Abstract
A molecular survey including 16,057 mosquitoes captured in Southwest Germany during the summer of 2009 showed the presence of Batai virus (BATV) in Anopheles maculipennis sensu lato. Until this survey, there was no evidence for circulation of BATV in Germany. Analysis of partial S, M, and L segments showed that the sequences from all three segments were most closely related to BATV, indicating that the virus has not undergone reassortment. Phylogenetic analysis revealed a close relationship of the isolated BATV strain from Germany with strains from Slovakia, Ukraine, and Russia.
Batai virus (BATV) belongs to the genus Orthobunyavirus of the family Bunyaviridae and is an arthropod-borne (arbo) single-stranded RNA virus that is widely distributed in Africa, Asia, and Europe.1 In Europe, BATV is transmitted by the zoophilic mosquitoes Anopheles maculipennis s.l. and An. claviger and less often, by Ochlerotatus spp.2 Orthobunyaviruses are able to extend their genetic diversity by reassortment of genome segments during a mixed infection.3 Ngari virus (NRIV) was shown to be a genetic reassortant with S and L RNA segments from Bunyamwera virus and an M RNA segment from BATV.3,4 In contrast to BATV, which is the etiologic agent of rather mild febrile illness in humans and animals,1 the reassortant NRIV was found to be associated with hemorrhagic fever outbreaks in East Africa.5 Until now, there was no evidence for circulation of BATV in Germany.6
Mosquitoes were trapped during the summer of 2009 at three sites (Kühkopf: 49°49′N 8°24′E; Waghäusel: 49°15′N 8°31′E; Weinheim: 49°33′N 8°40′E) in Southwest Germany with CO2-baited encephalitis vector surveillance traps (BioQuip, Compton, CA) and gravid traps (John W. Hook Company, Gainesville, FL). Mosquito identification (species and sex) as well as virus isolation and RNA extraction was performed as described recently.7 Extracted RNA was analyzed by a newly designed BATV real-time reverse transcription polymerase chain reaction (RT-PCR) using the primers BATAI-Fwd (5-GCTGGAAGGTTACTGTATTTAATAC-3; nucleotide [nt] positions 268–292 according to GenBank accession number X73464) and BATAI-Rev (5-CAAGGAATCCACTGAGTCTGTG-3; nt positions 345–366) and probe BATAI-P (5-FAM-AACAGTCCAGTTCCAGACGATGGTC-BHQ-1-3; nt positions 312–336 [FAM = 6-carboxyfluorescein; BHQ-1 = black hole quencher 1]). The target was a 99-nt-long region of the S segment. Real-time RT-PCR was performed with a Quanti-Tect probe RT-PCR kit according to the manufacturer's protocol (Qiagen, Hilden, Germany).
There were 16,057 female mosquitoes7 captured and pooled according to species (25 mosquitoes per pool). BATV RNA was detected by real-time RT-PCR in 1 of 643 pools tested (pool number 52.3). This pool included mosquitoes of the species complex An. maculipennis s.l. Inoculation of Vero cells with pool 52.3 caused cytopathic effect (CPE) after 48 hours, and BATV-specific RNA was detected by real-time RT-PCR in the supernatant of the infected cell culture after five passages. Moreover, electron microscopy of the infected cell culture showed enveloped viral particles measuring ~90 nm in diameter. For phylogenetic analysis, partial S, M, and L segments (838, 3,152, and 200 nt, respectively) of the isolate (called BATV strain 53.2) were amplified by RT-PCR and sequenced using the primers listed in Table 1. The sequences from all three segments were most closely related to BATV (Figure 1A–C), indicating that the virus has not undergone reassortment. The nucleotide identity between the German BATV strain and BATV strains from other regions ranged from 91.9% to 92.1% in the S segment, from 84.8% to 96.2% in the M segment, and from 84.3% to 93.8% in the L segment. Phylogenetic analysis by Bayesian inference revealed a close relationship of BATV strain 53.2 from Germany with strain 184 from Slovakia (Figure 1B), strain 499 from Ukraine, and strain 42 from Russia (Figure 1C).
Table 1.
Primers used to amplify and sequence the partial BATV S, M, and L segments
| Segment primer | Sequence (5′ to 3′) | Nucleotide position* | Amplicon size (nt) | Temp. (°C) |
|---|---|---|---|---|
| S segment primer | ||||
| BATAIS1F | TGGAATTCAATGATGTCGCTGCTAAC | 79–104 | 344 | 60 |
| BATAIS1R | TATAATCAATTTTTCCGGGTCACTCACTTT | 393–422 | 344 | 60 |
| BATAIS2F | TTGGGGGCTGGAAGGTTACTGT | 262–283 | 363 | 58 |
| BATAIS2R | TATATCTTTGGCGCATGGTCTTCTCC | 599–624 | 363 | 60 |
| BATAIS3F | CTGGGCAGATGGGGAGGAG | 467–485 | 451 | 56 |
| BATAIS3R | AAACTGCAATGCTTCAAAAACAAT | 894–917 | 451 | 54 |
| M segment primer | ||||
| BATAIM2F | TGTGGCCTAGCATATCACCCTTTCA | 793–817 | 1,407 | 60 |
| BATAIM2R | AGACCGGTGATGATGATCTGTAACCTCTA | 2,171–2,199 | 1,407 | 60 |
| BATAIM3F | CCTGGGGAAGCATTGTGATTACT | 1,704–1,726 | 525 | 56 |
| BATAIM3R | CTAGCCAGCGACTCTTGCCTTCC | 2,206–2,228 | 525 | 60 |
| BATAIM4F | GTCGCTGGCTAGTGCTACCTCTGG | 2,217–2,240 | 510 | 60 |
| BATAIM4R | CTGATTATTGTCGGATTTATTGGGAACCT | 2,698–2,726 | 510 | 60 |
| BATAIM5F | AAAGGTTCCCAATAAATCCGACAA | 2,696–2,719 | 525 | 57 |
| BATAIM5R | CAAATTCTTCACATCCCCAACGACTA | 3,195–3,220 | 525 | 59 |
| BATAIM6F | AGAATTTGGGTGCCTTGCTGTCA | 3,213–3,235 | 874 | 59 |
| BATAIM6R | AGATGTTTGGTCCCCTGTGCTTATTT | 4,061–4,086 | 874 | 59 |
| L segment primer | ||||
| BATAIL1F | GATGGCGATTTCCTGATTAT | 283–302 | 711 | 48 |
| BATAIL1R | TGACCCCAAGAGTTTCCTATTAT | 971–993 | 711 | 51 |
Figure 1.
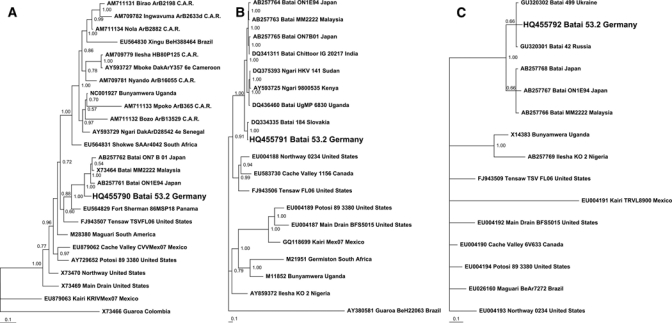
Bayesian phylogenetic tree of selected orthobunyaviruses based on partial S (A; length = 838 nucleotides), M (B; length = 3,152 nucleotides), and L segment (C; length = 200 nucleotides) sequences. Each sequence is identified by GenBank accession number, virus name, strain designation, and strain origin. Phylogenetic analysis was performed using MrBayes 3.0 program.8 Three heated chains and a single cold chain were used in all Markov Chain Monte Carlo (MCMC) analyses, which were run for 1,000,000 generations, sampling one tree every 100 generations. Trees obtained before convergent and stable likelihood values were discarded (i.e., a 2,500 tree burn-in). Four independent runs, each started from different randomly chosen trees, were performed to assess convergence. Posterior probabilities for nodes were assembled from all post–burn-in trees (i.e., 30,004 trees per analysis). Posterior probabilities are shown on each branch. Scale bar indicates number of nucleotide substitutions per site.
In conclusion, a molecular survey in mosquitoes showed the occurrence of BATV in the south of Germany. The BATV infection rate in the mosquito population seems to be low, because only 1 of 19 An. maculipennis s.l. pools tested positive. The infection rate was even lower when pools of other known BATV vectors, An. claviger and Ochlerotatus spp., were included in the analysis. BATV circulates in a mosquito to mammal cycle in agro-ecosystems.1 The Waghäusel trapping site is indeed a typical agro-ecosystem, with pig, sheep, and horse farms. BATV may cause a mild illness among sheep, but also, stillbirth and congenital abnormalities have been reported in association with BATV infections.9 In addition, BATV was isolated from a sentinel cattle herd in Okinawa, Japan.3 Moreover, the virus may cause human disease with influenza-like symptoms.10 Therefore, further studies have to be conducted to estimate the veterinary and medical importance of BATV in the affected area.
Footnotes
Authors' addresses: Hanna Jöst and Norbert Becker, German Mosquito Control Association (KABS), Waldsee, Germany, E-mails: hanna.joest@gmx.de and NorbertFBecker@web.de. Alexandra Bialonski, Christel Schmetz, Stephan Günther, and Jonas Schmidt-Chanasit, Bernhard Nocht Institute for Tropical Medicine, Department of Virology, Hamburg, Germany, E-mails: bialonski@bni-hamburg.de, schmetz@bni-hamburg.de, guenther@bni-hamburg.de, and jonassi@gmx.de.
References
- 1.Hubalek Z. Mosquito-borne viruses in Europe. Parasitol Res. 2008;103:S29–S43. doi: 10.1007/s00436-008-1064-7. [DOI] [PubMed] [Google Scholar]
- 2.Becker N, Petric D, Zgomba M, Boase C, Dahl C, Lane J, Kaiser A. Mosquitoes and Their Control. 1st ed. New York, NY: Kluwer; 2003. [Google Scholar]
- 3.Yanase T, Kato T, Yamakawa M, Takayoshi K, Nakamura K, Kokuba T, Tsuda T. Genetic characterization of Batai virus indicates a genomic reassortment between orthobunyaviruses in nature. Arch Virol. 2006;151:2253–2260. doi: 10.1007/s00705-006-0808-x. [DOI] [PubMed] [Google Scholar]
- 4.Briese T, Bird B, Kapoor V, Nichol ST, Lipkin WI. Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa. J Virol. 2006;80:5627–5630. doi: 10.1128/JVI.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen MD, Trappier SG, Sanchez AJ, Meyer RF, Goldsmith CS, Zaki SR, Dunster LM, Peters CJ, Ksiazek TG, Nichol ST. A reassortant bunyavirus isolated from acute hemorrhagic fever cases in Kenya and Somalia. Virology. 2001;291:185–190. doi: 10.1006/viro.2001.1201. [DOI] [PubMed] [Google Scholar]
- 6.Pfeffer M, Modlmaier M, Lundström JO. Lack of evidence for the presence of mosquito-borne arboviruses in the upper Rhine Valley, Germany, in 1999 to 2000. J Clin Microbiol. 2010;48:3457–3458. doi: 10.1128/JCM.01156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jöst H, Bialonski A, Storch V, Günther S, Becker N, Schmidt-Chanasit J. Isolation and phylogenetic analysis of Sindbis viruses from mosquitoes in Germany. J Clin Microbiol. 2010;48:1900–1903. doi: 10.1128/JCM.00037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ronquist F, Huelsenbeck JP. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 9.Singh KR, Pavri KM. Isolation of Chittoor virus from mosquitoes and demonstration of serological conversions in sera of domestic animals at Manjri, Poona, India. Indian J Med Res. 1966;54:220–224. [PubMed] [Google Scholar]
- 10.Nashed NW, Olson JG, el-Tigani A. Isolation of Batai virus (Bunyaviridae:Bunyavirus) from the blood of suspected malaria patients in Sudan. Am J Trop Med Hyg. 1993;48:676–681. doi: 10.4269/ajtmh.1993.48.676. [DOI] [PubMed] [Google Scholar]


