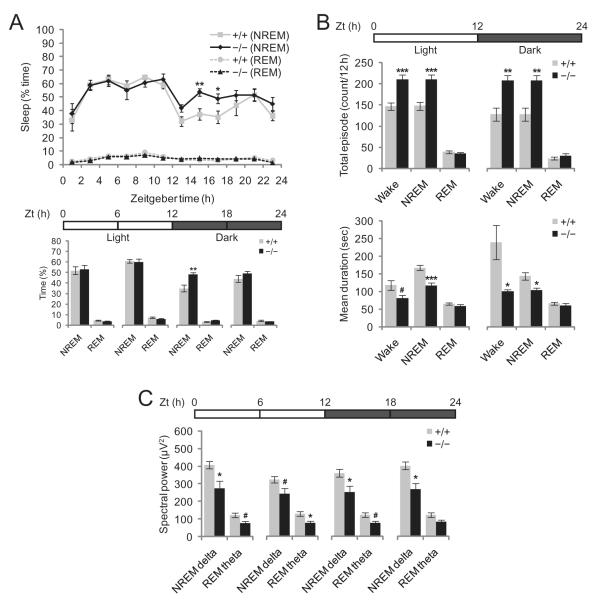
In an effort to assess whether neuronal signalling and/or survival pathways are disrupted throughout the entire hypothalamus in Ubb−/− mice, we decided to extend our investigation to other hypothalamic functions. Because several hypothalamic neuropeptides, including OreA and MCH, affect both sleep and feeding behaviours [23], we considered the possibility that reduced Ub levels in the hypothalamus of Ubb−/− mice [19] might also alter sleep behaviour. To evaluate this hypothesis, we analysed the sleep stages of adult (5- to 7-month-old) Ubb−/− mice and wild-type littermates via electroencephalographic/electromyographic recording. During the 12-h light period [Zeitgeber time (Zt) = 0–12 h], NREM and REM sleep state patterns were quite similar between Ubb−/− mice and their wild-type littermates (Figure 2A). During the first half of the 12-h dark period (Zt = 12–18 h), however, the amount of NREM sleep evidenced by the Ubb−/− mice was significantly increased over that of the wild-type littermates. The number of episodes of wakefulness and NREM sleep of shorter durations was also significantly increased (Figure 2B and Figure S1) over the entire 24-h period, thereby suggesting profound sleep/wake fragmentation and inability to consolidate wakefulness in the Ubb−/− mice. Increased sleep/wake fragmentation is a characteristic associated with OreA neurone-ablated or narcoleptic mice [24]. However, we rarely observed abnormal direct transitions from wakefulness to REM sleep (DREM), a typical characteristic of narcolepsy, in Ubb−/− mice (data not shown). In order to determine whether sleep quality was altered in Ubb−/− mice, analysis of the spectrum of EEG power in the NREM delta and REM theta bands revealed that spectral EEG power was reduced significantly in the Ubb−/− mice in both the light and dark periods (Figure 2C). Therefore, adult Ubb−/− mice show abnormal sleep behaviour with increased sleep/wake fragmentation and reduced sleep quality. We were not able to evaluate sleep behaviour in young mice, as the surgical implantation of EEG/EMG recording devices was conducted on adult mice only to maximize survival after surgery.
Figure 2.
Altered sleep behaviour in Ubb−/− mice. (A) Diurnal fluctuation of sleep in wild-type and Ubb−/− mice. The amount of sleep in the first half of the dark cycle (Zt = 12–18 h) was significantly increased in the Ubb−/− mice as compared with the wild-type mice (+/+, n = 10; −/−, n = 7). Bar graphs represent the total amount of sleep during the 6-h period as indicated by the horizontal bar. The amount of sleep is expressed as the percentage of time spent in sleep. (B) The number of total episodes and mean durations of sleep in wild-type and Ubb−/− mice. There were significantly more episodes of wake/non-rapid eye movement (NREM) sleep with shorter durations in the Ubb−/− mice as compared with the wild-type mice in both light and dark phases (+/+, n = 10; −/−, n = 7). (C) Spectral power analysis of sleep in wild-type and Ubb−/− mice. Power of electroencephalography spectra at 0.25–4 Hz (NREM delta) and 4–9 Hz (rapid eye movement [REM] theta) were reduced significantly in the Ubb−/− mice as compared with the wild-type mice (+/+, n = 10; −/−, n = 7). Average spectral power is shown during the 6-h period as indicated by the horizontal bar. All data are expressed as the means ± SEM from the indicated number of 5- to 7-month-old mice. Horizontal bars indicate light and dark phases. *P < 0.05; **P < 0.01; ***P < 0.001; #P < 0.1 vs. wild-type (+/+) mice.
Disrupted NREM sleep homeostasis in adult Ubb−/− mice
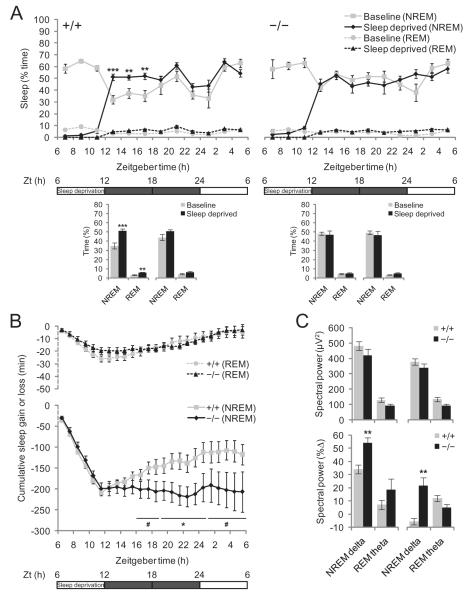
In an effort to evaluate the role of Ubb in sleep homeostasis, we monitored sleep behaviour in Ubb−/− and wild-type mice that had been sleep-deprived for 6 h. We initially deprived the mice of sleep during the first half of the light cycle, but sleep recovery was not observed during the next 6-h period (second half of light cycle), even in the wild-type mice, followed by a slight increase in amount of sleep during the first half of the dark cycle (P = 0.094) (Figure S2). No sleep recovery was observed in Ubb−/− mice during either of the 6-h periods. The lack of immediate sleep rebound in both genotypes may have been attributable to the high amount of baseline sleep during the second half of the light cycle. Therefore, we repeated the experiment and induced sleep deprivation in these mice during the second half of the light cycle. Under these conditions, a significant sleep rebound was noted in the wild-type mice (Figure 3A). However, Ubb−/− mice failed to exhibit the compensatory sleep ‘rebound’ characteristic of wild-type mice in response to sleep deprivation. Although wild-type mice proved able to recover approximately 50% of the NREM sleep they lost within 18 h of recovery, the Ubb−/− mice did not recover at all even after 18 h (Figure 3B). We also noted that EEG power density after sleep deprivation was not significantly different in the wild-type and Ubb−/− mice (Figure 3C, top). However, because the baseline (without sleep deprivation) NREM delta power was lower in the Ubb−/− mice than in the wild-type mice (see Figure 2C), the percentage increase of NREM delta power from the baseline after sleep deprivation was considerably higher in the Ubb−/− mice as compared with the wild-type mice (Figure 3C, bottom). An increase in sleep propensity, when it is defined as an increase in NREM delta power from baseline, was evident in the Ubb−/− mice, but, quite unexpectedly, this was not reflected in the amounts of recovery sleep. Therefore, disturbances in the mechanisms intrinsic to the regulation of NREM sleep homeostasis, resulting in an unusual dissociation of sleep propensity and sleep rebound, was apparent in Ubb−/− mice.
Figure 3.
Defective regulation of non-rapid eye movement (NREM) sleep homeostasis in Ubb−/− mice. (A) Recovery of NREM and rapid eye movement (REM) sleep after 6 h of sleep deprivation during the second half of the light cycle in wild-type and Ubb−/− mice (+/+, n = 10; −/−, n = 7). Time course of sleep amount (as percent of time spent) after sleep deprivation (top) and the amount of sleep during the two 6-h periods of dark cycle following sleep deprivation are shown (bottom). Sleep recovery was observed only in the wild-type mice. (B) Cumulative NREM and REM sleep loss and gain during the sleep deprivation and subsequent recovery period in wild-type and Ubb−/− mice (+/+, n = 10; −/−, n = 7). The amounts of sleep lost and gained over baseline levels were accumulated during the 6-h sleep deprivation and 18-h recovery period. It was noted that, even in the wild-type mice, despite a large amount of accumulated NREM sleep loss, NREM sleep gained during the recovery period did not reach the original levels. (C) Spectral power analysis of sleep during the two 6-h periods of dark cycle following sleep deprivation (Zt = 6–12 h) in wild-type and Ubb−/− mice (+/+, n = 10; −/−, n = 7). Average spectral power (top) and the percent increase from the baseline spectral power (bottom) during the 6-h period are shown as indicated by the horizontal bar. We noted no significant differences in average spectral EEG power at 0.25–4 Hz (NREM delta) and 4–9 Hz (REM theta) between wild-type and Ubb−/− mice; however, the percentage increase of NREM delta power from the baseline spectral power was significantly higher in the Ubb−/− mice. All data are expressed as the means ± SEM from the indicated number of 5- to 7-month-old mice. *P < 0.05; **P < 0.01; ***P < 0.001; #P < 0.1 vs. wild-type (+/+) mice.
Core body temperature and circadian rhythmicity in Ubb−/− mice
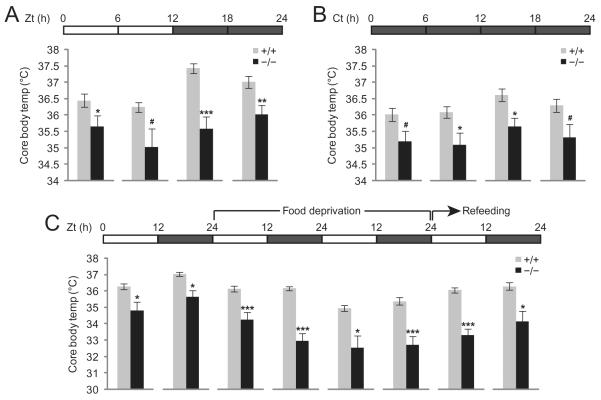
Generally, core body temperature in mammals is under circadian control, and is increased during the dark phase, in which the mice are more active. This trend was evident for wild-type mice, but was sharply blunted in Ubb−/− mice (Figure 4A). Moreover, we noted that core body temperature was reduced sharply in the Ubb−/− mice as compared with the wild-type controls, most significantly during the first half of the dark cycle (35.5°C vs. 37.5°C). A similar reduction in core body temperature was noted in Ubb−/− mice as compared with the wild-type controls under 24-h dark (constant dark) conditions (Figure 4B). This reduced core body temperature in the Ubb−/− mice might be a secondary manifestation of the reduced basal metabolic rate in the mutants, or might reflect a primary lesion in hypothalamic function. In order to discriminate between these two possibilities, mice were deprived of food for 48 h and refed (Figure 4C). After food deprivation, core body temperatures dropped by 1.5°C and 3°C from baseline in the wild-type and Ubb−/− mice, respectively. During the 24 h of refeeding, core body temperatures increased by 1°C and 1.5°C in the wild-type and Ubb−/− mice, respectively. Thus, although the Ubb−/− mice have chronically lower core body temperatures due to reduced metabolic rates, the induction of thermogenesis in response to food intake was intact in the Ubb−/− mice [25].
Figure 4.
Reduced core body temperature in Ubb−/− mice. (A) Mean core body temperature in wild-type and Ubb−/− mice (+/+, n = 9; −/−, n = 6). Mean core body temperature is shown during the 6-h period as indicated by the horizontal bar. (B) Mean core body temperature in wild-type and Ubb−/− mice (+/+, n = 4; −/−, n = 3) under constant dark (free-running) conditions. Mice were maintained under constant dark conditions for 2 weeks and mean core body temperature was measured during the last 24-h period as indicated by the horizontal bar. Ct, circadian time. (C) Mean core body temperatures in wild-type and Ubb−/− mice (+/+, n = 9; −/−, n = 6) are shown during the food deprivation and refeeding periods. In both the wild-type and Ubb−/− mice, core body temperatures were reduced after food deprivation and recovered during refeeding. Ubb−/− mice consistently exhibited lower core body temperatures, reaching down to 32.5°C during the second day of food deprivation, whereas the wild-type mice maintained their core body temperature at 35°C during the same period. All data are expressed as the means ± SEM from the indicated number of 5- to 7-month-old mice. In (B), the results are average of two independent experiments. Horizontal bars indicate light and dark phases. *P < 0.05; **P < 0.01; ***P < 0.001; #P < 0.1 vs. wild-type (+/+) mice.
Altered circadian rhythmicity could also contribute to the sleep abnormalities noted in the Ubb−/− mice. Therefore, we monitored the locomotor activity of mice under constant dark conditions for 2 weeks to assess whether the circadian rhythm control and circadian period length were intact in the Ubb−/− mice. Under these conditions, although the wild-type mice displayed clear changes in locomotor activity every 12 h with a 24-h circadian period length, the Ubb−/− mice exhibited less distinct locomotor activity patterns, thus suggesting that circadian rhythmicity is altered in Ubb−/− mice. However, we noted no differences in the circadian period length between the two genotypes (data not shown).
Ubb knockout leads to widespread early-onset reactive gliosis in the absence of neurodegeneration in lateral hypothalamic area
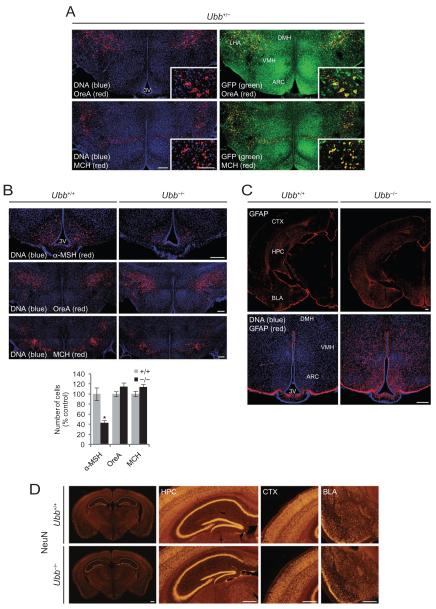
In an effort to determine the spatial distribution of Ubb expression in the hypothalamus, we directly visualized GFP fluorescence from GFP-Puror fusion protein knocked in to the Ubb locus (Figure 5A). These data showed that Ubb was abundantly expressed in all hypothalamic regions, including the arcuate nucleus, the ventromedial hypothalamus, the dorsomedial hypothalamus and to a lesser degree in the lateral hypothalamic area. Neurones expressing OreA and MCH were highly colocalized with neurones expressing high Ubb levels (Figure 5A, inset). However, cell counting revealed no evidence of a loss of these neurones in adult Ubb−/− mice (Figure 5B). Although the number of hypothalamic OreA/MCH-expressing neurones was not affected significantly by the loss of Ubb, we noted a 50% reduction in the number of neurones that express α-MSH in the arcuate nucleus of adult Ubb−/− mice, which is consistent with our previous report indicating the adult-onset neurodegeneration of AgRP-expressing neurones in the arcuate nucleus (Figure 5B). Thus, the sleep abnormalities noted in Ubb−/− mice cannot be principally due to the loss of OreA or MCH neurones, but might instead be attributable to the consequence of impaired neuronal function and/or signalling or to the loss of α-MSH-expressing neurones from the arcuate nucleus.
Figure 5.
No evidence of loss of orexin (OreA) or melanin-concentrating hormone (MCH) neurones, but early-onset reactive gliosis in Ubb−/− mice. (A) Colocalization of OreA and MCH immunoreactive neurones in the lateral hypothalamic area to Ubb-expressing neurones. Coronal brain sections were stained for OreA and MCH, DNA was visualized using TO-PRO-3 iodide, and green fluorescent protein (GFP) fluorescence was visualized directly from GFP-Puror protein. Representative confocal images from 4-month-old male Ubb+/− mice (n = 3) are shown. Scale bar, 200 μm; inset, 100 μm. (B) Loss of α-melanocyte-stimulating hormone (α-MSH), but not OreA and MCH, immunoreactive neurones in the hypothalamic sections of Ubb−/− mice. Sleep abnormalities in Ubb−/− mice are not attributable to the loss of OreA or MCH neurones. For quantification of neurones, cell counts were obtained from three coronal sections (200 μm interval) generated from each brain and compared with wild-type (+/+) controls. Images are representative of 5-month-old male mice for each group (+/+, n = 3; −/−, n = 3). Representative sections are from bregma −1.75 mm for α-MSH (second set), −1.6 mm for OreA (first set) and −1.85 mm for MCH (second set). Scale bar, 200 μm. Data are expressed as mean ± SEM from the indicated number of mice. *P < 0.05 vs. wild-type (+/+) mice. (C) Coronal brain sections were stained for glial fibrillary acidic protein (GFAP) and DNA was visualized using TO-PRO-3 iodide. Representative confocal images from 1-month-old wild-type and Ubb−/− mice (n = 3 per genotype) are shown. GFAP immunoreactivity was increased throughout the brain sections from the Ubb−/− mice, except for the subcortical regions (top). In the hypothalamus, GFAP immunoreactivity was most prominent in the arcuate nucleus (bottom). Scale bar, 200 μm. (D) Coronal brain sections were stained for NeuN to visualize neurones. Representative images from 4-month-old wild-type and Ubb−/− mice (n = 3 per genotype) are shown. NeuN-positive neurone number is similar between two genotypes in various brain regions such as HPC, CTX, and BLA. Scale bar, 500 μm. 3V, third ventricle; ARC, arcuate nucleus; BLA, basal amygdala; CTX, cerebral cortex; DMH, dorsomedial hypothalamus; HPC, hippocampus; LHA, lateral hypothalamic area; VMH, ventromedial hypothalamus.
In order to determine whether neuronal injury or inflammation accompanies the behavioural phenotypes in Ubb−/− mice and precedes neuronal loss from the arcuate nucleus [26], we utilized immunofluorescence to determine the expression of GFAP in the brains of the mice. The data revealed significantly increased GFAP immunoreactivity in multiple brain regions, including the hypothalamus and basal amygdala, in 1-month-old Ubb−/− mice (Figure 5C) that persisted through adulthood (data not shown). In the hypothalamus, elevated GFAP immunoreactivity was noted in the lateral hypothalamic area, in addition to arcuate nucleus, ventromedial hypothalamus and dorsomedial hypothalamus. Because, at 1 month of age, there is no evidence of neurodegeneration in Ubb−/− mice [19], these data indicate that neuronal stress occurs well prior to the onset of neurodegeneration, but coincides with the onset of behavioural abnormalities.
We investigated whether behavioural abnormalities with early-onset reactive gliosis and adult-onset neurodegeneration restricted to arcuate nucleus of hypothalamus in Ubb−/− mice accompanies any pathological abnormalities in brain. However, we did not observe any gross morphological changes in hypothalamus (Figure S3A) or in other brain regions, such as brain stem that also plays an important role in sleep regulation (Figure S3B). In accordance with this observation, we were not able to observe any pathological abnormalities or obvious loss of neurones in other brain areas that we examined, such as cerebral cortex, hippocampus and basal amygdala (Figure 5D). These data do not, however, exclude the possibility that other pathological changes may be present in other brain regions that we did not investigate.
Discussion
In this study we report that the homozygous disruption of Ubb results in an early-onset syndrome characterized by profoundly reduced locomotor activity and metabolic rates, impaired body temperature regulation and disturbed sleep homeostasis. These phenotypes are accompanied by widespread reactive gliosis throughout the hypothalamus prior to the onset of detectable neuronal loss in the arcuate nucleus, revealing the essential role of Ub supplied by Ubb for the regulation of the hypothalamic control of metabolic and sleep behaviours.
Body weight homeostasis and sleep regulation are tightly linked functions, which are controlled by anatomically distinct but overlapping hypothalamic regions. For example, OreA-deficient mice exhibit fragmented sleep patterns, reduced energy expenditure and obesity [24,27] in addition to reduced arousal responses during fasting [28]. Ubb−/− mice also suffer from disturbed metabolic homeostasis and sleep behaviour. However, Ubb−/− mice do not exhibit DREM or cataplexy-like behaviours, nor do they exhibit the dissociated REM sleep phenomena observed typically in OreA-deficient narcoleptic mice. Moreover, the disruption of NREM sleep homeostasis in Ubb−/− mice, which was assessed by evaluating sleep recovery and NREM delta EEG power after sleep deprivation, was not apparent in the narcolepsy models [29]. Therefore, the absence of OreA/MCH neuronal loss in Ubb−/− mice, coupled with the fact that the OreA/MCH systems are more critically involved in REM sleep regulation than NREM sleep homeostasis, suggests that OreA/MCH systems may not be involved directly in the sleep abnormalities observed in Ubb−/− mice. Thus, it is possible that metabolic changes in Ubb−/− mice influence the alteration of sleep behaviour through OreA/MCH systems affecting the function of these neurones, in addition to other hypothalamic mechanisms. Alternatively, sleep abnormalities in Ubb−/− mice could, at least in part, be due to the altered circadian rhythmicity with less distinct locomotor activity patterns. It is possible that reduced activity, especially during dark phase, could simply increase the likelihood to fall asleep, which is supported by the observation that Ubb−/− mice exhibit increased number of episode of NREM sleep. In addition, the concurrence of reactive gliosis and decreased locomotor activity has also been observed in β-amyloid precursor protein-deficient mice [30]. Interestingly, like Ubb−/− mice, no histological abnormalities were observed in this mouse model, implying that impaired neuronal function with reactive gliosis may be sufficient to exhibit abnormal behavioural phenotypes. It is also possible that other neuronal or humoral mechanisms, including the regulation of sleep patterns by cytokines and hormones [31,32], contribute to the sleep abnormalities noted in Ubb−/− mice. In this regard, the observed reduction of α-MSH-expressing neurones in Ubb−/− mice is interesting, as it has been reported previously that i.c.v. injection of α-MSH, which inhibits the activity of cytokine interleukin-1 β (IL-1), caused a reduction in the amount of NREM sleep [33]. Conversely, the injection of IL-1 caused an increase in the amount of NREM sleep [34]. IL-1 is a key molecule in NREM sleep regulation and sleep homeostasis [35], and has been demonstrated to be released from astroglial cells in an ATP (adenosine triphosphate)-dependent fashion [36,37]. IL-1 levels are up-regulated by feeding, sleep deprivation or neuronal activity (reviewed in [38]) and may also be released by reactive astrocytes in the Ubb−/− brain. Further studies will be necessary to determine whether the loss of α-MSH-expressing neurones could alter IL-1 release from astroglial cells, thus resulting in abnormal NREM sleep regulation in Ubb−/− mice.
Because Ubb is one of four genes in the mouse that encode for Ub, it is anticipated that all of the phenotypes observed in Ubb−/− mice must arise either directly or indirectly from a deficiency of this protein – and, indeed, we have previously reported that total Ub levels are reduced in hypothalamic tissues from Ubb−/− mice [19]. Ubb, similarly to the other three Ub-encoding genes, is abundantly expressed in neurones, astrocytes and oligodendrocytes [39], thereby suggesting that some of the phenotypes observed in these mice may arise from the impairment of glial, as well as neuronal, functions. Indeed, the direct visualization of GFP fluorescence in a series of Ubb+/− sagittal brain sections indicates that Ubb is widely expressed throughout the brain, including in the olfactory bulb, hippocampus, hypothalamus, cerebral cortex and cerebellum, although it appears that Ubb expression levels in hypothalamic neurones are relatively higher (K.-Y. R. and R.R.K., unpub. data). Therefore, the observed phenotypes in Ubb−/− mice are most likely attributable to principal defects in hypothalamic function. However, we cannot exclude the possibility that defects in other brain regions might contribute to the observed phenotypes. Interestingly, Ubb is also highly expressed in locus coeruleus (K.-Y. R. and R.R.K., unpub. data) which mediates sympathetic effects upon stress. Therefore, it is possible that Ub levels in this brain region is reduced in Ubb−/− mice and the activation of locus coeruleus by stress might be impaired, leading to abnormal behaviours including sleep. Detailed study is underway to directly demonstrate that Ubb plays an important role in locus coeruleus. In order to determine whether the effects of Ubb disruption on neuronal signalling and survival reported here are cell-autonomous will require a conditional, cell-type specific knockout of this gene. Nevertheless, our data establish a fundamental requirement for the adequate maintenance of Ub levels for proper neuronal function and viability.
Supplementary Material
Acknowledgements
We thank our colleagues in Kopito lab for helpful discussions and comments, and thank our colleagues in Nishino lab; especially Humberto Garcia for his help in mouse surgery and sleep deprivation; and Atsushi Soya, You Hwi Song, and Mari Matsumura for their help in sleep deprivation study. This work was funded in part by a grant from the National Institutes of Health (NS42842) to R.R.K., and a grant from the Korea Healthcare technology R&D Project, Ministry for Health, Welfare & Family Affairs, Korea (A084520) to K.-Y.R.
Footnotes
Supporting information Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–39. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 3.Kimura Y, Yashiroda H, Kudo T, Koitabashi S, Murata S, Kakizuka A, Tanaka K. An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell. 2009;137:549–59. doi: 10.1016/j.cell.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 4.Haas AL, Bright PM. The dynamics of ubiquitin pools within cultured human lung fibroblasts. J Biol Chem. 1987;262:345–51. [PubMed] [Google Scholar]
- 5.Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–94. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–61. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker RT, Board PG. The human ubiquitin gene family: structure of a gene and pseudogenes from the Ub B subfamily. Nucleic Acids Res. 1987;15:443–63. doi: 10.1093/nar/15.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker RT, Board PG. The human ubiquitin-52 amino acid fusion protein gene shares several structural features with mammalian ribosomal protein genes. Nucleic Acids Res. 1991;19:1035–40. doi: 10.1093/nar/19.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finley D, Bartel B, Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 10.Lund PK, Moats-Staats BM, Simmons JG, Hoyt E, D’Ercole AJ, Martin F, Van Wyk JJ. Nucleotide sequence analysis of a cDNA encoding human ubiquitin reveals that ubiquitin is synthesized as a precursor. J Biol Chem. 1985;260:7609–13. [PubMed] [Google Scholar]
- 11.Redman KL, Rechsteiner M. Identification of the long ubiquitin extension as ribosomal protein S27a. Nature. 1989;338:438–40. doi: 10.1038/338438a0. [DOI] [PubMed] [Google Scholar]
- 12.Wiborg O, Pedersen MS, Wind A, Berglund LE, Marcker KA, Vuust J. The human ubiquitin multigene family: some genes contain multiple directly repeated ubiquitin coding sequences. EMBO J. 1985;4:755–9. doi: 10.1002/j.1460-2075.1985.tb03693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saigoh K, Wang YL, Suh JG, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, Wada K. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;23:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–18. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 15.Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, Householder DB, Fletcher CF, Miller RJ, Copeland NG, Jenkins NA. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet. 2002;32:420–5. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- 16.Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, Wilson SM. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem. 2005;95:724–31. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- 17.Crimmins S, Jin Y, Wheeler C, Huffman AK, Chapman C, Dobrunz LE, Levey A, Roth KA, Wilson JA, Wilson SM. Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J Neurosci. 2006;26:11423–31. doi: 10.1523/JNEUROSCI.3600-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryu KY, Sinnar SA, Reinholdt LG, Vaccari S, Hall S, Garcia MA, Zaitseva TS, Bouley DM, Boekelheide K, Handel MA, Conti M, Kopito RR. The mouse polyubiquitin gene Ubb is essential for meiotic progression. Mol Cell Biol. 2008;28:1136–46. doi: 10.1128/MCB.01566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryu KY, Garza JC, Lu XY, Barsh GS, Kopito RR. Hypothalamic neurodegeneration and adult-onset obesity in mice lacking the Ubb polyubiquitin gene. Proc Natl Acad Sci USA. 2008;105:4016–21. doi: 10.1073/pnas.0800096105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerkerian L, Pelletier G. Effects of monosodium l-glutamate administration on neuropeptide Y-containing neurons in the rat hypothalamus. Brain Res. 1986;369:388–90. doi: 10.1016/0006-8993(86)90557-3. [DOI] [PubMed] [Google Scholar]
- 21.Olney JW. Brain lesions, obesity, and other disturbances in mice treated with monosodium glutamate. Science. 1969;164:719–21. doi: 10.1126/science.164.3880.719. [DOI] [PubMed] [Google Scholar]
- 22.Garza JC, Guo M, Zhang W, Lu XY. Leptin increases adult hippocampal neurogenesis in vivo and in vitro. J Biol Chem. 2008;283:18238–47. doi: 10.1074/jbc.M800053200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adamantidis A, de Lecea L. Physiological arousal: a role for hypothalamic systems. Cell Mol Life Sci. 2008;65:1475–88. doi: 10.1007/s00018-008-7521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara J, Beuckmann CT, Nambu T, Willie JT, Chemelli RM, Sinton CM, Sugiyama F, Yagami K, Goto K, Yanagisawa M, Sakurai T. Genetic ablation of orexin neurons in mice results in narcolepsy, hypophagia, and obesity. Neuron. 2001;30:345–54. doi: 10.1016/s0896-6273(01)00293-8. [DOI] [PubMed] [Google Scholar]
- 25.Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–60. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- 26.Castillo-Ruiz MM, Campuzano O, Acarin L, Castellano B, Gonzalez B. Delayed neurodegeneration and early astrogliosis after excitotoxicity to the aged brain. Exp Gerontol. 2007;42:343–54. doi: 10.1016/j.exger.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S, Zeitzer JM, Sakurai T, Nishino S, Mignot E. Sleep/wake fragmentation disrupts metabolism in a mouse model of narcolepsy. J Physiol. 2007;581:649–63. doi: 10.1113/jphysiol.2007.129510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamanaka A, Beuckmann CT, Willie JT, Hara J, Tsujino N, Mieda M, Tominaga M, Yagami K, Sugiyama F, Goto K, Yanagisawa M, Sakurai T. Hypothalamic orexin neurons regulate arousal according to energy balance in mice. Neuron. 2003;38:701–13. doi: 10.1016/s0896-6273(03)00331-3. [DOI] [PubMed] [Google Scholar]
- 29.Mochizuki T, Crocker A, McCormack S, Yanagisawa M, Sakurai T, Scammell TE. Behavioral state instability in orexin knock-out mice. J Neurosci. 2004;24:6291–300. doi: 10.1523/JNEUROSCI.0586-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng H, Jiang M, Trumbauer ME, Sirinathsinghji DJ, Hopkins R, Smith DW, Heavens RP, Dawson GR, Boyce S, Conner MW, Stevens KA, Slunt HH, Sisoda SS, Chen HY, Van der Ploeg LH. beta-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell. 1995;81:525–31. doi: 10.1016/0092-8674(95)90073-x. [DOI] [PubMed] [Google Scholar]
- 31.Krueger JM, Fang J, Hansen MK, Zhang J, Obal F., Jr Humoral regulation of sleep. News Physiol Sci. 1998;13:189–94. doi: 10.1152/physiologyonline.1998.13.4.189. [DOI] [PubMed] [Google Scholar]
- 32.Krueger JM, Obal F, Jr, Fang J. Humoral regulation of physiological sleep: cytokines and GHRH. J Sleep Res. 1999;8(Suppl. 1):53–9. doi: 10.1046/j.1365-2869.1999.00009.x. [DOI] [PubMed] [Google Scholar]
- 33.Opp MR, Obal F, Jr, Krueger JM. Effects of alpha-MSH on sleep, behavior, and brain temperature: interactions with IL 1. Am J Physiol. 1988;255:R914–22. doi: 10.1152/ajpregu.1988.255.6.R914. [DOI] [PubMed] [Google Scholar]
- 34.Krueger JM, Walter J, Dinarello CA, Wolff SM, Chedid L. Sleep-promoting effects of endogenous pyrogen (interleukin-1) Am J Physiol. 1984;246:R994–9. doi: 10.1152/ajpregu.1984.246.6.R994. [DOI] [PubMed] [Google Scholar]
- 35.Krueger JM, Rector DM, Churchill L. Sleep and cytokines. Sleep Med Clin. 2007;2:161–9. doi: 10.1016/j.jsmc.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bianco F, Pravettoni E, Colombo A, Schenk U, Moller T, Matteoli M, Verderio C. Astrocyte-derived ATP induces vesicle shedding and IL-1 beta release from microglia. J Immunol. 2005;174:7268–77. doi: 10.4049/jimmunol.174.11.7268. [DOI] [PubMed] [Google Scholar]
- 37.Sanz JM, Di Virgilio F. Kinetics and mechanism of ATP-dependent IL-1 beta release from microglial cells. J Immunol. 2000;164:4893–8. doi: 10.4049/jimmunol.164.9.4893. [DOI] [PubMed] [Google Scholar]
- 38.Krueger JM, Obal FJ, Fang J, Kubota T, Taishi P. The role of cytokines in physiological sleep regulation. Ann N Y Acad Sci. 2001;933:211–21. doi: 10.1111/j.1749-6632.2001.tb05826.x. [DOI] [PubMed] [Google Scholar]
- 39.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.