Genetically encoded unnatural amino acids (UAAs) enable novel chemical and physical properties to be selectively introduced into proteins directly in live cells, which provides great potential for addressing biological questions at the molecular and cellular level in native settings.[1-3] UAAs have been genetically incorporated into proteins in mammalian cells using orthogonal tRNA-codon-synthetase sets,[4-6] yet the current low incorporation efficiency hinders their effective application. Efforts to improve efficiency have focused on optimizing the expression and activity of the orthogonal tRNA and synthetase,[5-7] whereas the bioavailability of the UAA inside mammalian cells, a prerequisite for incorporation, has not been addressed. In addition, there are many UAAs that have not been genetically incorporated into proteins successfully, such as glycosylated and phosphorylated amino acids. These amino acids can be invaluable for studying the contribution of posttranslational modifications to protein function and the role of a target protein in cellular signal transduction. Among many reasons, the inability of the UAA to enter cells prevents evolving a mutant synthetase specific for the UAA using cell-based selections or screens.[2] Here we show that an UAA structurally deviating from the canonical amino acids in side chain could not be efficiently transported into mammalian cells, but masking the carboxyl group of the UAA as an ester greatly increased the rate of cellular uptake and intracellular concentration of the UAA. This resulted in a significant increase in the incorporation of this UAA into proteins in mammalian cells. Among three esters tested, acetoxymethyl ester (AME) yielded the highest UAA incorporation efficiency with a concomitant reduction of the UAA required in the growth media.
UAAs with side chains similar to canonical amino acids can be transported into cells by endogenous amino acid transporters,[3, 8] which are relatively nonspecific for substrates.[9] However, UAAs significantly deviating from canonical amino acids in side chain structure may not be recognized by these transporters. Cell membranes are more permeable to neutral than charged molecules. As a zwitterion, UAA has only a very small proportion present in the neutral form at the physiological pH, making it difficult to cross the cell membrane. Esters have been widely used in prodrugs to derivatize carboxyl, hydroxyl and thio functionalities to promote membrane permeability of the parent drugs.[10] In particular, Tsien et al. pioneered the use of AME to enhance cellular uptake of various charged molecules such as carboxylate-containing Ca2+ chelators and phosphate-containing second messengers.[11-12] Inspired by these examples, we reasoned that masking the carboxyl group of the UAA with an ester would convert the UAA into a protonated weak base, which has a higher percentage of neutral form and increased lipophilicity to translocate the membrane. Once inside the cell, intracellular esterases can cleave the ester to regenerate the original UAA for incorporation.
We demonstrated this strategy with UAA 2-amino-3-(5-(dimethylamino)naphthalene-1-sulfonamide)propanoic acid (DanAla, 1), which does not resemble and is bulkier in size than any canonical amino acid. Its fluorescence property also makes it an attractive candidate to develop optical reporters for imaging protein activities in live cells. We synthesized the methyl, ethyl and acetoxymethyl ester of DanAla using methods shown in Scheme 1b. Compound 5 was synthesized from Boc-Dap-OH and dansylchloride according to the known procedure.[13] Alkylation of 5 with iodomethane and iodoethane gave intermediates 6 and 7, which afforded DanAla-OMe (2) and DanAla-OEt (3) after deprotection, respectively. However, in the presence of bromomethyl acetate and DIPEA, compound 5 was transformed to the undesired cyclization product 8, instead of Boc-DanAla-OAM. Therefore, the carboxyl group and the sulfonamide group were protected with the benzyl group and the Boc group,[14] respectively. After debenzylation, carboxylic acid 10 was transformed to acetoxymethyl ester 11,[15] which furnished DanAla-OAM (4) after deprotection of the Boc groups.
Scheme 1.
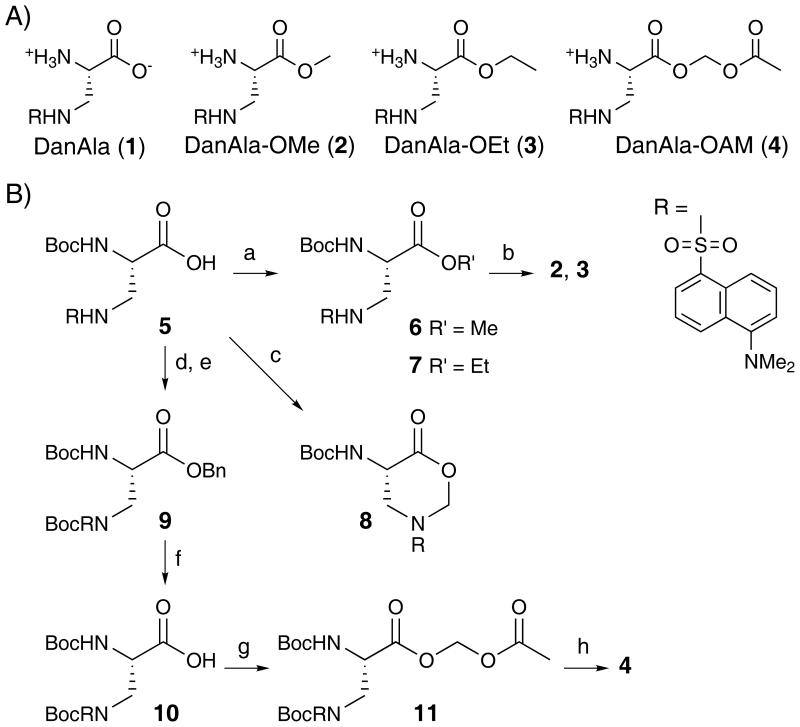
To determine if the DanAla esters could enhance the incorporation of DanAla into proteins in mammalian cells, we used an in cellulo fluorescence assay to evaluate the incorporation efficiency of DanAla.[6-7] The incorporation of DanAla into the green fluorescent protein (GFP) was measured using a stable clonal HeLa cell line, in which the GFP gene containing a premature UAG stop codon at a permissive site (Tyr182) was integrated into the genome. The cells were transfected with the orthogonal tRNA-synthetase pair specific for DanAla,[13] and DanAla or DanAla ester was added in the growth media. The incorporation of DanAla at the UAG182 position results in full-length, fluorescent GFP, whereas no incorporation results in truncated, nonfluorescent GFP. The total fluorescence intensity of cells was measured by flow cytometry. As shown in Figure 1a, the cell fluorescence intensity increased when more DanAla was added to the growth media, but reached a plateau from 0.25 mM to 1.00 mM. The plateau of DanAla incorporation suggests that either the concentration of DanAla inside cells has saturated the synthetase or the cellular availability of DanAla is limiting. When DanAla-OMe was used, the cell fluorescence intensity doubled in comparison to cells incubated with same concentrations of DanAla, indicating that the cellular availability of DanAla was the limiting factor. Similar results were also obtained for DanAla-OEt. Nonetheless, there was no significant increase in cell fluorescence intensity when the DanAla-OMe or DanAla-OEt was increased from 0.10 mM to 0.25 mM. In contrast, DanAla-OAM doubled the fluorescence intensity at 0.10 mM and quadrupled it at 0.25 mM. In comparison to 1.00 mM of DanAla, the amount often used in UAA incorporation, the DanAla-OAM increased the cell fluorescence intensity 4 fold in addition to requiring 75% less compound in the growth media (0.25 mM). Western blot analysis of the cell lysates confirmed the increase in the amount of GFP produced by the AME modification (Figure 1b). These results suggest that all three ester modifications were able to increase DanAla incorporation into proteins with a concomitant reduction of the extracellular supply of UAA. Furthermore we identified DanAla-OAM as the most effective out of the three esters tested.
Figure 1.
(A) Flow cytometric analysis of the incorporation efficiency of DanAla into GFP with different compounds added in the growth media. Reporter cells transfected with the orthogonal suppressor tRNA and the wild type LeuRS were used as the positive control to normalize the total fluorescence intensity. Error bars represent s.e.m., n = 3. (B) Western blot analysis of the GFP protein with a GFP-specific antibody. Same number of cells were used in lane 2 - 5.
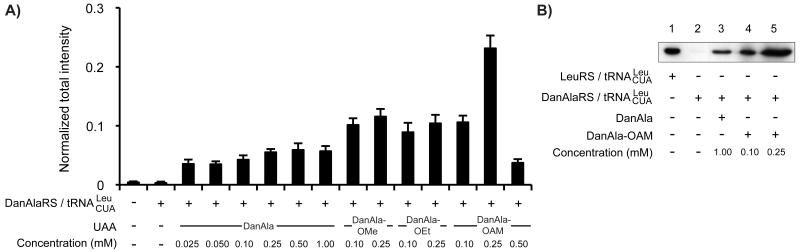
To understand how DanAla-OAM increases DanAla incorporation, we first monitored the uptake of DanAla and DanAla-OAM into cells using fluorescence microscopy. We added 0.10 mM of DanAla or DanAla-OAM to the media of HEK293T cells and measured the cytosolic fluorescence intensity of cells at sequential time points using confocal microscopy (Figure 2a). For cells incubated with DanAla, the cytosolic fluorescence intensity was almost flat in the range of 620-750 AU. A striking difference was observed in the DanAla-OAM incubated sample. Within 5 mins, the intracellular fluorescence intensity rapidly rose to 1100 AU, a 48% increase from the DanAla sample. The intensity increased for 3 hrs peaking at 1613 AU, which was 2-fold of the DanAla peak intensity. The intensity of DanAla-OAM treated cells then gradually dropped, possibly due to the depletion of the extracellular DanAla-OAM in combination with the equilibration of converted DanAla in and outside of cells. The AME modification thus significantly accelerates the cellular uptake rate of the compound.
Figure 2.
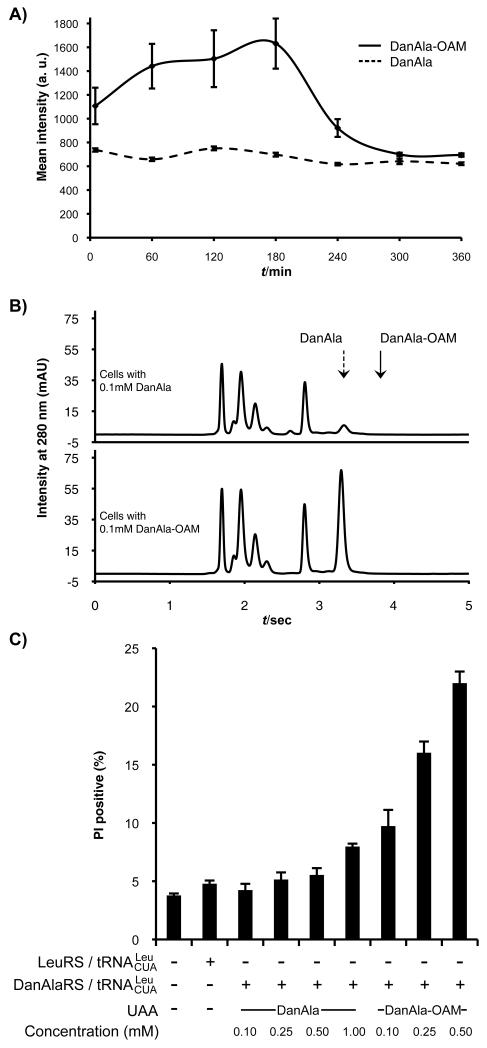
(A) Cytosolic fluorescence intensity of cells incubated with 0.10 mM DanAla or DanAla-OAM. Error bars represent s.d. (B) HPLC analysis of cell extracts from cells incubated with 0.10 mM DanAla or DanAla-OAM. Arrows indicate the peak positions for DanAla and DanAla-OAM determined using pure compounds. (C) PI staining analysis of cell health at indicated conditions. Error bars represent s.e.m., n = 3.
Next we quantified the intracellular concentrations of DanAla and DanAla-OAM using HPLC. HEK293T cells were incubated with 0.10 mM of the compounds for 1 hr, and cell contents were extracted. Small molecules in the cell extracts were separated by HPLC, and peaks corresponding to DanAla and DanAla-OAM were verified using pure compounds and mass spectrometry. Surprisingly, after only 1 hr incubation, cells incubated with DanAla-OAM showed no peak for DanAla-OAM but a large peak for DanAla (Figure 2b), indicating that all intracellular DanAla-OAM had been hydrolyzed to DanAla. The peak area for DanAla was used to determine its intracellular concentration. Cells incubated with DanAla and DanAla-OAM had 0.28 mM and 8.9 mM intracellular concentration of DanAla, respectively. The AME modification dramatically increased the intracellular concentration of DanAla by 31-fold.
When the concentration of DanAla-OAM was further increased to 0.50 mM, the cell fluorescence intensity decreased unexpectedly (Figure 1a). We used propidium iodide (PI) staining to assess the health of cells incubated with DanAla or DanAla-OAM (Figure 2c). For cells treated with DanAla from 0.10 mM to 0.50 mM, the percentage of PI positive cells was similar to that of cells without UAA. A slight increase of PI positive cells was observed at 1.00 mM of DanAla. In contrast, cells treated with DanAla-OAM showed significantly higher percentage of PI positive cells, especially at the concentration of 0.50 mM. DanAla-OAM hydrolysis releases formaldehyde, which will negatively affect cell health at high concentration.[11] High intracellular concentration of DanAla may be toxic to cells as well. Therefore, an optimal concentration of the AME ester should be used to achieve highest UAA incorporation efficiency with minimal negative effect to cell health.
In summary, we demonstrated that masking the carboxyl group of DanAla with AME increased the uptake rate and intracellular concentration of the UAA in mammalian cells. This resulted in a higher DanAla incorporation efficiency accompanied by the added benefit of reducing the amount of UAA in the growth media. Efficient UAA incorporation would prove valuable for the effective application of genetically encoded UAAs in studying various biological processes in mammalian cells. Modification of UAAs with AME may be generally applicable to other UAAs that are difficult to enter mammalian cells. To test this hypothesis, more unnatural amino acids need to be modified and their incorporation efficiency determined. However, for these candidate unnatural amino acids there are currently no orthogonal tRNA-synthetase available for their incorporation, because a synthetase cannot be evolved for an UAA that cannot readily enter the cell. We are applying the esterification strategy to highly polar amino acids such as glycosylated and phosphorylated amino acids to overcome this problem with the final goal to genetically incorporate these biologically important amino acids into proteins.
Experimental Section
Synthesis of DanAla and DanAla esters
All reactions were carried out under a nitrogen atmosphere with dry solvents in anhydrous conditions, unless otherwise noted. Dry N,N-dimethylformamide (DMF), methanol (MeOH), methylene chloride (CH2Cl2) and acetonitrile (CH3CN) were obtained by passing commercially available pre-dried, oxygen-free formulations through activated alumina columns. Yields refer to chromatographically homogeneous materials. Reagents were purchased from Sigma-Aldrich and used without further purification. Reactions were monitored by thin-layer chromatography (TLC) carried out on 0.25 mm Silicycle silica gel plates (60F-254) using UV light for visualization and an ethanolic solution of phosphomolybdic acid and cerium sulfate for developing under heat. Silicycle silica gel (60, particle size 0.040–0.063 mm) was used for flash column chromatography. NMR spectra were recorded on Varian Oxford AS500 instrument and calibrated using residual undeuterated solvent or tetramethylsilane as an internal reference. LCMS experiments were performed on an Agilent 1100 Series LC/MSD instrument with Phenomenex Synergi 4u Fusion-RP 80A column (150 × 4.6 mm). DanAla was synthesized as described.[13]
NMR (1H and 13C) and HRMS data for compounds are given in the Supporting Information.
Compound 5. Boc-Dap-OH (2.0 g, 9.8 mmol) was added in one portion to the stirring solution of dansyl chloride (2.4 g, 8.9 mmol) and Et3N (2.6 mL) in CH2Cl2 (50 mL) at 0 °C. The reaction mixture was allowed to warm to room temperature. After stirring for 12 hours, the reaction mixture was concentrated under reduced pressure. The residue was purified by flash chromatography (MeOH/CH2Cl2 (v/v) = 7/93) to give compound 5 (3.675 g, 94 %) as yellow solid. Rf = 0.22 (SiO2, MeOH/CHCl3 (v/v) = 1/8).
Compound 6. Compound 5 (218.8 mg, 0.50 mmol) was dissolved in DMF (2 mL) and cooled to 0 °C. DIPEA (0.096 mL, 0.55 mmol) and iodomethane (0.062 mL, 1.00 mmol) was added dropwise to the solution. The reaction mixture was allowed to warm to room temperature and stirred for 12 hours. Water (20 mL) was added and the solution was extracted with ethyl acetate (20 mL x3). The organic phase was washed with brine, dried with sodium sulfate and concentrated under reduced pressure. The residue was purified by flash chromatography (ethyl acetate/hexanes (v/v) = 3/7) to give compound 6 (197.4 mg, 87%) as yellow solid.
Compound 7. Compound 5 (218.8 mg, 0.50 mmol) was dissolved in DMF (2 mL) and cooled to 0 °C. DIPEA (0.096 mL, 0.55 mmol) and iodoethane (0.080 mL, 1.00 mmol) was added dropwise to the solution. The reaction mixture was allowed to warm to room temperature and stirred for 12 hours. Water (20 mL) was added and the solution was extracted with ethyl acetate (20 mL x3). The organic phase was washed with brine, dried with sodium sulfate and concentrated under reduced pressure. The residue was purified by flash chromatography (ethyl acetate/hexanes (v/v) = 3/7) to give compound 7 (174.1 mg, 75%) as yellow solid.
Compound 2. Compound 6 (45.2 mg, 0.10 mmol) was dissolved in CH2Cl2 (0.9 mL) and cooled to 0 °C. Trifluoroacetic acid (0.3 mL) was added dropwise to the solution. The reaction mixture was allowed to warm to room temperature and stirred for 4 hours. The reaction mixture was concentrated under reduced pressure. After being under vacuum for 48 hours, the residue was dissolved in DMSO (1.0 mL) for use.
Compound 3. Compound 7 (46.6 mg, 0.10 mmol) was dissolved in CH2Cl2 (0.9 mL) and cooled to 0 °C. Trifluoroacetic acid (0.3 mL) was added dropwise to the solution. The reaction mixture was allowed to warm to room temperature and stirred for 4 hours. The reaction mixture was concentrated under reduced pressure. After being under vacuum for 48 hours, the residue was dissolved in DMSO (1.0 mL) for use.
Compound 9. Compound 5 (569.4 mg, 1.30 mmol) was dissolved in DMF (10 mL) and cooled to 0 °C. DIPEA (0.25 mL, 1.43 mmol) and benzyl bromide (0.31 mL, 2.60 mmol) was added dropwise to the solution. The reaction mixture was allowed to warm to room temperature and stirred for 24 hours. Water (70 mL) was added and the solution was extracted with ethyl acetate (50 mL). The organic phase was washed with brine, dried with sodium sulfate and concentrated under reduced pressure. The residue was purified by flash chromatography (ethyl acetate/hexanes (v/v) = 1/3 to 1/2) to give Boc-DanAla-OBn (522.1 mg, 76%) as yellow solid. Rf = 0.25 (SiO2, ethyl acetate/hexanes (v/v) = 1/2). Boc-DanAla-OBn (466.7 mg, 0.885 mmol) and DMAP (21.6 mg, 0.177 mmol) were dissolved in CH2Cl2 (4 mL) and cooled to 0 °C. To this stirring mixture, Et3N (0.136 mL, 0.974 mmol) and Boc2O (289.8 mg, 1.328 mmol) in CH2Cl2 (2 mL) were added dropwise. The reaction mixture was allowed to warm to room temperature and stirred for 4 hours. Saturated aqueous NH4Cl solution was added to the mixture. The aqueous phase was separated and washed with CH2Cl2 (10 mL) for 3 times. The combined organic phase was washed with brine, dried with sodium sulfate and concentrated under reduced pressure. The residue was purified by flash chromatography (ethyl acetate/hexanes (v/v) = 1/3) to give compound 9 (538.6 mg, 97%). Rf = 0.34 (SiO2, ethyl acetate/hexanes (v/v) = 1/2).
Compound 10. To a solution of compound 9 (515.0 mg, 0.82 mmol) in methanol was added Pd/C (87.3 mg). The reaction mixture was stirred under hydrogen atmosphere for 2 hours at room temperature. The reaction mixture was passed through a short plug of celite and elute with ethyl acetate. The filtrate was concentrated to give compound 10 (467.3 mg) as yellow green solid. Rf = 0.22 (SiO2, MeOH/CHCl3 (v/v) = 1/10).
Compound 11. Compound 10 (434.9 mg, 0.809 mmol) was dissolved in CH3CN and cooled to 0 °C. DIPEA (0.56 mL, 3.236 mmol) and bromomethyl acetate (0.24 mL, 2.427 mmol) was added dropwise to the solution. The reaction mixture was allowed to warm to room temperature and stirred for 12 hours. The reaction mixture was concentrated under reduced pressure and the residue was purified by flash chromatography (ethyl acetate/hexanes (v/v) = 1/3) to give compound 11 (389.5 mg, 84 % over 2 steps) as yellow green solid. Rf = 0.31 (SiO2, ethyl acetate/hexanes (v/v) = 1/2).
Compound 4. Compound 11 (61.0 mg, 0.10 mmol) was dissolved in CH2Cl2 (0.9 mL) and cooled to 0 °C. Trifluoroacetic acid (0.3 mL) was added dropwise to the solution. The reaction mixture was allowed to warm to room temperature and stirred for 6 hours. The reaction mixture was concentrated under reduced pressure. After being under vacuum for 48 hours, the residue was dissolved in DMSO (1.0 mL) for use.
Cell lines and culture conditions
HEK293T cells were used for determining UAA intracellular concentration and imaging experiments. A stable clonal HeLa cell line containing the GFP gene with Tyr182UAG mutation was previously established in this lab and was used here for assaying UAA incorporation efficiency.[6] HEK293T and HeLa-GFP(Tyr182UAG) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Mediatech) supplemented with 10% FBS.
Fluorescence imaging
HEK293T cells (3 × 105) were seeded into an imaging dish supplemented with DMEM with 10% FBS and without phenol red. After 18-24 hrs, DanAla or DanAla-OAM was added to the media to a final concentration of 0.10 mM. Cells were immediately imaged using Olympus IX81 spinning disc microscope with a Hamamatsu EM-CCD under same conditions. The excitation wavelength was 360 ± 30 nm and the emission wavelength was 535 ± 20 nm. After each time point, cells were placed back into the incubator. Images were analyzed in Slidebook 4.2 using the masking tool and average intensity function. Cells not treated with any compound were used as control to determine background fluorescence, which was subtracted from the measured intensities of samples to yield to final net intensities.
HPLC analysis of intracellular concentration
HEK293T cells (8.5 × 106) were seeded into a 10 cm tissue culture dish supplemented with DMEM containing 10% FBS. After 18-24 hrs, DanAla or DanAla-OAM was added to the media to a final concentration of 0.1 mM. The compounds were incubated with cells for 1hr. Cells were then washed with 1 mL of phosphate buffer saline (PBS) several times. After trypsinization with 1 mL of 0.05% trypsin (Mediatech), cells were centrifuged and media was removed. Cells were washed again with 1 mL of water and centrifuged. The supernatant was removed and cells were re-suspended in 250 μL of water. Toluene (50 μL) was added and the cells were placed in a 37 °C shaking incubator for 30 mins. Cells were centrifuged at 20,000 rcf for 10 mins. The aqueous layer was placed into a Microcon Ultracel YM10 column (Millipore) and spun for 30 mins at 12,000 rcf. The flow-through was analyzed by HPLC-MS (Agilent 1100 Series LC/MSD instrument): 5 μL of the aqueous layer was separated on a Phenomenex Synergi 4u Fusion-RP 80A column (150 × 4.6 mm) with a gradient of aqueous 0.1% formic acid/acetonitrile: 0.1% formic acid (75:25 to 25:75) over 10 mins. DanAla and DanAla-OAM were identified by extracting their MW (+1) from the total ion mass spectrum. Pure DanAla and DanAla-OAM were also added into the flow-though prepared from cells untreated with compounds to verify the peak positions of DanAla (3.3 min) and DanAla-OAM (3.7 min). Calculation of DanAla concentrations in each sample was based on the extracted ion area of DanAla and the standard curve. The standard curve was made by extracting the area of DanAla in solutions with different concentrations (0.10. 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80 0.90, 1.00 mM). Cells not treated with any compound were used to measure the background area at 3.3 min and 3.7 min. The intracellular concentration was given by the total molar amount of DanAla divided by the total volume of cells, using the volume of a single cell in reference 16.[16]
Flow cytometry
Stable HeLa-GFP(Tyr182TAG) clonal cells were seeded in a 3.5 cm culture dish and transfected with 5 μg of pELcua-LeuRS or pELcua-DanAlaRS plasmid DNA using Lipofectamine 2000.[6] HeLa-GFP(Tyr182TAG) cells that were not transfected with a tRNA-synthetase pair were used as a negative control. DanAla and DanAla-OAM were added 24 hrs post transfection and flow cytometry was carried out 24 hrs after the addition of the compounds. Cells were trypsinized and washed with PBS twice. Samples were centrifuged and re-suspended in 1.0 mL PBS and 5 μL of propidium iodide. Samples were analyzed with a FACScan (Becton & Dickinson). The excitation wavelength was 488 nm, and the emission filter was 530/30 nm. The excitation light (488 nm) excites GFP but not DanAla. For each sample the total fluorescence intensity of 30,000 cells was recorded, and was normalized to the total fluorescence intensity of cells transfected with pELcua-LeuRS. To determine cell viability using flow cytometry, propidium iodide (PI, 5 μg/mL) was used (Sigma-Aldrich). Cells were trypsinized and incubated with PI for 15 min at room temperature, and then analyzed by using FACScan (Becton & Dickinson).
Western blot
HeLa-GFP(Tyr182TAG) clonal cells were transfected and incubated with appropriate DanAla or DanAla ester as previously described for flow cytometry. Cells were trypsinized, washed with PBS, and cell number was counted using a hemocytometer. Samples were centrifuged and re-suspended with 20 μL of PBS containing the EDTA-free protease inhibitor cocktail (Roche) and DNase I (Roche). These samples were lysed by flash freezing in liquid nitrogen and thawing by sonication. The lysis procedure was repeated three times to ensure complete lysis. Loading buffer was added and samples were boiled. Prepared samples were loaded onto a 12% SDS-PAGE gel. For the pELcua-LeuRS sample, 104 cells were loaded while 7.5 × 104 cells were loaded for all other samples. The primary antibody (Anti-GFP Monoclonal Antibody 7G9, BioPioneer) and HRP conjugated secondary antibody (Goat Anti-Mouse IgG-HRP Conjugate, Santa Cruz Biotechnology) were used to detect GFP proteins.
Supplementary Material
Acknowledgements
We thank Dr. Michael Burkart for help with HRMS. This work was supported by the Ray Thomas Edwards Foundation, Beckman Young Investigator Program, March of Dimes Foundation (#5-FY08-110), California Institute for Regenerative Medicine (RN1-00577-1) and National Institutes of Health (1DP2OD004744-01).
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org.
References
- [1].Wang L, Brock A, Herberich B, Schultz PG. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- [2].Wang L, Schultz PG. Angew. Chem. Int. Ed. Engl. 2004;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- [3].Wang Q, Parrish AR, Wang L. Chem. Biol. 2009;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sakamoto K, Hayashi A, Sakamoto A, Kiga D, Nakayama H, Soma A, Kobayashi T, Kitabatake M, Takio K, Saito K, Shirouzu M, Hirao I, Yokoyama S. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu W, Brock A, Chen S, Chen S, Schultz PG. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- [6].Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee KF, Slesinger PA, Wang L. Nat. Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Takimoto JK, Adams KL, Xiang Z, Wang L. Mol Biosyst. 2009;5:931–934. doi: 10.1039/b904228h. [DOI] [PubMed] [Google Scholar]
- [8].Wang L, Xie J, Schultz PG. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- [9].Malandro MS, Kilberg MS. Annu. Rev. Biochem. 1996;65:305–336. doi: 10.1146/annurev.bi.65.070196.001513. [DOI] [PubMed] [Google Scholar]
- [10].Rautio J, Kumpulainen H, Heimbach T, Oliyai R, Oh D, Jarvinen T, Savolainen J. Nat. Rev. Drug Discov. 2008;7:255–270. doi: 10.1038/nrd2468. [DOI] [PubMed] [Google Scholar]
- [11].Tsien RY. Nature. 1981;290:527–528. doi: 10.1038/290527a0. [DOI] [PubMed] [Google Scholar]
- [12].Schultz C, Vajanaphanich M, Harootunian AT, Sammak PJ, Barrett KE, Tsien RY. J. Biol. Chem. 1993;268:6316–6322. [PubMed] [Google Scholar]
- [13].Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Neustadt BR. Tetrahedron Lett. 1994;35:379–380. [Google Scholar]
- [15].Meijler MM, Arad-Yellin R, Cabantchik ZI, Shanzer A. J. Am. Chem. Soc. 2002;124:12666–12667. doi: 10.1021/ja027013s. [DOI] [PubMed] [Google Scholar]
- [16].Thomas P, Smart TG. J. Pharmacol. Toxicol. Methods. 2005;51:187–200. doi: 10.1016/j.vascn.2004.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.