Abstract
Genetic linkage studies in bipolar disorder (BPD) suggest that a susceptibility locus exists on chromosome 18p11. The metallophosphoesterase (MPPE1) gene maps to this region. Dysregulation of protein phosphorylation and subsequent abnormal cellular signaling has been postulated to be involved in neuropsychiatric disorders thus making MPPE1 a plausible biological candidate gene for BPD. In this study, we hypothesized that genetic variation in the MPPE1 gene contributes to BPD. We tested this hypothesis by genotyping 4 SNPs (rs871044; rs3974590; rs593713; rs602201) in BPD patients (n=570) and healthy controls (n=725). Genotypes and allele frequencies were compared between groups using Chi square contingency analysis. Linkage disequilibrium (LD) between markers was calculated and estimated haplotype frequencies were compared between groups. Single marker analysis revealed an association of rs3974590 with BPD (p=0.009; permutation corrected p=0.046). Haplotype analysis did not show any significant association with disease after permutation correction. Our results provide evidence of an association between a polymorphism in the MPPE1 gene and BPD. Additional studies are necessary to confirm and elucidate the role of MPPE1 as a susceptibility gene for BPD on chromosome 18p.
Keywords: linkage, chromosome 18, association, polymorphism, MPPE1, metallophosphoesterase 1
Bipolar disorder (BPD) is a chronic psychiatric disease, which affects approximately 1% of the general population and is characterized by episodes of mania and depression. Family, adoption and twin studies show that BPD has a strong genetic component (Craddock and Jones 1999). Linkage studies in BPD suggest that a susceptibility locus exists on chromosome 18p11 (Bennett and others 2002; Berrettini and others 1994; Bowen and others 1999; Detera-Wadleigh and others 1999; McInnes and others 1996; Mukherjee and others 2006; Nothen and others 1999; Stine and others 1995). Several genes in that region have been investigated previously but results are inconclusive and no functional variations of major effect have been identified so far (Berrettini and others 1998; Chavarria-Siles and others 2007; Corradi and others 2005; Esterling and others 1997; Evans and others 2008; Ishiguro and others 2001; Lohoff and Berrettini 2005; Lohoff and others 2008a; Lohoff and others 2008b; Mulle and others 2007; Ohnishi and others 2007; Reyes and others 2002; Rojas and others 2000; Tsiouris and others 1996; Washizuka and others 2004; Washizuka and others 2003; Weller and others 2006; Yoshikawa and others 2000; Yoshikawa and others 1997). In addition, recent whole genome association analyses failed to show a significant association between BPDs and the chromosome 18p region (Baum and others 2008; Ferreira and others 2008; Sklar and others 2008; TheWelcomeTrustCaseControlConsortium 2007).
Difficulties in elucidating BPD susceptibility factors in the 18p region might be due to the complex mode of inheritance, clinical and locus heterogeneity and use of underpowered samples. Another hypothesis is that several genes with small effects might contribute to the linkage peak, complicating the detection of risk alleles by the classic single candidate gene approach. In an attempt to further investigate the 18p region in BPD, we identified and investigated the metallophosphoesterase (MPPE1) gene as another candidate gene (Vuoristo and Ala-Kokko 2001).
MPPE1 encodes a metallophosphoesterase protein that is widely brain expressed (Vuoristo and Ala-Kokko 2001) and is a member of the calcineurin-like phosphoesterase superfamily. Phosphoesterases are involved in a variety of diverse biochemical reactions, including protein phosphorylation-dephosphorylation processes that modulate functional properties of proteins. Despite the large number of metallophosphoesterases that have been biochemically characterized, the function of MPPE1 remains unknown and no natural substrate has been identified so far (Miller and others 2007; Vogel and others 2002). Nevertheless, variation in the MPPE1 gene might lead to an altered enzyme with downstream effects on protein phosphorylation involved in cellular signaling. In fact, emergent data suggest that alterations in protein phosphorylation play a critical role in dopaminergic neurotransmission implicated in BPD and schizophrenia, as shown for the phosphoprotein DARPP-32 (Greengard and others 1999; Liu and others 2005). Dysregulation of protein phosphorylation and subsequent abnormal cellular signaling might contribute to the etiology of neuropsychiatric disorders thus making MPPE1 a plausible biological candidate gene for BPD. Based on the chromosomal location and the biological function of the MPPE1 gene we hypothesize that genetic variants might increase susceptibility to neuropsychiatric syndromes. To test this hypothesis, we designed a case-control association study using BPD patients and healthy controls of European descent and genotyped four genetic markers distributed across the MPPE1 gene.
DNA samples from five hundred and seventy unrelated BPD type I patients were collected at centers involved in the National Institute of Mental Health (NIMH) Genetics Initiative on BPD (http://zork.wustl.edu/nimh/bp.html). The diagnosis of BPD type I was in accordance with the DSM-IV criteria. Background, inclusion and exclusion criteria as well as detailed methodology for the NIMH Genetics Initiative are described elsewhere (Dick and others 2003). All subjects were assessed with the Diagnostic Instrument for Genetic Studies (DIGS) (Nurnberger and others 1994). Family history information was obtained through the Family Interview for Genetic Studies (FIGS), and medical records were requested. Final best estimate diagnosis was made using all available information including medical records, information from relatives, and the DIGS interview, by two independent senior diagnosticians adhering to DSM-IV criteria. The patient group consisted of 38% males and 62% females. The average age at recruitment was 41.6 years. Psychotic symptoms were present in 66% of the probands at some point during their illness. Psychosis was defined as presence of auditory/visual hallucinations and/or paranoid or bizarre delusions. Seven hundred and twenty-five DNA samples from unrelated controls were collected by the NIMH Genetic Initiative (http://www.nimhgenetics.org). Control participants were screen online using a self-report-based survey on the Composite International Diagnostic Interview Short-Form (CIDI-SF) (Kessler and others 1998). Accepted controls had no history of psychiatric or chronic neurological disease and consisted of 50% males and 50% females with an average age of 51.8 years at recruitment. All cases and controls were of European descent. Informed consent was obtained from all individuals in accordance with Institutional Review Board (IRB) procedures. DNA was extracted from peripheral leukocytes using standard protocol.
The MPPE1 gene is located on chromosome 18p11.21 and is flanked by the G protein Golf alpha (GNAL) gene and the myo-inositol monophosphatase gene (IMPA2). Interestingly, MPPE1 and GNAL are oriented tail-to-tail with partially overlapping 3′UTRs (Vuoristo and Ala-Kokko 2001). MPPE1 contains 12 exons and spans 25,090 bp (Ensembl Human Exon View accession OTTHUMG00000131661). SNPs for genotyping were selected using the tagging SNP algorithm based on available HapMap data with a minor allele frequency > 0.25 in the Caucasian European population and a pairwise linkage disequilibrium (LD) r2 cutoff of > 0.8 (SNP1: rs871044; SNP2: rs3974590; SNP3: rs593713; SNP4: rs602201). SNP genotyping was performed using Applied Biosystems Inc. (ABI) (Foster City, CA, USA) ‘Assays-on-demand’ as per manufacturer protocol. Quality control was maintained by genotyping 10% duplicates for cases and controls.
Genotype and allele frequencies were compared between groups using X2 contingency analysis. A two-tailed type I error rate of 5% was chosen for the analysis. Linkage disequilibrium (LD) and haplotype frequencies were estimated using the Haploview software (version 4.1). Haplotype blocks were identified using the solid spine of LD method in Haploview (Barrett and others 2005). Correction for multiple testing was performed using permutation correction by the Haploview program (Barrett and others 2005). This approach corrects for multiple testing but takes into account the correlation between markers. Permutation correction is thus less conservative than the Bonferroni correction but is appropriate for independent tests with multiple markers (Camargo and others 2008). For the single-marker analysis, 10,000 permutations were carried out to estimate the significance of the best results, correcting for the four loci tested. Haplotype analysis was performed using the Haploview software and P-values were corrected by permutation analysis as described above. Hardy-Weinberg equilibrium (HWE) was calculated separately for cases and controls. Our sample size had reasonable power to detect a disease association at a P value less than or equal to 0.05, assuming an odds ratio of 1.5 and a minor allele frequency (MAF) of 25% (99% for a log additive mode of inheritance, 92% for a dominant and 43% for a recessive mode of inheritance). Power analysis was performed using the Quanto program (Gauderman, 2002).
None of the genotype distributions deviated significantly from those expected by HWE for cases or controls. LD measures and haplotype blocks across the MPPE1 gene are shown in Fig. 1. We observed weak LD across the gene with the exception of SNP2 and SNP3 that are in strong LD. Single marker analysis (Table 1) revealed a statistically significant association of rs3974590 with BPD (SNP2: allelic p=0.009; permutation corrected p=0.046) and a trend towards association for SNP3 (p=0.051). Haplotype analysis of all four SNP configurations did not reveal an association with BPD (Table 2). However, two-marker haplotype analysis, involving the two markers in strong LD (SNP2 and SNP3), revealed an association between the GA haplotype and BPD (Table 3). Genotyping success rates were between 97.1% and 98.2%. The concordance rate for all the markers was 100% with respect to the 10% of samples that were genotyped twice for quality control. Allelic frequencies were consistent with those reported in the HapMap database for Utah residents with Northern and Western Europe ancestry from the CEPH collection.
Figure 1.
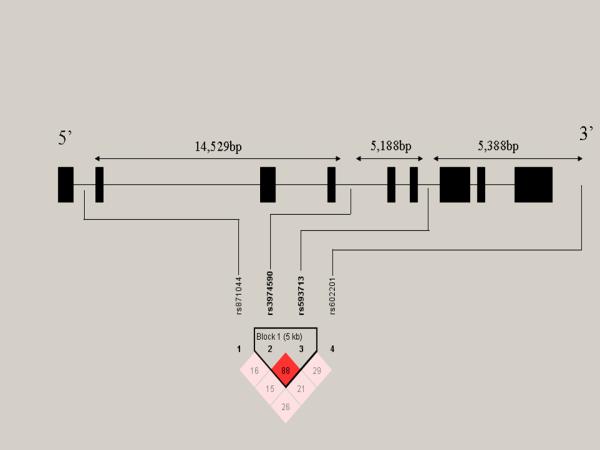
Linkage disequilibrium measures (D’) across the MPPE1 gene. From left to right, the SNPs are aligned from 5′ to 3′. Black boxes represent exons. LD patterns and haplotype blocks were defined by the “solid spine of LD” using the Haploview software. A standard color scheme is used to display LD pattern, with dark red for very strong LD, white for no LD, and shades of red for intermediate LD. Increasing intensity of red indicates increasing degrees of LD.
Table 1.
Genotype and Allele frequencies of variations in the MPPE1 gene
| SNP | Sample | n | Genotype frequency | P* | Allele frequency |
P** | ||
|---|---|---|---|---|---|---|---|---|
| rs871044 | G/G | G/A | A/A | f(G) | ||||
| Bipolar | 554 | 0.354 | 0.478 | 0.168 | 0.498 | 0.593 | 0.704 | |
| Controls | 722 | 0.348 | 0.506 | 0.147 | 0.600 | |||
| rs3974590 | G/G | G/A | A/A | f(G) | ||||
| Bipolar | 567 | 0.467 | 0.441 | 0.092 | 0.022 | 0.688 | 0.009a | |
| Controls | 724 | 0.544 | 0.381 | 0.075 | 0.735 | |||
| rs593713 | A/A | A/G | G/G | f(A) | ||||
| Bipolar | 563 | 0.472 | 0.435 | 0.092 | 0.122 | 0.690 | 0.051 | |
| Controls | 719 | 0.530 | 0.391 | 0.079 | 0.725 | |||
| rs602201 | T/T | T/A | A/A | f(T) | ||||
| Bipolar | 565 | 0.405 | 0.469 | 0.126 | 0.946 | 0.640 | 0.739 | |
| Controls | 712 | 0.397 | 0.472 | 0.131 | 0.633 | |||
P values for comparison of genotype frequencies between bipolar individuals and controls
P values for comparison of allele frequencies between bipolar individuals and controls.
Permutation correction p-value: 0.046
Table 2.
Analysis of four-marker haplotypes in the MPPE1 gene
| Haplotype block |
Case frequencies |
Control frequencies |
Chi square |
P Value |
Permutation P value |
|---|---|---|---|---|---|
| Block 1 | |||||
| GGAT | 0.278 | 0.294 | 0.790 | 0.374 | 0.962 |
| AGAT | 0.192 | 0.206 | 0.761 | 0.383 | 0.966 |
| GGAA | 0.146 | 0.157 | 0.533 | 0.465 | 0.991 |
| GAGA | 0.095 | 0.091 | 0.131 | 0.717 | 1.000 |
| AAGT | 0.095 | 0.077 | 2.495 | 0.114 | 0.503 |
| AAGA | 0.053 | 0.047 | 0.415 | 0.520 | 0.997 |
| AGAA | 0.044 | 0.051 | 0.680 | 0.410 | 0.977 |
| GAGT | 0.034 | 0.033 | 0.651 | 0.420 | 0.981 |
| GAAT | 0.017 | 0.009 | 2.786 | 0.095 | 0.431 |
Table 3.
Analysis of two-marker haplotypes in the MPPE1 gene
| Haplotype block |
Case frequencies |
Control frequencies |
Chi square |
P Value |
Permutation P value |
|---|---|---|---|---|---|
| Block 1 | |||||
| GA | 0.661 | 0.708 | 6.589 | 0.010 | 0.0495 |
| AG | 0.282 | 0.249 | 3.607 | 0.058 | 0.259 |
| GG | 0.027 | 0.026 | 0.054 | 0.816 | 1.000 |
| AA | 0.030 | 0.017 | 4.475 | 0.034 | 0.170 |
In the present study, we show a statistically significant association between polymorphisms in the MPPE1 gene on chromosome 18p and BPD. Interestingly, both associated markers (SNP2 and SNP3) are in strong LD (D’:0.88; r2: 0.77); however, only SNP2 remains statistically significant after correction for multiple testing. Our results indicate a protective effect of the major allele for SNP2 and SNP3. Haplotype analysis of these 2 markers further confirms a protective effect, with the frequency of the major allele diplotype (G-A) in the control group being significantly increased when compared to cases (70% versus 66%; corrected p-value: 0.04).
Although there are no data available in the literature that would suggest that either of the associated SNPs has functional effects, the MPPE1 protein contains metal binding and active sites similar to serine/threonine phosphoprotein phosphatase catalytic subunits. These phospatases are involved in a variety of cellular processes, including gene expression, cell growth and cell differentiation. Dysregulation of protein phosphorylation and subsequent abnormal cellular signaling might play a significant role in the pathophysiology of neuropsychiatric disorders (Greengard and others 1999; Liu and others 2005), thus making MPPE1 a plausible biological candidate gene for BPD as well as other phenotypes such as schizophrenia. Even though our study provides evidence for a possible association between variation in the MPPE1 gene and BPD, it could be possible that other variations which are in LD with these SNPs might contribute to the observed association or that a haplotype confers risk rather than a single polymorphism. In fact, several studies investigating the adjacent genes GNAL and IMPA2 documented some positive and negative associations of genetic polymorphisms with BPD (Corradi and others 2005; Ohnishi and others 2007; Sjoholt and others 2004; Sjoholt and others 2000; Tsiouris and others 1996; Vuoristo and others 2000; Yoshikawa and others 1997). Together, these studies suggest genetic heterogeneity or that multiple genes of small effect on chromosome 18p confer risk or protective properties to BPD. Additional genetic (including sequencing), computational and biological studies are necessary to further evaluate the role of the MPPE1, GNAL and IMPA2 in this BPD linkage region.
There are several limitations to our study that should be mentioned. First, it is possible that our finding might be a false positive result due to population stratification. All cases and controls in this study were of European descent; however, undetected differences in population structure might contribute to false positive results in association studies (Freedman and others 2004; Pritchard and Donnelly 2001). Furthermore, cases and controls were not matched for age, gender or regional European ancestry which might also contribute to population stratification effects. Possible strategies to control for these stratification issues are the use of genomic controls (Bacanu and others 2000; Devlin and Roeder 1999) and/or the use of a family-based association design, a method that matches the genotype of an affected offspring with parental alleles not inherited by the offspring (Spielman and Ewens 1996). In addition to population stratification issues, spurious positive association findings remain a valid concern as shown recently in a statistical simulation study of the COMT gene by Sullivan (2007). Thus, our results should be interpreted with caution and ultimately require careful replication and confirmation in an independent population of patients and controls.
Our candidate gene was selected based on prior linkage evidence for BPD on chromosome 18p and suggestive evidence of phosphorylation and signal transduction dysregulation in affective disorders (Newberg and others 2008). This candidate gene approach has the advantage of taking prior research into account and provides adequate power for genes of small to moderate effect sizes. While recent technological advances have made it possible to conduct genome-wide association studies (GWAS) in complex diseases like BPD (Baum and others 2008; TheWelcomeTrustCaseControlConsortium 2007), the initial results have been limited with a lack of breakthrough findings and little or no replication between several datasets. Although the concept of a “hypothesis-free” design in GWAS is appealing as an approach to discover new genes and pathways involved in neuropsychiatric disease, it ignores previous advances in neuroscience research and disregards a priori biological relevance. It is furthermore likely that much larger sample sizes are needed for GWAS, in the ten-thousands, in order to achieve adequate power in the face of multiple testing (Frayling 2007). Candidate gene approaches, including cluster analyses of genes involved in neurobiological pathways, are thus still a reasonable strategy to investigate genetic factors of complex disorders (Serretti and Mandelli 2008). Although our SNP selection was based on available HapMap data and four tagging SNPs were selected in order to cover the majority of the gene region, it is possible that we missed additional markers that might have been associated with disease. As indicated by the relatively weak observed LD, additional genotyping of markers might improve gene coverage; however, ultimately future studies will also have to include deep sequencing of this genomic region in order to detect rare variants that might confer risk to disease.
In conclusion, we show a positive association between polymorphisms in the MPPE1 gene and BPD. While the study requires replication in an independent sample, it provides additional information on the 18p genomic region and serves to further characterize this region that has been implicated in BPD. Additional experiments, including subphenotype and endophenotypic analyses, are necessary to elucidate the genetic factors contributing to the linkage peak on 18p11.
Acknowledgments
This work was supported by the Center for Neurobiology and Behavior, Department of Psychiatry, University of Pennsylvania. Financial support is gratefully acknowledged from National Institutes of Health grants K08MH080372 (F.W.L.), a grant from the Tzedakah Foundation (W.H.B.) and a grant from Philip and Marcia Cohen (W.H.B.). We thank Candice Schwebel, Aleksandra Nall and Rachel Hodge for technical assistance. We thank the families who have participated in and contributed to these studies. Most importantly, we thank the subjects who have participated in and contributed to these studies.
Data and biomaterials utilized in this study were collected as part of ten projects that participated in the National Institute of Mental Health (NIMH) Bipolar Disorder Genetics Initiative. From 1999-03, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, R01 MH59545, John Nurnberger, M.D., Ph.D., Marvin Miller, M.D., Elizabeth Bowman, M.D., N. Leela Rau, M.D, P. Ryan Moe, M.D., Nalini Samavedy, M.D., Rif El-Mallakh, M.D, (at University of Louisville), Husseini Manji, M.D. (at Wayne State University), Debra A. Blitz, M.D (at Wayne State University), Eric T. Meyer, M.S., Carrie Smiley, R.N., Tatiana Foroud, Ph.D., Leah Flury, M.S., Danielle M. Dick, Ph.D., Howard Edenberg, Ph.D.; Washington University, St. Louis, MO, R01 MH059534, John Rice, Ph.D., Theodore Reich, M.D., Allison Goate, Ph.D., Laura Bierut, M.D.; Johns Hopkins University, Baltimore, MD, R01 MH59533, Melvin McInnis, M.D., J. Raymond DePaulo, Jr., M.D., Dean F. MacKinnon, M.D., Francis M. Mondimore, M.D., James B. Potash, M.D., Peter P. Zandi, Ph.D, Dimitrios Avramopoulos, Jennifer Payne; University of Pennsylvania, PA, R01 MH59553, Wade Berrettini, M.D., Ph.D.; University of California at Irvine, CA, R01 MH60068, William Byerley, M.D. and Mark Vawter, M.D.; University of Iowa, IA, R01 MH059548, William Coryell, M.D., Raymond Crowe, M.D.; University of Chicago, IL, R01 MH059535, Elliot Gershon, M.D., Judith Badner, Ph.D., Francis McMahon, M.D., Chunyu Liu, Ph.D., Alan Sanders, M.D., Maria Caserta, Steven Dinwiddie, M.D., Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567, John Kelsoe, M.D., Rebecca McKinney, B.A.; Rush University, IL, R01 MH059556, William Scheftner, M.D., Howard M. Kravitz, D.O., M.P.H., Diana Marta, B.S., Annette Vaughn-Brown, M.S.N., R.N., Laurie Bederow, M.A.; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH02810-01, Francis J. McMahon, M.D., Layla Kassem, PsyD., Sevilla Derta-Wadleigh, Ph.D., Lisa Austin, Ph.D., Dennis L. Murphy, M.D.
References
- Bacanu SA, Devlin B, Roeder K. The power of genomic control. Am J Hum Genet. 2000;66(6):1933–44. doi: 10.1086/302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, Schulze TG, Cichon S, Rietschel M, Nothen MM. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13(2):197–207. doi: 10.1038/sj.mp.4002012. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P, Segurado R, Jones I, Bort S, McCandless F, Lambert D, Heron J, Comerford C, Middle F, Corvin A. The Wellcome trust UK-Irish bipolar affective disorder sibling-pair genome screen: first stage report. Mol Psychiatry. 2002;7(2):189–200. doi: 10.1038/sj.mp.4000957. others. [DOI] [PubMed] [Google Scholar]
- Berrettini WH, Ferraro TN, Goldin LR, Weeks DE, Detera-Wadleigh S, Nurnberger JI, Jr., Gershon ES. Chromosome 18 DNA markers and manic-depressive illness: evidence for a susceptibility gene. Proc Natl Acad Sci U S A. 1994;91(13):5918–21. doi: 10.1073/pnas.91.13.5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrettini WH, Vuoristo J, Ferraro TN, Buono RJ, Wildenauer D, Ala-Kokko L. Human G(olf) gene polymorphisms and vulnerability to bipolar disorder. Psychiatr Genet. 1998;8(4):235–8. doi: 10.1097/00041444-199808040-00006. [DOI] [PubMed] [Google Scholar]
- Bowen T, Kirov G, Gill M, Spurlock G, Vallada HP, Murray RM, McGuffin P, Collier DA, Owen MJ, Craddock N. Linkage studies of bipolar disorder with chromosome 18 markers. Am J Med Genet. 1999;88(5):503–9. [PubMed] [Google Scholar]
- Camargo A, Azuaje F, Wang H, Zheng H. Permutation - based statistical tests for multiple hypotheses. Source Code Biol Med. 2008;3(1):15. doi: 10.1186/1751-0473-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Siles I, Walss-Bass C, Quezada P, Dassori A, Contreras S, Medina R, Ramirez M, Armas R, Salazar R, Leach RJ. TGFB-induced factor (TGIF): a candidate gene for psychosis on chromosome 18p. Mol Psychiatry. 2007;12(11):1033–41. doi: 10.1038/sj.mp.4001997. others. [DOI] [PubMed] [Google Scholar]
- Corradi JP, Ravyn V, Robbins AK, Hagan KW, Peters MF, Bostwick R, Buono RJ, Berrettini WH, Furlong ST. Alternative transcripts and evidence of imprinting of GNAL on 18p11.2. Mol Psychiatry. 2005;10(11):1017–25. doi: 10.1038/sj.mp.4001713. [DOI] [PubMed] [Google Scholar]
- Craddock N, Jones I. Genetics of bipolar disorder. J Med Genet. 1999;36(8):585–94. doi: 10.1136/jmg.36.8.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, Badner JA, Berrettini WH, Yoshikawa T, Goldin LR, Turner G, Rollins DY, Moses T, Sanders AR, Karkera JD. A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc Natl Acad Sci U S A. 1999;96(10):5604–9. doi: 10.1073/pnas.96.10.5604. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Foroud T, Flury L, Bowman ES, Miller MJ, Rau NL, Moe PR, Samavedy N, El-Mallakh R, Manji H. Genomewide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73(1):107–14. doi: 10.1086/376562. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterling LE, Cox Matise T, Sanders AR, Yoshikawa T, Overhauser J, Gershon ES, Moskowitz MT, Detera-Wadleigh SD. An integrated physical map of 18p11.2: a susceptibility region for bipolar disorder. Mol Psychiatry. 1997;2(6):501–4. doi: 10.1038/sj.mp.4000317. [DOI] [PubMed] [Google Scholar]
- Evans LM, Akiskal HS, Greenwood TA, Nievergelt CM, Keck PE, Jr., McElroy SL, Sadovnick AD, Remick RA, Schork NJ, Kelsoe JR. Suggestive linkage of a chromosomal locus on 18p11 to cyclothymic temperament in bipolar disorder families. Am J Med Genet B Neuropsychiatr Genet. 2008;147(3):326–32. doi: 10.1002/ajmg.b.30601. [DOI] [PubMed] [Google Scholar]
- Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40(9):1056–8. doi: 10.1038/ng.209. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8(9):657–62. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36(4):388–93. doi: 10.1038/ng1333. others. [DOI] [PubMed] [Google Scholar]
- Greengard P, Allen PB, Nairn AC. Beyond the dopamine receptor: the DARPP-32/protein phosphatase-1 cascade. Neuron. 1999;23(3):435–47. doi: 10.1016/s0896-6273(00)80798-9. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Ohtsuki T, Okubo Y, Kurumaji A, Arinami T. Association analysis of the pituitary adenyl cyclase activating peptide gene (PACAP) on chromosome 18p11 with schizophrenia and bipolar disorders. J Neural Transm. 2001;108(7):849–54. doi: 10.1007/s007020170034. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Andrews G, Mroczek D, Ustun B, Wittchen H. The World Health Organization Composite International Diagnostic Interview short-form (CIDI-SF) International Journal of Methods in Psychiatric Research. 1998;7(4):171–185. [Google Scholar]
- Liu QR, Gong JP, Uhl GR. Families of protein phosphatase 1 modulators activated by protein kinases a and C: focus on brain. Prog Nucleic Acid Res Mol Biol. 2005;79:371–404. doi: 10.1016/S0079-6603(04)79008-X. [DOI] [PubMed] [Google Scholar]
- Lohoff FW, Berrettini WH. Lack of association between variations in the melanocortin 5 receptor gene and bipolar disorder. Psychiatr Genet. 2005;15(4):255–8. doi: 10.1097/00041444-200512000-00007. [DOI] [PubMed] [Google Scholar]
- Lohoff FW, Bloch PJ, Weller AE, Ferraro TN, Berrettini WH. Association analysis of the pituitary adenylate cyclase-activating polypeptide (PACAP/ADCYAP1) gene in bipolar disorder. Psychiatr Genet. 2008a;18(2):53–8. doi: 10.1097/YPG.0b013e3282f60320. [DOI] [PubMed] [Google Scholar]
- Lohoff FW, Weller AE, Bloch PJ, Nall AH, Ferraro TN, Berrettini WH. Association between polymorphisms in the vesicle-associated membrane protein-associated protein A (VAPA) gene on chromosome 18p and bipolar disorder. J Neural Transm. 2008b;115(9):1339–45. doi: 10.1007/s00702-008-0093-9. [DOI] [PubMed] [Google Scholar]
- McInnes LA, Escamilla MA, Service SK, Reus VI, Leon P, Silva S, Rojas E, Spesny M, Baharloo S, Blankenship K. A complete genome screen for genes predisposing to severe bipolar disorder in two Costa Rican pedigrees. Proc Natl Acad Sci U S A. 1996;93(23):13060–5. doi: 10.1073/pnas.93.23.13060. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Shuvalova L, Evdokimova E, Savchenko A, Yakunin AF, Anderson WF. Structural and biochemical characterization of a novel Mn2+-dependent phosphodiesterase encoded by the yfcE gene. Protein Sci. 2007;16(7):1338–48. doi: 10.1110/ps.072764907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee O, Meera P, Ghosh S, Kubendran S, Kiran K, Manjunath KR, Subhash MN, Benegal V, Brahmachari SK, Majumder PP. Evidence of linkage and association on 18p11.2 for psychosis. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(8):868–873. doi: 10.1002/ajmg.b.30363. others. [DOI] [PubMed] [Google Scholar]
- Mulle JG, Fallin MD, Lasseter VK, McGrath JA, Wolyniec PS, Pulver AE. Dense SNP association study for bipolar I disorder on chromosome 18p11 suggests two loci with excess paternal transmission. Mol Psychiatry. 2007;12(4):367–75. doi: 10.1038/sj.mp.4001916. [DOI] [PubMed] [Google Scholar]
- Newberg AR, Catapano LA, Zarate CA, Manji HK. Neurobiology of bipolar disorder. Expert Rev Neurother. 2008;8(1):93–110. doi: 10.1586/14737175.8.1.93. [DOI] [PubMed] [Google Scholar]
- Nothen MM, Cichon S, Rohleder H, Hemmer S, Franzek E, Fritze J, Albus M, Borrmann-Hassenbach M, Kreiner R, Weigelt B. Evaluation of linkage of bipolar affective disorder to chromosome 18 in a sample of 57 German families. Mol Psychiatry. 1999;4(1):76–84. doi: 10.1038/sj.mp.4000454. others. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr., Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51(11):849–59. doi: 10.1001/archpsyc.1994.03950110009002. discussion 863-4. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Yamada K, Ohba H, Iwayama Y, Toyota T, Hattori E, Inada T, Kunugi H, Tatsumi M, Ozaki N. A promoter haplotype of the inositol monophosphatase 2 gene (IMPA2) at 18p11.2 confers a possible risk for bipolar disorder by enhancing transcription. Neuropsychopharmacology. 2007;32(8):1727–37. doi: 10.1038/sj.npp.1301307. others. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Donnelly P. Case-control studies of association in structured or admixed populations. Theoretical Population Biology. 2001;60(3):227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- Reyes GD, Esterling LE, Corona W, Ferraren D, Rollins DY, Padigaru M, Yoshikawa T, Monje VD, Detera-Wadleigh SD. Map of candidate genes and STSs on 18p11.2, a bipolar disorder and schizophrenia susceptibility region. Mol Psychiatry. 2002;7(4):337–9. doi: 10.1038/sj.mp.4001000. [DOI] [PubMed] [Google Scholar]
- Rojas K, Liang L, Johnson EI, Berrettini WH, Overhauser J. Identification of candidate genes for psychiatric disorders on 18p11. Mol Psychiatry. 2000;5(4):389–95. doi: 10.1038/sj.mp.4000737. [DOI] [PubMed] [Google Scholar]
- Serretti A, Mandelli L. The genetics of bipolar disorder: genome ‘hot regions,’ genes, new potential candidates and future directions. Mol Psychiatry. 2008;13(8):742–71. doi: 10.1038/mp.2008.29. [DOI] [PubMed] [Google Scholar]
- Sjoholt G, Ebstein RP, Lie RT, Berle JO, Mallet J, Deleuze JF, Levinson DF, Laurent C, Mujahed M, Bannoura I. Examination of IMPA1 and IMPA2 genes in manic-depressive patients: association between IMPA2 promoter polymorphisms and bipolar disorder. Mol Psychiatry. 2004;9(6):621–9. doi: 10.1038/sj.mp.4001460. others. [DOI] [PubMed] [Google Scholar]
- Sjoholt G, Gulbrandsen AK, Lovlie R, Berle JO, Molven A, Steen VM. A human myo-inositol monophosphatase gene (IMPA2) localized in a putative susceptibility region for bipolar disorder on chromosome 18p11.2: genomic structure and polymorphism screening in manic-depressive patients. Mol Psychiatry. 2000;5(2):172–80. doi: 10.1038/sj.mp.4000681. [DOI] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, Nimgaonkar VL, McQueen MB, Faraone SV, Kirby A. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13(6):558–69. doi: 10.1038/sj.mp.4002151. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, Ewens WJ. The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet. 1996;59(5):983–9. [PMC free article] [PubMed] [Google Scholar]
- Stine OC, Xu J, Koskela R, McMahon FJ, Gschwend M, Friddle C, Clark CD, McInnis MG, Simpson SG, Breschel TS. Evidence for linkage of bipolar disorder to chromosome 18 with a parent-of-origin effect. Am J Hum Genet. 1995;57(6):1384–94. others. [PMC free article] [PubMed] [Google Scholar]
- TheWelcomeTrustCaseControlConsortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiouris SJ, Breschel TS, Xu J, McInnis MG, McMahon FJ. Linkage disequilibrium analysis of G-olf alpha (GNAL) in bipolar affective disorder. Am J Med Genet. 1996;67(5):491–4. doi: 10.1002/(SICI)1096-8628(19960920)67:5<491::AID-AJMG11>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Vogel A, Schilling O, Niecke M, Bettmer J, Meyer-Klaucke W. ElaC encodes a novel binuclear zinc phosphodiesterase. J Biol Chem. 2002;277(32):29078–85. doi: 10.1074/jbc.M112047200. [DOI] [PubMed] [Google Scholar]
- Vuoristo JT, Ala-Kokko L. cDNA cloning, genomic organization and expression of the novel human metallophosphoesterase gene MPPE1 on chromosome 18p11.2. Cytogenet Cell Genet. 2001;95(1-2):60–3. doi: 10.1159/000057018. [DOI] [PubMed] [Google Scholar]
- Vuoristo JT, Berrettini WH, Overhauser J, Prockop DJ, Ferraro TN, Ala-Kokko L. Sequence and genomic organization of the human G-protein Golfalpha gene (GNAL) on chromosome 18p11, a susceptibility region for bipolar disorder and schizophrenia. Mol Psychiatry. 2000;5(5):495–501. doi: 10.1038/sj.mp.4000758. [DOI] [PubMed] [Google Scholar]
- Washizuka S, Iwamoto K, Kazuno AA, Kakiuchi C, Mori K, Kametani M, Yamada K, Kunugi H, Tajima O, Akiyama T. Association of mitochondrial complex I subunit gene NDUFV2 at 18p11 with bipolar disorder in Japanese and the National Institute of Mental Health pedigrees. Biol Psychiatry. 2004;56(7):483–9. doi: 10.1016/j.biopsych.2004.07.004. others. [DOI] [PubMed] [Google Scholar]
- Washizuka S, Kakiuchi C, Mori K, Kunugi H, Tajima O, Akiyama T, Nanko S, Kato T. Association of mitochondrial complex I subunit gene NDUFV2 at 18p11 with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2003;120(1):72–8. doi: 10.1002/ajmg.b.20041. [DOI] [PubMed] [Google Scholar]
- Weller AE, Dahl JP, Lohoff FW, Ferraro TN, Berrettini WH. Analysis of variations in the NAPG gene on chromosome 18p11 in bipolar disorder. Psychiatr Genet. 2006;16(1):3–8. doi: 10.1097/01.ypg.0000180678.88169.b0. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Padigaru M, Karkera JD, Sharma M, Berrettini WH, Esterling LE, Detera-Wadleigh SD. Genomic structure and novel variants of myo-inositol monophosphatase 2 (IMPA2) Mol Psychiatry. 2000;5(2):165–71. doi: 10.1038/sj.mp.4000688. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Turner G, Esterling LE, Sanders AR, Detera-Wadleigh SD. A novel human myo-inositol monophosphatase gene, IMP.18p, maps to a susceptibility region for bipolar disorder. Mol Psychiatry. 1997;2(5):393–7. doi: 10.1038/sj.mp.4000325. [DOI] [PubMed] [Google Scholar]


