Abstract
This study was conducted to determine which genotypes were present in southwestern Korea. Nested polymerase chain reaction (PCR) and DNA sequence analysis targeting the Orientia tsutsugamushi-specific 56-kDa protein gene was performed with samples of blood and eschar. Of the 69 PCR-positive samples, 61 clustered with the Boryong previously isolated in Korea. CUH 4-6 had sequence homology of 100% with Kato, and CUH 4-3 had homology of 99.8% with Kato and formed the Kato cluster. CUH 4-57, CUH 4-31, CUH 4-142, and CUH 4-324 formed a Kawasaki cluster. CUH 4-271 had sequence homology of 100% with Jecheon and formed a Karp cluster. CUH 4-117 had homology of 99.8% with Neimeng-65, and Gilliam cluster. The most common genotype of O. tsutsugamushi in the southwestern part of Korea is the Boryong genotype. We also identified O. tsutsugamushi of the Kato, Neimeng-65 and Kawasaki genotypes, which had not been identified before in Korea.
Introduction
Scrub typhus is an acute febrile disease caused by Orientia tsutsugamushi (O. tsutsugamushi), which is widely distributed in rural areas of Southeast Asia.1–3 Orientia tsutsugamushi characteristically has many variants, and eight scrub typhus antigen genes (Sta), namely Sta 150, 110, 72, 58, 56, 49, 47, and 20, have been identified.4,5
The 56-kDa type-specific antigen is a major outer membrane protein located on the surface of Orientia species that may be involved in penetration into host cells.4 This 56-kDa antigen is an immunodominant antigen that induces strong humoral immunity. In addition, it contains both group-specific and type-specific epitopes, which are useful for the diagnosis of scrub typhus.6 Ohashi and others7 have demonstrated that the 56-kDa protein gene has variable domains that differ among strains. Three prototypes, the Karp, Kato, and Gilliam strains, account for the majority of isolates, and other serotypes such as Shimokoshi, Kuroki, Kawasaki, and Boryong, are also found because of variability of the main antigens.8 Although the 56-kDa type-specific antigen differs between strains, its basic structure is constant and consists of ~1,600 base pairs.4,7,9 It has been reported that the virulence of O. tsutsugamushi differs between strains depending on their serotype.10 In the pre-antibiotic era, mortality was reported to be 45% in a certain region and 20% in another region.11 This remarkable difference in mortality rate may be explained by regional strain differences, which implies that the virulence of O. tsutsugamushi is related to its serotype or genotype. Thus, it is important to investigate the regional distribution of serotypes and genotypes. Because each serotype of O. tsutsugamushi classified by differences in the antigen is associated with a specific variation of nucleotide sequences, further detailed genotypes may exist.
This study was conducted to determine which genotypes are present in the southwestern part of Korea by amplifying and sequencing the gene encoding the 56-KDa protein by means of the polymerase chain reaction (PCR).
Materials and Methods
Patients.
This study included adult patients ≥ 18 years of age with acute febrile disease who presented at our university hospital between September 1 and December 31, 2004, because of the presence of an eschar or maculopapular skin rash along with at least two of the following clinical symptoms: headache, generalized weakness, myalgia, cough, nausea, and abdominal discomfort. At that time, we collected and stored the samples of blood and eschar. In a previous study,12 we performed PCR on eschars and blood buffy coats, together with indirect immunofluorescence assays (IFAs) on the sera, and we used the remaining specimens in this study. Scrub typhus was confirmed on the basis of either a single indirect immunofluorescent-specific immunoglobulin M (IgM) titer of ≥ 1:10 againt O. tsutsugamushi or a 4-fold or greater rise in the IFA IgG titer.12
Nested PCR.
Because Orientia is an intracellular microorganism, the white blood cell buffy coat of whole blood was purified using a QIAamp DNA mini kit (Qiagen, Hilden, Germany).12
Nested PCR was performed by a modification of the method described by Furuya and others.13 Nucleotide primers were based on the nucleotide sequence of the gene encoding the 56-kDa antigen of the Gilliam strain of O. tsutsugamushi.13 Primers 34 (5-TCA AGC TTA TTG CTA GTG CAA TGT CTGC-3) and 55 (5-AGG GAT CCC TGC TGC TGT GCT TGC TGCG-3) were used in the first PCR. Nested PCR primers 10 (5-GAT CAA GCT TCC TCA GCC TAC TAT AAT GCC-3) and 11 (5-CTA GGG ATC CCG ACA GAT GCA CTA TTA GGC-3) were used in the second PCR amplification of the resulting 483 bp fragment (Figure 1). The primers amplified variable domains II and III of the 56-kD gene (Figure 1). All primers were purchased from Bioneer (Daejeon, Korea). The first round of PCR amplification was performed in a 20-μL reaction volume containing 2 μL of DNA, 1 μL of each 5 pmol/μL primer (forward and reverse primers), 10 μL of 2X EXCEL-Tag PreMix (Tag polymerase 2 U, 400 μm dNTP, 2.0 mM MgCl2 KCL, Tris-Cl; Corebio, Seoul., Korea) and sterile triple distilled water. The PCR reactions were carried out under the following conditions: initial denaturation at 94°C for 10 minutes, followed by 35 cycles, each consisting of denaturation at 94°C for 1 minute, 61°C for 1 minute and 72°C for 1 minute, and final extension at 72°C for 10 minutes. The second round of PCR amplification was conducted using 1 μL of the first PCR product as template DNA under the following conditions: initial denaturation at 94°C for 10 minutes, followed by 30 cycles, each consisting of denaturation at 94°C for 30 seconds, 63°C for 30 seconds and 72°C for 1 minute, and extension at 72°C for 7 minutes. The PCR was performed with a Biosystems Veriti 96-well thermal cycler (Applied Biosystems, Foster city, CA). The amplified PCR product was electrophoresed for 40 minutes on a 1.2% agarose gel (Seakem LE agarose) containing 0.5 ng/mL ethidium bromide at 100 volts (0.5X TAE, Bioneer) using a Bioneer electrophoresis machine.
Figure 1.
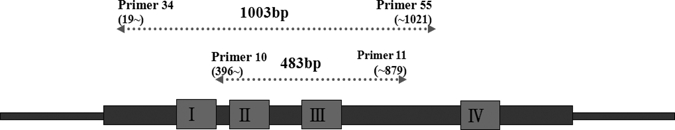
Positions of the primers used for 56-kDa gene amplification. The open reading frame of the gene is represented by the heavy line, and boxes I, II, III, and IV show the positions of the variable domains.
Nucleotide sequences and phylogenetic analysis.
The amplified PCR product was purified using a QIAquick gel extraction kit (Qiagen) and sent to Genotech (Daejeon, Korea) for sequencing with a 3730x1 DNA analyzer (Applied Biosystems, Foster, CA). The retrieved sequences were made with the basic local alignment search tool (BLAST) using database from the National Center for Biotechnology Information (NCBI) to determine the closest relatives, and newly identified nucleotide sequences were aligned with their closest relatives with the CLUSTAL X software program. The neighbor-joining method was used for phylogenetic analysis, and bootstrap was performed 1,000 times to increase phylogenetic reliability. The phylogenetic analysis was performed with the nucleotide sequences of the O. tsutsugamushi 56-kDa gene registered in Genbank that are distributed in many countries including Korea (Table 1).7,14–20 The Tree Explorer was used to find the strain pedigree, and the LaserGene Program (DNASTAR, Madison, WI) was used to compare homologies. The clusters of genotypes in the strain pedigree were grouped by a modification of the method described by Tamura and others15 and Fournier and others.21
Table 1.
Strains of Orientia tsutsugamushi used in this study
| Cluster* | Strain | Source (when available) | Geographical origin | GenBank accession no. | Reference |
|---|---|---|---|---|---|
| Karp | 402I | Human | Japan | AF173047 | 14 |
| Karp | Kamimoto | Human | Japan | AF173046 | 14 |
| Karp | Mori | Human | Japan | AF173044 | 14 |
| Karp | Okazaki | Human | Japan | AF173045 | 14 |
| Karp | CMM1 | Rodent | Japan | AF302986 | 15 |
| Karp | KNP1 | Rodent | Japan | AF302987 | 15 |
| Karp | KNP2 | Rodent | Japan | AF302988 | 15 |
| Karp | Hirahata | Chiggers | Japan | AF173176 | 16 |
| Karp | R39 | Chiggers | Japan | AF201836 | 16 |
| Karp | R9 | Chiggers | Japan | AF201837 | 16 |
| Karp | Jecheon | Human | Korea | AF430143 | Unpublished |
| Karp | TW261 | Rodent | Taiwan | AY222636 | 17 |
| Karp | Karp | Human | New Guinea | AY956315 | Direct Submission |
| Karp | Matsuzawa | Human | Japan | AF173043 | 14 |
| Karp | Yeojoo | Human | Korea | AF430144 | Unpublished |
| TW73R | Rodent | Taiwan | AY222628 | 17 | |
| Saitama | TW121 | Rodent | Taiwan | AY222639 | 17 |
| Saitama | TW141 | Rodent | Taiwan | AY222638 | 17 |
| Saitama | TW441 | Rodent | Taiwan | AY222634 | 17 |
| Saitama | FAR1 | Rodent | Japan | AF302989 | 15 |
| Saitama | HSB1 | Rodent | Japan | AF302983 | 15 |
| Saitama | HSB2 | Rodent | Japan | AF302984 | 15 |
| Saitama | UAP1 | Rodent | Japan | AF302991 | 15 |
| Saitama | UAP2 | Rodent | Japan | AF302992 | 15 |
| Saitama | UAP4 | Rodent | Japan | AF302993 | 15 |
| Saitama | UAP7 | Rodent | Japan | AF302995 | 15 |
| Saitama | Pajoo | Human | Korea | AF430142 | Direct Submission |
| Saitama | Yongworl | Human | Korea | AF430141 | Direct Submission |
| LA-1 | Chiggers | Thailand | AF173049 | 14 | |
| TW45R | Rodent | Taiwan | AY222632 | 17 | |
| TW201 | Rodent | Taiwan | AY222637 | 17 | |
| TWyu81 | Chiggers | Taiwan | AY222640 | 17 | |
| Boryong | Kuroki | Human | Japan | M63380 | 7 |
| Boryong | Nishino | Human | Japan | AF173048 | 14 |
| Boryong | Boryong | Human | Korea | AM494475 | 18 |
| Kato | Kato | Human | Japan | M63382 | 7 |
| Kato | Akita-7 | Rodent | Japan | AF173041 | 14 |
| Kato | Omagari | Rodent | Japan | AF173040 | 14 |
| LF-1 | Chiggers | Thailand | AF173050 | 14 | |
| Kawasaki | Kawasaki | Human | Japan | M63383 | 7 |
| Kawasaki | Kanda | Human | Japan | AF173039 | 14 |
| Kawasaki | Oishi | Human | Japan | AF173037 | 14 |
| Kawasaki | Taguchi | Human | Japan | AF173038 | 14 |
| Gilliam | Gilliam | Taiwan | DQ485289 | Unpublished | |
| Gilliam | Neimeng-65 | China | AF140143 | Unpublished | |
| Gilliam | TW461 | Rodent | Taiwan | AY222631 | 17 |
| Gilliam | 405S | Human | Japan | AF173036 | 14 |
| Gilliam | Ikeda | Human | Japan | AF173033 | 14 |
| Gilliam | Iwataki-1 | Rodent | Japan | AF173035 | 14 |
| Gilliam | LP-1 | Chiggers | Japan | AF173034 | 14 |
| Gilliam | FAR2 | Rodent | Japan | AF302990 | 15 |
| Gilliam | HSB3 | Rodent | Japan | AF302985 | 15 |
| Gilliam | UAP6 | Rodent | Japan | AF302994 | 15 |
| Gilliam | Yonchon | Human | Korea | U19903 | 19 |
| Gilliam | SXH951 | Human | China | AF050669 | Unpublished |
| Shimokoshi | Human | Japan | M63381 | 7 | |
| Fuji | Chiggers | Japan | AF201834 | 16 | |
| LX-1 | Chiggers | Japan | AF173042 | 14 | |
| TA678 | Rodent | Thailand | U19904 | 20 | |
| TW381 | Rodent | Taiwan | AY222635 | 17 | |
| TW521 | Rodent | Taiwan | AY222630 | 17 | |
| TA763 | Rodent | Thailand | U80636 | 20 | |
| TA686 | Rodent | Thailand | U80635 | 20 | |
| TA716 | Rodent | Thailand | U19905 | 20 | |
| TW44R | Rodent | Taiwan | AY222633 | 17 | |
| TW62R | Rodent | Taiwan | AY222629 | 17 | |
| TWyu11 | Chiggers | Taiwan | AY222641 | 17 |
Results
The PCR was positive from the blood or eschar of 69 patients who were confirmed with scrub typhus. A phylogenetic tree was created of the 483 bps of 56-kDa genes of 67 O. tsutsugamushi strains registered in GenBank and their nucleotide sequences. With regard to nucleotide sequences, CUH 4-117 had the lowest similarity of 66.9% to LX-1 and TA686, whereas CUH4-271 was identical to the Jecheon strain, and CUH-4-6 was identical to the Kato and Omagari strains (Table 2). Sixty-one of the 69 genotypes found in this study, including CUH4-134 and CUH4-485, clustered together with strains related to the Boryong type. The 61 amplicons that belong to the Boryong cluster showed a minimum homology of 99.5% (CUH4-134) to the Boryong strain (accession no. AM 494475) and a maximum homology of 99.8% (CUH4-485) (Table 2). CUH4-6 was identical to the Kato and Omagari strains, and CUH4-3 had 99.8% homology to both the Omagari and Kato strains and clustered together with the Kato strains. The CUH4-31, CUH4-142, and CUH4-324 genotypes were 100% homologous to each other, and 99.8% homologous to both the Oishi and Taguchi strains and 99.5% each to Kawasaki and Kanda strains. CUH4-57 showed a homology of 99.8% to the Kawasaki and Kanda strains. CUH4-57, CUH4-31, CUH4-142, and CUH4-324 clustered together with the Kawasaki strains (Table 2, Figure 2). CUH4-271 had 100% homology to the Jecheon strain isolated in Korea and formed the Karp cluster together with the Hirahata, Kamimoto, and Matsuzawa strains. CUH4-117 had homology of 99.8% to the Neimeng-65 strain and clustered together with the Gilliam strain (Figure 2).
Table 2.
Homology of 56kDa gene nucleotide sequences among Orientia tsutsugamushi strains
| CUH 4-134 | CUH 4-485 | CUH 4-271 | CUH 4-3 | CUH 4-6 | CUH 4-117 | CUH 4-57 | CUH 4-31 | CUH 4-142 | CUH 4-324 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Hirahata | 90.6 | 90.9 | 99.1 | 70.8 | 71 | 71.5 | 69.4 | 69.6 | 69.6 | 69.6 |
| Mori | 89.7 | 90 | 97.9 | 70.1 | 70.3 | 70.5 | 69.2 | 69.4 | 69.4 | 69.4 |
| Kamimoto | 89.7 | 90 | 97.9 | 70.1 | 70.3 | 70.5 | 69.2 | 69.4 | 69.4 | 69.4 |
| Okazaki | 89.7 | 90 | 97.9 | 70.1 | 70.3 | 70.5 | 69.2 | 69.4 | 69.4 | 69.4 |
| Jecheon | 90.6 | 90.9 | 100 | 70.3 | 70.5 | 71.2 | 69.2 | 69.4 | 69.4 | 69.4 |
| Karp | 91.1 | 91.3 | 99.1 | 70.5 | 70.8 | 71.2 | 69.9 | 70.1 | 70.1 | 70.1 |
| Matsuzawa | 89.5 | 89.7 | 96.8 | 70.8 | 71 | 70.8 | 69.9 | 70.1 | 70.1 | 70.1 |
| Yeojoo | 91.1 | 91.3 | 98.6 | 71 | 71.2 | 70.8 | 69.2 | 69.4 | 69.4 | 69.4 |
| Yongworl | 90.6 | 90.9 | 93.6 | 71 | 71.2 | 71.2 | 69.9 | 70.1 | 70.1 | 70.1 |
| Pajoo | 91.8 | 92 | 94.7 | 70.5 | 70.8 | 71.7 | 70.5 | 70.8 | 70.8 | 70.8 |
| LA-1 | 90.6 | 90.9 | 93.6 | 70.8 | 71 | 72.1 | 71 | 71.2 | 71.2 | 71.2 |
| Nishino | 98.2 | 98.4 | 90.1 | 72.2 | 72.4 | 71 | 69.9 | 70.1 | 70.1 | 70.1 |
| Boryong | 99.5 | 99.8 | 91.3 | 71.7 | 72 | 71.7 | 70.6 | 70.8 | 70.8 | 70.8 |
| Kuroki | 99.5 | 99.8 | 91.3 | 71.7 | 72 | 71.7 | 70.6 | 70.8 | 70.8 | 70.8 |
| Akita-7 | 73.2 | 73.5 | 72.3 | 99.5 | 99.8 | 75.6 | 75.6 | 75.4 | 75.4 | 75.4 |
| Omagari | 73.5 | 73.7 | 72.5 | 99.8 | 100 | 75.8 | 75.8 | 75.6 | 75.6 | 75.6 |
| Kato | 73.5 | 73.7 | 72.5 | 99.8 | 100 | 75.8 | 75.8 | 75.6 | 75.6 | 75.6 |
| LF-1 | 73.9 | 74.2 | 73.2 | 94.6 | 94.8 | 78.9 | 77.2 | 77 | 77 | 77 |
| Kanda | 74.5 | 74.7 | 73.5 | 78.1 | 78.3 | 91.7 | 99.8 | 99.5 | 99.5 | 99.5 |
| Kawasaki | 74.5 | 74.7 | 73.5 | 78.1 | 78.3 | 91.7 | 99.8 | 99.5 | 99.5 | 99.5 |
| Oishi | 74.7 | 74.9 | 73.7 | 77.9 | 78.1 | 92 | 99.5 | 99.8 | 99.8 | 99.8 |
| Taguchi | 74.7 | 74.9 | 73.7 | 77.9 | 78.1 | 92 | 99.5 | 99.8 | 99.8 | 99.8 |
| Neimeng-65 | 76.2 | 76.4 | 76.2 | 78.6 | 78.8 | 99.8 | 92.2 | 92.5 | 92.5 | 92.5 |
| Gilliam | 76.4 | 76.6 | 76.4 | 79.1 | 79.3 | 99.3 | 91.7 | 92 | 92 | 92 |
| Ikeda | 73.2 | 73.5 | 73.2 | 77.9 | 78.1 | 94.2 | 91.2 | 91 | 91 | 91 |
| Iwataki-1 | 73.2 | 73.5 | 73.2 | 77.9 | 78.1 | 94.2 | 91.2 | 91 | 91 | 91 |
| Yonchon | 73.2 | 73.5 | 73.2 | 77.9 | 78.1 | 94.2 | 91.2 | 91 | 91 | 91 |
| Shimokoshi | 81 | 81.2 | 80.5 | 74.8 | 74.8 | 74.1 | 74.3 | 74.6 | 74.6 | 74.6 |
| Fuji | 74.5 | 74.7 | 72.1 | 70.2 | 70.4 | 70.2 | 71.2 | 70.9 | 70.9 | 70.9 |
| LX-1 | 75.5 | 75.8 | 74.1 | 69.9 | 70.2 | 66.9 | 67.4 | 67.6 | 67.6 | 67.6 |
| TA678 | 74.1 | 74.3 | 71.8 | 72.7 | 72.9 | 68.5 | 67.6 | 67.8 | 67.8 | 67.8 |
| TA763 | 78.3 | 78.6 | 75.2 | 73.8 | 74 | 73.8 | 73.6 | 73.3 | 73.3 | 73.3 |
| TA686 | 78.5 | 78.8 | 74.9 | 67.6 | 67.8 | 66.9 | 67.6 | 67.4 | 67.4 | 67.4 |
| TA716 | 76.5 | 76.7 | 73.4 | 76.5 | 76.7 | 69.5 | 69.1 | 69.3 | 69.3 | 69.3 |
Figure 2.
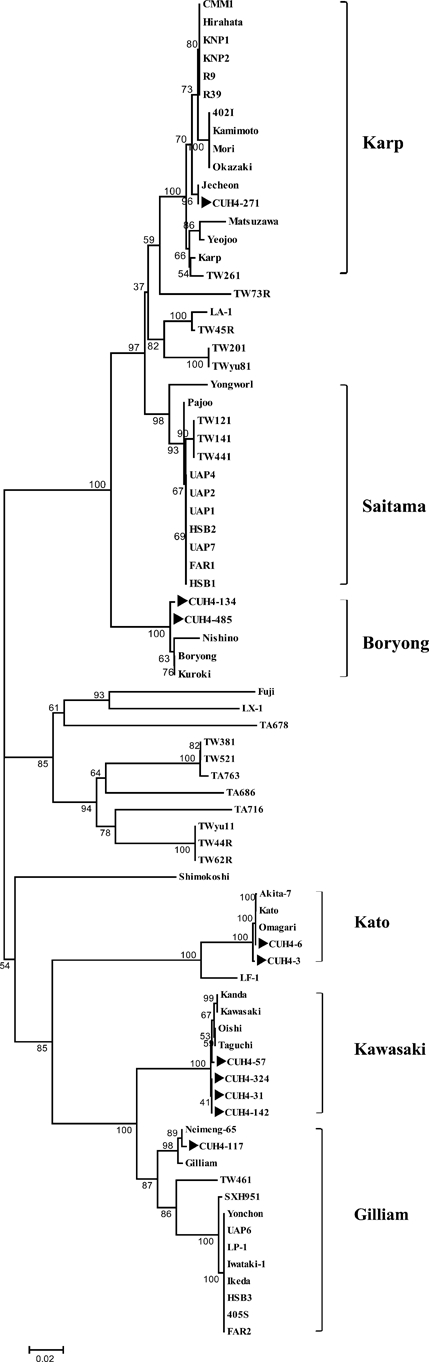
Phylogenetic tree based on the nucleotide sequences of Orientia tsutsugamushi 56-kDa genes.
Discussion
It has been reported that O. tsutsugamushi has a variety of serotypes that differ between endemic areas.22–25 In Korea, the Boryong serotype was reported to be distributed throughout the country except for Cheju Island.26 The Karp and Gilliam serotypes were prevalent in Taiwan, whereas the Gilliam serotype was prevalent in China.27
Thus, determining the serotypes in endemic areas is important for basic research on the classification of O. tsutsugamushi, the development of vaccines, and definitive diagnosis of scrub typhus. Expanding the panel of antigens used to test scrub typhus and to take into account local antigenic diversity would improve sensitivity of serologic diagnosis.28 However, even though O. tsutsugamushi infection can be identified by serologic diagnosis, patient sera often cross-react with the antigens from different strains.27 In our study, cross-reactions among Boryong, Gilliam, Karp, and Kato antigen were observed in most patients.
Genotypic identification using nested PCR and DNA sequencing methods could be useful.
The 16s rRNA gene of O. tsutsugamushi and its 56-kDa protein gene have been used for the diagnosis of scrub typhus. Because the 16s rRNA genes of the Gilliam, Karp, Kato, and Kuroki strains are ≥ 98.4% homologous,29 this gene is useful for differentiating between Orientia and other genera/species, but not between O. tsutsugamushi strains. It has been reported that the 56-kDa protein gene is more useful for differentiating between O. tsutsugamushi strains than the 16s rRNA gene.14
In this study, we amplified a 483 bp region containing VDII and VDIII of the O. tsutsugamushi-specific 56-kDa genes by nested PCR, because most genotypes can be differentiated at these loci because of the great variation.30 The DNA sequences were compared with the nucleotide sequences of 67 O. tsutsugamushi registered in GenBank. The Boryong genotype was found to prevail in the southwestern part of Korea, which is consistent with previous reports, indicating that it is more prevalent in areas south to Choongnam Province of South Korea, whereas the Karp and Gilliam genotypes are prevalent in Kyounggi and Kangwon Provinces.31,32 In the southwestern part of Korea, we identified CUH4-271 (Karp cluster), which had homology of 100% to the Jecheon genotype, and CUH4-3/CUH4-6 (Kato cluster), CUH4-31/CUH4-57/CUH4-142/CUH4-324 (Kawasaki cluster), and CUH4-117 (Gilliam cluster similar to Neimeng-65). However, further studies are needed to ascertain if there is a significant difference in the clinical manifestations and severity of scrub typhus according to genotype.
In conclusion, the results of this study confirmed that the Boryong genotype is most common in the southwestern part of Korea. In addition we identified Kato, Neimeng-65, and Kawasaki genotypes, which had not been encountered before in Korea. Thus, the results of this study confirm that various genotypes including the Boryong, Kato, Neimeng-65, Kawasaki, and Gilliam strains are present in this area.
Disclaimer: The authors do not have any commercial interest or other association that might pose a conflict of interest.
Footnotes
Authors' addresses: Yu-Mi Lee, Dong-Min Kim, and Ganesh Prasad Neupane, Division of Infectious Diseases, Department of Internal Medicine, Chosun University, School of Medicine, Gwangju, Republic of Korea, E-mails: moksha1001@hanmail.net, drongkim@chosun.ac.kr, and neupaneganesh@yahoo.com. Mi-Sun Jang, Division of Infectious Diseases Department of Internal Medicine and Department of Public Health, Chosun University, School of Medicine, Gwangju, Republic of Korea, E-mail: sangkim3507@nate.com. Seung-Hyun Lee, Department of Pediatrics, Seonam University, College of Medicine, nam-won, Republic of Korea, E-mail: 31gmlakd@hanmail.net.
Reprint requests: Dong-Min Kim, Division of Infectious Diseases, Department of Internal Medicine, Chosun University College of Medicine, 588 Seosuk-dong, Dong-gu, Gwangju, 501-717, Republic of Korea, E-mail: drongkim@chosun.ac.kr.
References
- 1.Leelarasamee A, Chupaprawan C, Chenchittikul M, Udompanthurat S. Etiologies of acute undifferentiated febrile illness in Thailand. J Med Assoc Thai. 2004;87:464–472. [PubMed] [Google Scholar]
- 2.Phongmany S, Rolain JM, Phetsouvanh R, Blacksell SD, Soukkhaseum V, Rasachack B, Phiasakha K, Soukkhaseum S, Frichithavong K, Chu V, Keolouangkhot V, Martinez-Aussel B, Chang K, Darasavath C, Rattanavong O, Sisouphone S, Mayxay M, Vidamaly S, Parola P, Thammavong C, Heuangvongsy M, Syhavong B, Raoult D, White NJ, Newton PN. Rickettsial infections and fever, Vientiane, Laos. Emerg Infect Dis. 2006;12:256–262. doi: 10.3201/eid1202.050900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suttinont C, Losuwanaluk K, Niwatayakul K, Hoontrakul S, Intaranongpai W, Silpasakorn S, Suwancharoen D, Panlar P, Saisongkorh W, Rolain JM, Raoult D, Suputtamongkol Y. Causes of acute, undifferentiated, febrile illness in rural Thailand: results of a prospective observational study. Ann Trop Med Parasitol. 2006;100:363–370. doi: 10.1179/136485906X112158. [DOI] [PubMed] [Google Scholar]
- 4.Stover CK, Marana DP, Carter JM, Roe BA, Mardis E, Oaks EV. The 56-kilodalton major protein antigen of Rickettsia tsutsugamushi: molecular cloning and sequence analysis of the sta56 gene and precise identification of a strain-specific epitope. Infect Immun. 1990;58:2076–2084. doi: 10.1128/iai.58.7.2076-2084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oaks EV, Stover CK, Rice RM. Molecular cloning and expression of Rickettsia tsutsugamushi genes for two major protein antigens in Escherichia coli. Infect Immun. 1987;55:1156–1162. doi: 10.1128/iai.55.5.1156-1162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohashi N, Nashimoto H, Ikeda H, Tamura A. Cloning and sequencing of the gene (tsg56) encoding a type-specific antigen from Rickettsia tsutsugamushi. Gene. 1990;91:119–122. doi: 10.1016/0378-1119(90)90171-m. [DOI] [PubMed] [Google Scholar]
- 7.Ohashi N, Nashimoto H, Ikeda H, Tamura A. Diversity of immunodominant 56-kDa type-specific antigen (TSA) of Rickettsia tsutsugamushi. Sequence and comparative analyses of the genes encoding TSA homologues from four antigenic variants. J Biol Chem. 1992;267:12728–12735. [PubMed] [Google Scholar]
- 8.Kawamori F, Akiyama M, Sugieda M, Kanda T, Akahane S, Yamamoto S, Ohashi N, Tamura A. Two-step polymerase chain reaction for diagnosis of scrub typhus and identification of antigenic variants of Rickettsia tsutsugamushi. J Vet Med Sci. 1993;55:749–755. doi: 10.1292/jvms.55.749. [DOI] [PubMed] [Google Scholar]
- 9.Ohashi N, Tamura A, Ohta M, Hayashi K. Purification and partial characterization of a type-specific antigen of Rickettsia tsutsugamushi. Infect Immun. 1989;57:1427–1431. doi: 10.1128/iai.57.5.1427-1431.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ree HI, Kim TE, Lee IY, Jeon SH, Hwang UW, Chang WH. Determination and geographical distribution of Orientia tsutsugamushi serotypes in Korea by nested polymerase chain reaction. Am J Trop Med Hyg. 2001;65:528–534. doi: 10.4269/ajtmh.2001.65.528. [DOI] [PubMed] [Google Scholar]
- 11.Hara Y, Abe T. The influence of chemotherapy on the mortality rates of tsutsugamushi disease in northern Japan, and some other statistical information. Am J Trop Med Hyg. 1956;5:218–223. doi: 10.4269/ajtmh.1956.5.218. [DOI] [PubMed] [Google Scholar]
- 12.Kim DM, Yun NR, Yang TY, Lee JH, Yang JT, Shim SK, Choi EN, Park MY, Lee SH. Usefulness of nested PCR for the diagnosis of scrub typhus in clinical practice: a prospective study. Am J Trop Med Hyg. 2006;75:542–545. [PubMed] [Google Scholar]
- 13.Furuya Y, Yoshida Y, Katayama T, Yamamoto S, Kawamura A., Jr Serotype-specific amplification of Rickettsiaa tsutsugamushi DNA by nested polymerase chain reaction. J Clin Microbiol. 1993;31:1637–1640. doi: 10.1128/jcm.31.6.1637-1640.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enatsu T, Urakami H, Tamura A. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol Lett. 1999;180:163–169. doi: 10.1111/j.1574-6968.1999.tb08791.x. [DOI] [PubMed] [Google Scholar]
- 15.Tamura A, Yamamoto N, Koyama S, Makisaka Y, Takahashi M, Urabe K, Takaoka M, Nakazawa K, Urakami H, Fukuhara M. Epidemiological survey of Orientia tsutsugamushi distribution in field rodents in Saitama Prefecture, Japan, and discovery of a new type. Microbiol Immunol. 2001;45:439–446. doi: 10.1111/j.1348-0421.2001.tb02643.x. [DOI] [PubMed] [Google Scholar]
- 16.Tamura A, Makisaka Y, Kadosaka T, Enatsu T, Okubo K, Koyama S, Yu Q, Urakami H. Isolation of Orientia tsutsugamushi from Leptotrombidium fuji and its characterization. Microbiol Immunol. 2000;44:201–204. doi: 10.1111/j.1348-0421.2000.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 17.Qiang Y, Tamura A, Urakami H, Makisaka Y, Koyama S, Fukuhara M, Kadosaka T. Phylogenetic characterization of Orientia tsutsugamushi isolated in Taiwan according to the sequence homologies of 56-kDa type-specific antigen genes. Microbiol Immunol. 2003;47:577–583. doi: 10.1111/j.1348-0421.2003.tb03420.x. [DOI] [PubMed] [Google Scholar]
- 18.Cho NH, Kim HR, Lee JH, Kim SY, Kim J, Cha S, Kim SY, Darby AC, Fuxelius HH, Yin J, Kim JH, Kim J, Lee SJ, Koh YS, Jang WJ, Park KH, Andersson SG, Choi MS, Kim IS. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc Natl Acad Sci USA. 2007;104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seong SY, Park SG, Huh MS, Jang WJ, Choi MS, Chang WH, Kim IS. T-track PCR fingerprinting for the rapid detection of genetic polymorphism. FEMS Microbiol Lett. 1997;152:37–44. doi: 10.1111/j.1574-6968.1997.tb10406.x. [DOI] [PubMed] [Google Scholar]
- 20.Elisberg BL, Campbell JM, Bozeman FM. Antigenic diversity of Rickettsia tsutsugamushi: epidemiologic and ecologic significance. J Hyg Epidemiol Microbiol Immunol. 1968;12:18–25. [PubMed] [Google Scholar]
- 21.Fournier PE, Siritantikorn S, Rolain JM, Suputtamongkol Y, Hoontrakul S, Charoenwat S, Losuwanaluk K, Parola P, Raoult D. Detection of new genotypes of Orientia tsutsugamushi infecting humans in Thailand. Clin Microbiol Infect. 2008;14:168–173. doi: 10.1111/j.1469-0691.2007.01889.x. [DOI] [PubMed] [Google Scholar]
- 22.Traub R, Wisseman CL., Jr The ecology of chigger-borne rickettsiosis (scrub typhus) J Med Entomol. 1974;11:237–303. doi: 10.1093/jmedent/11.3.237. [DOI] [PubMed] [Google Scholar]
- 23.Tamura A, Takahashi K, Tsuruhara T, Urakami H, Miyamura S, Sekikawa H, Kenmotsu M, Shibata M, Abe S, Nezu H. Isolation of Rickettsia tsutsugamushi antigenically different from Kato, Karp, and Gilliam strains from patients. Microbiol Immunol. 1984;28:873–882. doi: 10.1111/j.1348-0421.1984.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto S, Kawabata N, Tamura A, Urakami H, Ohashi N, Murata M, Yoshida Y, Kawamura A., Jr Immunological properties of Rickettsia tsutsugamushi, Kawasaki strain, isolated from a patient in Kyushu. Microbiol Immunol. 1986;30:611–620. doi: 10.1111/j.1348-0421.1986.tb02988.x. [DOI] [PubMed] [Google Scholar]
- 25.Shirai A, Coolbaugh JC, Gan E, Chan TC, Huxsoll DL, Groves MG. Serologic analysis of scrub typhus isolates from the Pescadores and Philippine Islands. Jpn J Med Sci Biol. 1986;35:255–259. doi: 10.7883/yoken1952.35.255. [DOI] [PubMed] [Google Scholar]
- 26.Choi MS, Chang WJ, Park SK, Huh MS, Kim HR, Han TH, Kim IS. Seroepidemiological survey of scrub typhus in Korea, 1995–1996. J Bacteriol Virol. 1997;32:219–226. [Google Scholar]
- 27.Manosroi J, Chutipongvivate S, Auwanit W, Manosroi A. Determination and geographic distribution of Orientia tsutsugamushi serotypes in Thailand by nested polymerase chain reaction. Diagn Microbiol Infect Dis. 2006;55:185–190. doi: 10.1016/j.diagmicrobio.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Parola P, Blacksell SD, Phetsouvanh R, Phongmany S, Rolain JM, Day NP, Newton PN, Raoult D. Genotyping of Orientia tsutsugamushi from humans with scrub typhus, Laos. Emerg Infect Dis. 2008;14:1483–1485. doi: 10.3201/eid1409.071259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohashi N, Fukuhara M, Shimada M, Tamura A. Phylogenetic position of Rickettsia tsutsugamushi and the relationship among its antigenic variants by analyses of 16S rRNA gene sequences. FEMS Microbiol Lett. 1995;125:299–304. doi: 10.1111/j.1574-6968.1995.tb07372.x. [DOI] [PubMed] [Google Scholar]
- 30.Tay ST, Rohani YM, Ho TM, Shamala D. Sequence analysis of the hypervariable regions of the 56 kDa immunodominant protein genes of Orientia tsutsugamushi strains in Malaysia. Microbiol Immunol. 2005;49:67–71. doi: 10.1111/j.1348-0421.2005.tb03641.x. [DOI] [PubMed] [Google Scholar]
- 31.Chang WH, Kang JS, Lee WK, Choi MS, Lee JH. Serological classification by monoclonal antibodies of Rickettsia tsutsugamushi isolated in Korea. J Clin Microbiol. 1990;28:685–688. doi: 10.1128/jcm.28.4.685-688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim IS, Seong SY, Woo SG, Choi MS, Chang WH. High-level expression of a 56-kilodalton protein gene (bor56) of Rickettsia tsutsugamushi Boryong and its application to enzyme-linked immunosorbent assays. J Clin Microbiol. 1993;31:598–605. doi: 10.1128/jcm.31.3.598-605.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]


