Abstract
Polymerase chain reaction (PCR) assays for intestinal parasites are increasingly being used on fecal DNA samples for enhanced specificity and sensitivity of detection. Comparison of these tests against microscopy and copro-antigen detection has been favorable, and substitution of PCR-based assays for the ova and parasite stool examination is a foreseeable goal for the near future. One challenge is the diverse list of protozoan and helminth parasites. Several existing real-time PCR assays for the major intestinal parasites—Cryptosporidium spp., Giardia intestinalis, Entamoeba histolytica, Ancylostoma duodenale, Ascaris lumbricoides, Necator americanus, and Strongyloides stercoralis—were adapted into a high throughput protocol. The assay involves two multiplex PCR reactions, one with specific primers for the protozoa and one with specific primers for the helminths, after which PCR products are hybridized to beads linked to internal oligonucleotide probes and detected on a Luminex platform. When compared with the parent multiplex real-time PCR assays, this multiplex PCR-bead assay afforded between 83% and 100% sensitivity and specificity on a total of 319 clinical specimens. In conclusion, this multiplex PCR-bead protocol provides a sensitive diagnostic screen for a large panel of intestinal parasites.
Introduction
Expertise in stool microscopy is waning and multiple sampling, species-specific concentration and staining methods are needed to improve its performance. Meanwhile, polymerase chain reaction (PCR) methods for detecting intestinal parasites are increasingly available and exhibit excellent sensitivity and specificity compared with conventional methods such as microscopy.1–5 Therefore, in our view shifting to a molecular approach for the common intestinal parasites is desirable as an adjunct or alternative for stool ova and parasite examination. One challenge, however, is that infection with several pathogens is common and assays directed at individual organisms may routinely miss the diversity of infecting parasites. Although dependent on the setting, the major intestinal parasites considered are Entamoeba histolytica, Giardia intestinalis, Cryptosporidium spp., hookworms, Ascaris lumbricoides, and Strongyloides stercoralis. We have recently developed multiplexed real-time PCR assays for these common intestinal parasites. These parent multiplex real-time PCR assays have been extensively validated against conventional techniques including enzyme-linked immunosorbent assay (ELISA) and microscopy and are being used in routine diagnostic settings and in epidemiological surveys.6–11 In this study, these methods were adapted into a combined bead-based Luminex assay as an alternative molecular diagnostic platform to capture each of these parasites in a single protocol.
Materials And Methods
Control material.
Positive control materials used in this study included axenic culture of ATCC strain 30459 E. histolytica HM-1:IMSS, purified Giardia lamblia cysts, and Cryptosporidium parvum oocysts (Waterborne Inc., New Orleans, LA). Control DNA was obtained from L3 larvae of S. stericoralis and Ancylostoma duodenale from coproculture. Control DNA for Necator americanus and A. lumbricoides was isolated from adult worms. Negative controls were water. For sensitivity testing, positive control materials were serially spiked into an identical stool sample from a healthy donor aliquoted into 200 mg volumes. We used an exogenous phocine herpes virus spiked into the lysis buffer for stool DNA extraction to provide a control for extraction and amplification.
Fecal specimens.
One hundred twenty-nine fecal DNA specimens were obtained from preschool-age or younger children from the International Centre for Diarrhoeal Diseases and Research, Bangladesh (ICDDR,B). These samples were tested by saline wet mount microscopy per routine. Furthermore, 190 DNA specimens were obtained from the Leiden University Medical Center (LUMC) clinical microbiology laboratory and several epidemiological surveys in different countries from several collaborative projects with the Department of Parasitology at the LUMC. For this validation exercise at LUMC, positive samples were all previously extracted and tested for either protozoa or helminths by real-time PCR, thus we were not able to explicitly evaluate different DNA extraction methods in this work or test incoming specimens in real-time. All PCR-positive samples from LUMC were also positive by microscopy. Both positive and negative stool extracted DNA samples were included to establish sensitivity and specificity. Specifically, we sought to obtain at least 20 PCR-positive samples per organism with the exception of A. duodenale for which positive samples were limited. All protocols were approved by the Ethics Committees of ICDDR,B and the institutional review boards of University of Virginia (UVA) or Leiden University.
DNA extraction.
The DNA was extracted using a slightly modified QIAamp DNA Stool Mini Kit protocol (Qiagen Inc., Valencia, CA) as described previously for specimens from ICDDR,B.3 The DNA extraction for the specimens from LUMC was performed using regular QIAamp mini Kit spin columns with modifications, which included a pretreatment with PVPP (polyvinylpyrrolidone) and boiling.12
Protozoa-positive controls were extracted using an automated nucleic acid isolation system, QuickGene-810, with QuickGene DNA tissue kit S (Fujifilm, Tokyo, Japan). The manufacturer's protocol was modified to accommodate a larger input stool volume of 200 mg. Briefly, 1 mL of the tissue lysis buffer MDT was added to the stool, and the suspension was then pretreated by bead beating with a tube of 0.15 mm garnet beads (MO-BIO Laboratories, Inc, Carlsbad, CA) for 2 min followed by boiling for 7 min before extraction. Other modifications included the addition of 100 μL of EDT solution (Proteinase K) from the kit and a longer incubation time following the addition of EDT solution for 90 min. All DNA samples were stored at −80°C until use in PCR.
Multiplex PCR.
Primer and probe sequences3,4,13–17 and the real-time PCR protocols have been described previously (Table 1). Target genes included the 18S ribosomal RNA (rRNA) gene for E. histolytica, G. lamblia, and S. stercoralis and COWP for Cryptosporidium spp. (see GenBank accession nos. X64142, M54878, AF279916, and AF248743, respectively). Furthermore, the internal transcribed spacer (ITS) 1 target was used for A. lumbricoides assay and the ITS2 for A. duodenale and N. americanus assays (GenBank accession nos. ALJ000895, AJ001594, and AJ001599, respectively).
Table 1.
Sequences for primers and probes used in the real-time polymerase chain reaction (PCR) and PCR-Luminex assays
| Organism | Target | Reference | Name | Sequence (5′ → 3′) |
|---|---|---|---|---|
| Cryptosporidium spp. | COWP | 13 | C151F C151R* C151P | CAAATTGATACCGTTTGTCCTTCTGGGCATGTCGATTCTAATTCAGCTTGCCATACATTGTTGTCCTGACAAATTGAAT |
| Entamoeba histolytica | 18S rRNA | 3 | Eh134F Eh134R* Eh134P | AACAGTAATAGTTTCTTTGGTTAGTAAAACTTAGAATGTCATTTCTCAATTCATATTAGTACAAAATGGCCAATTCATTCA |
| Giardia lamblia | 18S rRNA | 14 | G62FG62R*G62P | GACGGCTCAGGACAACGGTTTTGCCAGCGGTGTCCGCCCGCGGCGGTCCCTGCTAG |
| Ascaris lumbricoides | ITS1 | 15 | Alum96F*Alum183RAlum124P | GTAATAGCAGTCGGCGGTTTCTTGCCCAACATGCCACCTATTCTTGGCGGACAATTGCATGCGAT |
| Ancylostoma duodenale | ITS2 | 4 | Ad125FAd195R*Ad155P | GAATGACAGCAAACTCGTTGTTGATACTAGCCACTGCCGAAACGTATCGTTTACCGACTTTAG |
| Necator americanus | ITS2 | 4 | Na58F*Na158RNa81P | CTGTTTGTCGAACGGTACTTGCATAACAGCGTGCACATGTTGCCTGTACTACGCATTGTATAC |
| Strongyloides stercoralis | 18S rRNA | 17 | Stro18SFStro18SR*Stro18SP | GAATTCCAAGTAAACGTAAGTCATTAGCTGCCTCTGGATATTGCTCAGTTCACACACCGGCCGTCGCTGC |
| Extraction control (phocine herpes virus) | Glycoprotein B | 16 | PhHV267FPhHV337R*PhHV305P | GGGCGAATCACAGATTGAATCGCGGTTCCAAACGTACCAATTTTTATGTGTCCGCCACCATCTGGATC |
Biotinylated primer in the PCR-Luminex assay.
Briefly, real-time PCR amplification reaction for the 3-plex protozoa was performed in 25 uL volume with 12.5 μL of iQ Supermix (Bio-Rad, Hercules, CA), which contains dNTPs, 6 mM MgCl2, 50 U/mL iTaq DNA, additional 2 mM MgCl2, 0.4 μM E. histolytica primers, 0.6 μM Giardia primers, 1.0 μM Cryptosporidium primers, 0.08 μM E. histolytica probe, 0.16 μM Giardia probe, 0.4 μM Cryptosporidium probe, and 4 μL of sample DNA. The Taqman probes used for this assay were labeled with Yakima Yellow for E. histolytica, FAM for Giardia, and Texas Red for Cryptosporidium and purchased from Eurogentec (Fremont, CA). The PCR cycling condition consisted of an initial 3 min 95°C cycle followed by 40 cycles of 30 sec at 95°C, 30 sec at 55°C, and 30 sec at 72°C with a final extension for 7 min at 72°C. Amplification reaction for the helminth real-time assay was performed in 25 μL volume consisting of 12.5 μL HotStarTaq Master Mix (Qiagen Inc.), which contains 1.5 mM MgCl2 and 200 μM each dNTP, additional 3.5 mM MgCl2 for a final concentration of 5 mM MgCl2, 0.1 mg/mL of BSA, 0.2 μM Ancylostoma primers, 0.1 μM Ancylostoma probe, 0.2 μM Necator primers, 0.05 μM Necator probe, 0.08 μM Ascaris primers, 0.05 μM Ascaris probe, 0.1 μM Strongyloides primers, 0.05 μM Strongyloides probe, 0.15 μM PhHV primers, 0.05 PhHV probe, and 5 μL of sample DNA. Taqman Fluorophores for the Taqman probes included Cy5, FAM, Quasar705, Texas Red, and Yakima Yellow for the PhHV, Necator, Strongyloides, Ancylostoma, and Ascaris probes, respectively. The helminth primers and probes were synthesized by Biolegio (Nijmegen, The Netherlands). Amplification steps were as follows: 15 min at 95°C; 50 cycles of 30 sec at 94°C, 30 sec at 60°C, and 1 min at 72°C; final extension for 10 min at 72°C. All real-time PCRs were performed either on the Bio-Rad iCycler or the Bio-Rad CFX platform (Bio-Rad, Hercules, CA). Real-time PCR Ct was analyzed by the iCycler software version 3.1 and CFX96 Real-Time System by CFX software version 1.1 (Bio-Rad, Hercules, CA).
Multiplex PCR for the Luminex assays were prepared exactly as the real-time mixture except without the TaqMan probes, and either the forward or the reverse primers were biotinylated on the 5′-end. Internal probes were amine modified at the 5′-end and included 12 carbon spacers. All probes and primers were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Probes were coupled to carboxylated Luminex beads with EDC (1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride). Hybridization of beads to the PCR products was performed at 50°C for both the helminth and protozoa assays for 35 min using the Oligonucleotide Hybridization Protocol from Luminex Corporation.18 Detection of the amplicon was performed by the Bioplex 200 (Bio-Rad) or Luminex 100 (Luminex Corporation, Austin, TX) after addition of streptavidin PE as the reporter molecule. Negative control material (nuclease-free water) and positive controls were included with each run. All discrepant samples were retested by both modalities.
Analytical sensitivity.
Analytical sensitivity for the Luminex 3-plex protozoa assay was determined by serially spiking C. parvum oocysts, G. lamblia cysts, and E. histolytica trophozoites into a single common stool specimen followed by DNA extraction with the Fujifilm method. For Ancylostoma, Ascaris, Necator, and Strongyloides, where cultured materials were not available, we used serial dilutions of control DNA to establish analytical sensitivity.
Luminex data.
Luminex data were reported as mean fluorescence intensity (MFI). We calculated a corrected MFI (cMFI), which normalizes to background fluorescence as follows: cMFI = [MFI (sample) − MFI (background)]/MFI (background) to accommodate testing on different Luminex systems under both high and low photomultiplier tube voltages and different software (Star Station version 2.0, Applied Cytometry Systems, Sacramento, CA, and Bioplex software version 5.0, Bio-Rad, Hercules, CA).
Statistics.
The cycle threshold (Ct) and cMFI were compared using the Mann-Whitney test because all data sets were not normally distributed (SPSS Inc., Chicago, IL). To measure the degree of association between Ct values and cMFIs, Spearman's rank correlation test was used because the relationship between the variables was nonlinear. All P values were two-tailed. Data shown as mean + SEM unless otherwise indicated. We calculated 95% confidence intervals (CI) for sensitivity and specificity based on the sample size chosen for this study using the formula (CI = p ± 1.96 × [p(1 − p)/n]1/2)
where p = sensitivity or specificity and n = number of infected samples for sensitivity or uninfected samples for specificity determined by real-time PCR, the gold standard test.19
Results
Selection of PCR reactions.
In developing the Luminex assays, we found that biotinylating the primer closest to the probe yielded higher fluorescence for each amplicon (data not shown). Luminex-PCR reactions were all initially performed in singleplex reactions and then multiplexed. The sensitivity of this assay was highest if performed in two separate multiplex PCR reactions, one for the three protozoa and one for the four helminths plus an extraction control (data below).
Analytical sensitivity of the assays.
The Luminex protozoa assay showed a lower limit of detection of 103 Giardia cysts, 102 Cryptosporidium oocysts, and 101 E. histolytica trophozoites in 200 mg of a stool specimen (Table 2). Of note, this lower limit of detection was equivalent to or better than the parent real-time PCR assays, which detected 103 Giardia, 103 Cryptosporidium, and 101 E. histolytica (Table 2). The sensitivity of the multiplex Luminex assay was equivalent to that obtained when each of the targets was amplified and detected in singleplex, however the raw MFI values did decrease when amplified in multiplex (data not shown). Analytical sensitivity for Ancylostoma, Ascaris, Necator, and Strongyloides was also examined. This revealed that the Luminex helminth assay could detect Ancylostoma, Ascaris, Necator, and Strongyloides at 10−4, 10−6, 10−6, and 10−4 dilutions, respectively (Table 3). In general, the relationship between parasite quantity and real-time Ct appeared more linear than that of parasite quantity and cMFI, however the Luminex assay had better or equal lower limits of detection as compared with the real-time PCR (Tables 2 and 3).
Table 2.
Analytical sensitivity of serially spiked protozoa stool samples*
| Sample | 10n organisms | Cryptosporidium | Entamoeba histolytica | Giardia lamblia | |||
|---|---|---|---|---|---|---|---|
| qPCR Ct | Luminex cMFI | qPCR Ct | Luminex cMFI | qPCR Ct | Luminex cMFI | ||
| Cryptosporidium oocysts | 5 | 28.8 ± 0.1 | 91.4 ± 1.2 | nd | −0.1 ± 0.1 | nd | 0.0 ± 0.0 |
| 4 | 32.1 ± 0.1 | 84.1 ± 1.5 | nd | −0.1 ± 0.0 | nd | −0.1 ± 0.1 | |
| 3 | 34.5 ± 0.2 | 79.6 ± 0.4 | nd | 0.0 ± 0.1 | nd | −0.1 ± 0.0 | |
| 2 | nd | 12.0 ± 11.9 | nd | −0.1 ± 0.1 | nd | 0.0 ± 0.1 | |
| 1 | nd | 0.1 ± 0.0 | nd | 0.0 ± 0.1 | nd | −0.2 ± 0.0 | |
| Entamoeba histolytica trophozoites | 5 | nd | 0.1 ± 0.1 | 21.3 ± 0.0 | 84.3 ± 0.9 | nd | 0.0 ± 0.0 |
| 4 | nd | 0.3 ± 0.0 | 25.9 ± 0.1 | 84.5 ± 3.1 | nd | 0.0 ± 0.1 | |
| 3 | nd | 0.2 ± 0.0 | 28.0 ± 0.0 | 83.8 ± 0.0 | nd | 0.1 ± 0.1 | |
| 2 | nd | 0.0 ± 0.0 | 30.8 ± 0.1 | 81.1 ± 0.2 | nd | 0.2 ± 0.0 | |
| 1 | nd | 0.2 ± 0.0 | 34.3 ± 0.1 | 70.9 ± 3.3 | nd | 0.0 ± 0.0 | |
| Giardia lamblia cysts | 5 | nd | 0.0 ± 0.1 | nd | 0.0 ± 0.1 | 27.0 ± 0.1 | 48.7 ± 0.9 |
| 4 | nd | 0.1 ± 0.0 | nd | 0.0 ± 0.0 | 30.0 ± 0.1 | 37.2 ± 0.9 | |
| 3 | nd | 0.0 ± 0.0 | nd | −0.1 ± 0.1 | 34.1 ± 0.2 | 18.9 ± 0.8 | |
| 2 | nd | 0.2 ± 0.1 | nd | −0.1 ± 0.1 | nd | 0.3 ± 0.2 | |
| 1 | nd | 0.1 ± 0.1 | nd | 0.0 ± 0.1 | nd | 0.0 ± 0.0 | |
nd = not detected; qPCR = quantitative polymerase chain reaction; Ct = cycle threshold; cMFI = corrected mean fluorescence intensity.
Table 3.
Analytical sensitivity of serially diluted helminth DNA samples*
| Sample | Dilution factor | Ancylostoma | Ascaris | Necator | Strongyloides | ||||
|---|---|---|---|---|---|---|---|---|---|
| qPCR Ct | Luminex cMFI | qPCR Ct | Luminex cMFI | qPCR Ct | Luminex cMFI | qPCR Ct | Luminex cMFI | ||
| Ancylostoma | −1 | 27.6 | 13.0 | nd | −0.1 | nd | 0.2 | nd | −0.1 |
| −2 | 30.9 | 68.0 | nd | 0.0 | nd | −0.1 | nd | 0.0 | |
| −3 | 34.7 | 57.4 | nd | −0.1 | nd | −0.1 | nd | 0.0 | |
| −4 | 38.7 | 7.3 | nd | −0.2 | nd | 0.1 | nd | −0.3 | |
| −5 | nd | 0.2 | nd | −0.1 | nd | −0.1 | nd | 0.0 | |
| −6 | nd | −0.2 | nd | −0.2 | nd | 0.1 | nd | −0.2 | |
| Ascaris | −1 | nd | −0.1 | 17.7 | 87.6 | nd | 0.0 | nd | 0.0 |
| −2 | nd | 0.0 | 21.2 | 89.2 | nd | 0.0 | nd | −0.1 | |
| −3 | nd | 0.1 | 24.5 | 87.5 | nd | 0.1 | nd | 0.0 | |
| −4 | nd | 0.0 | 27.9 | 81.1 | nd | −0.1 | nd | 0.1 | |
| −5 | nd | −0.1 | 31.1 | 0.0 | nd | −0.1 | nd | 0.0 | |
| −6 | nd | 0.0 | 34.4 | 17.7 | nd | 0.0 | nd | −0.1 | |
| Necator | −1 | nd | 0.0 | nd | 0.2 | 20.6 | 60.1 | nd | 0.1 |
| −2 | nd | 0.0 | nd | −0.2 | 23.5 | 61.9 | nd | −0.1 | |
| −3 | nd | −0.2 | nd | −0.2 | 27.1 | 58.5 | nd | 0.0 | |
| −4 | nd | 0.0 | nd | −0.3 | 30.8 | 57.8 | nd | 0.0 | |
| −5 | nd | 0.0 | nd | 0.1 | 34.0 | 58.1 | nd | 0.1 | |
| −6 | nd | 0.0 | nd | −0.1 | 36.5 | 34.0 | nd | −0.2 | |
| Strongyloides | −1 | nd | 0.0 | nd | −0.1 | nd | 0.1 | 27.0 | 54.8 |
| −2 | nd | 0.0 | nd | 0.0 | nd | −0.1 | 30.5 | 25.0 | |
| −3 | nd | −0.1 | nd | 0.1 | nd | 0.0 | 33.9 | 16.7 | |
| −4 | nd | −0.3 | nd | 0.0 | nd | 0.1 | 38.8 | 5.8 | |
| −5 | nd | 0.1 | nd | −0.1 | nd | 0.1 | nd | 1.2 | |
| −6 | nd | 0.3 | nd | 0.0 | nd | 0.1 | nd | 0.0 | |
nd = not detected; qPCR = quantitative polymerase chain reaction; Ct = cycle threshold; cMFI = corrected mean fluorescence intensity.
Performance on fecal DNA specimens.
We then examined the performance of the assays on 319 specimens, which included 129 clinical specimens collected from preschool-age or younger children from ICDDR,B and 190 specimens obtained from LUMC. These specimens were selected because they had been previously characterized by real-time PCR. Specifically, 129 samples from ICDDR,B and 99 samples from LUMC were used to examine the protozoa assay and 91 samples from LUMC were used to examine the helminth assay. In all instances the PCR-Luminex assay was tested against the real-time PCR simultaneously. Using the real-time PCR result as the gold standard, cut-off threshold values were estimated by receiver operating characteristic (ROC) analysis and were ∼2.5 for all pathogens with the exception of Cryptosporidium and Strongyloides, which were 7.3 and 9.0, respectively, and we used 9.0 for E. histolytica to maximize specificity. Using these cutoffs the Luminex assay exhibited 83–100% sensitivity and specificity. In most instances discrepancies appeared to be of lower burden infections, for example Cryptosporidium, E. histolytica, and Giardia PCR−/Luminex+ results were of statistically lower cMFI than the respective PCR+/Luminex+ results (Table 4). Similarly, Giardia PCR+/Luminex− results were of statistically higher Ct than the corresponding PCR+/Luminex+ results. Mixed infections were common, in that the Luminex assay revealed 23 specimens that were positive for two or more protozoa and 9 specimens that were positive for two or more helminths. In terms of microscopy, this study was not designed to evaluate that aspect because the real-time PCR assays have been extensively validated against conventional methods.6–11 Indeed, all LUMC specimens were pre-selected as microscopy+/PCR+. However, Bangladesh specimens were not pre-selected, and saline wet-mount microscopy revealed only 28% sensitivity/99% specificity for Giardia, similar to previous observations.20
Table 4.
Performance of multiplex polymerase chain reaction (PCR)-Luminex vs. multiplex real-time PCR assays
| Organism | qPCR+ | qPCR− | Sensitivity | Specificity |
|---|---|---|---|---|
| CryptosporidiumPCR/Luminex+* | 35(Ct = 34.0 ± 2.3; cMFI = 122.3 ± 9.6†) | 7‡(cMFI = 53.2 ± 12.3) | 0.97(0.92 1.00)§ | 0.96(0.93 0.99)§ |
| CryptosporidiumPCR/Luminex− | 1‡(Ct = 38.68) | 185 | ||
| E. histolyticaPCR/Luminex+* | 33(Ct = 28.0 ± 0.9; cMFI = 93.9 ± 8.9†) | 4¶(cMFI = 19.8 ± 3.0) | 0.83(0.70 0.96)§ | 0.98(0.96 1.00)§ |
| E. histolyticaPCR/Luminex− | 7(Ct = 28.4 ± 3.2) | 184 | ||
| GiardiaPCR/Luminex+* | 56(Ct = 29.2 ± 0.7†; cMFI = 17.2 ± 2.3†) | 9‖(cMFI = 5.1 ± 0.8) | 0.93(0.87 0.99)§ | 0.95(0.92 0.98)§ |
| GiardiaPCR/Luminex− | 4(Ct = 35.0 ± 2.1) | 159 | ||
| AncylostomaPCR/Luminex+b | 11(Ct = 33.7 ± 1.4; cMFI = 27.7 ± 6.2) | 1(cMFI = 19.3) | 1.00 | 0.99(0.97 1.00)§ |
| AncylostomaPCR/Luminex−* | 0 | 79 | ||
| AscarisPCR/Luminex+* | 20(Ct = 35.6 ± 0.6; cMFI = 59.6 ± 6.5) | 0 | 1.00 | 1.00 |
| AscarisPCR/Luminex− | 0 | 71 | ||
| NecatorPCR/Luminex+* | 26(Ct = 36.0 ± 0.5; cMFI = 32.1 ± 3.0) | 3(cMFI = 19.7 ± 4.6) | 0.90(0.79 1.00)§ | 0.95(0.90 1.00)§ |
| NecatorPCR/Luminex− | 3(Ct = 35.5 ± 3.6) | 59 | ||
| StrongyloidesPCR/Luminex+* | 16(Ct = 30.7 ± 1.1; cMFI = 36.3 ± 4.3) | 2(cMFI = 18.1 ± 3.0) | 1.00 | 0.97(0.93 1.00)§ |
| StrongyloidesPCR/Luminex− | 0 | 73 | ||
| Extraction controlPCR/Luminex+* | 74(Ct = 35.1 ± 0.5; cMFI = 94.8 ± 1.9) | 2(cMFI = 63.1 ± 28.7) | 1.00 | 0.88(0.73 1.00)§ |
| Extraction controlPCR/Luminex− | 0 | 15 |
Luminex positive was defined as corrected mean fluorescence intensity (cMFI) ≥ 2.5 for all pathogens except Cryptosporidium, Strongyloides, and E. histolytica where we used 7.3, 9.0, and 9.0.
P < 0.05 comparing cMFI in quantitative polymerase reaction (qPCR)+ vs. qPCR− specimens; or comparing qPCR cycle threshold (Ct) in Luminex+ vs. Luminex− specimens.
The 95% confidence interval based on sample size tested.
Three of these 7 Luminex+/qPCR− samples were available and retested positive by a third PCR method. One Luminex−/qPCR+ sample was available and retested negative by a third method.
Three of these 4 Luminex+/qPCR− samples were available and retested negative by a third PCR method. One of these was positive with an E. dispar assay and was sequenced as E. dispar in the probe region and E. histolytica in the primer region, suggesting mispriming.
Four of these 9 Luminex+/qPCR− samples were available and two retested positive by a third PCR method.
Correlation between real-time PCR Ct values and Luminex cMFI.
The quantitative relationship between real-time PCR Ct values and Luminex cMFI in positive specimens is shown in Figure 1. Spearman's correlation was statistically significant for Cryptosporidium, Ascaris, Strongyloides, and Giardia (P < 0.05 for all, Spearman's ρ values of −0.45, −0.81, −0.49, and −0.39, respectively). Negative Spearman's ρ values indicate that as Ct values increase, cMFI values decrease or that the two variables are inversely proportional. For E. histolytica, Ancylostoma, and Necator no statistical correlation was observed.
Figure 1.
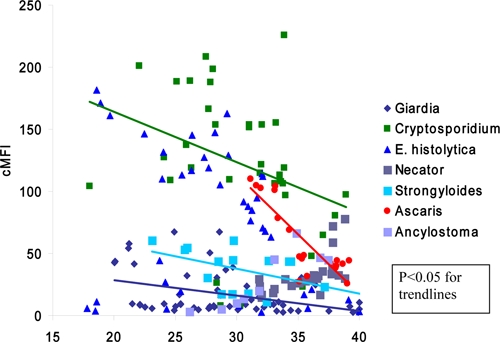
Correlation between multiplex real-time polymerase chain reaction (PCR) cycle threshold (Ct) and the multiplex PCR-Luminex corrected mean fluorescence intensity (cMFI) values on clinical specimens. Each of the quantitative PCR (qPCR) positive/Luminex-positive specimens of Table 4 was plotted for potential correlation. Trendlines indicate statistically significant Spearman's correlation, if present.
Discussion
The PCR-Luminex described in this study provides an alternative molecular detection system for the major intestinal parasites observed in stool microscopic examinations. The assay yields performance on par with the respective real-time PCR assays, which we know from previous studies are superior to microscopy or other methods.1–4 Detecting the major intestinal parasites is highly useful for a clinical laboratory, many of which are losing expertise in stool microscopy and sending stool samples out to referral laboratories. By contrast, expertise in the performance molecular assays is becoming more available. In this environment, the PCR-bead approach of this study can offer more sensitive detection than microscopy, save technician time by eliminating the need to perform several testing modalities (e.g., concentration methods for larvae, ova, and cysts, specific staining, or immunoassays), and give laboratories the capacity to perform their own tests in-house. Because equipment in clinical laboratories is variable, we developed this Luminex assay as an alternative to real-time PCR. Performance was overall comparable, and ultimately one's choice can depend on operational issues, such as availability of equipment and local reagent procurement realities. The protocol can be made high throughput with a liquid handler and test ∼96 extracted specimens in a few hours. The approach is modular—if the laboratory does not see certain parasites in their population or does not want to detect certain targets because not all tests were ordered, those reagents can be withheld from the assay. This assay can also be useful for research laboratories trying to understand the burden of intestinal parasites on global health. For instance, hookworm projects typically rely on detection of eggs by microscopy and do not gather species data that are implicit to our PCR approach and important for species-specific strategies such as vaccine development.
There were several limitations of this study. Initially, the intention was to develop one multiplex PCR that included all eight primer sets but we found that all targets were not efficiently amplified. Second, we attempted to hybridize the two described multiplex PCR reactions onto one bead cocktail. While it worked for some targets, it incurred a loss in signal for others. Although these issues may be surmountable with additional development, it will be challenging because the number of parasites is often low in stool and some parasites can be more difficult to extract DNA from than others. For instance, we have experienced much less difficulty in developing extensive multiplex PCR reactions for Escherichia coli or enteric viruses (data not shown). Finally, we did observe some discrepant results between the Luminex and real-time PCR assays, even though primers and probes were identical. Especially in the samples from the projects in parasite endemic areas it is not unlikely to have low-level mixed infections, which may be near the detection limit and yield some discrepancies. One of 10 samples that was positive with an Entamoeba dispar-specific PCR revealed low level positive fluorescence values with the E. histolytica Luminex assay. We are continuing to refine the cycling and hybridization conditions that should minimize this problem.
In summary, this multiplex PCR-bead protocol provides an alternative high-throughput molecular diagnostic platform for specific and sensitive detection of several major intestinal parasites and a potential alternative to microscopy for equipped laboratories.
ACKNOWLEDGMENTS
We thank all collaborators in several projects and programs with the department of Parasitology of the Leiden University Medical Center that have provided the control samples used. We also thank all the collaborators and the staff at the Parasitology Lab at ICDDR,B for providing DNA samples for this study.
Footnotes
Financial Support: This work was supported by National Institutes of Health grants U01 AI075396, AI043596, and the Bill and Melinda Gates Foundation.
Authors' addresses: Mami Taniuchi, William A. Petri Jr., and Eric R. Houpt, Department of Medicine, Division of Infectious Diseases and International Health, University of Virginia, Charlottesville, VA, E-mails: mt2f@virginia.edu, wap3g@virginia.edu, and erh6k@virginia.edu. Jaco J. Verweij and Lisette van Lieshout, Departments of Parasitology and Medical Microbiology (Clinical Microbiology Laboratory), Leiden University Medical Center, Leiden, The Netherlands, E-mails: J.J.Verweij@lumc.nl and LvanLieshout@lumc.nl. Zannatun Noor, Shihab U. Sobuz, and Rashidul Haque, International Centre for Diarrhoeal Disease Research (ICDDR,B), Centre for Health and Population Research, Mohakhali, Dhaka, Bangladesh, E-mails: zn5qh@virginia.edu, shihab119@icddrb.org, and rhaque@icddrb.org.
Reprint requests: Mami Taniuchi, Department of Medicine, Division of Infectious Diseases and International Health, University of Virginia, 345 Crispell Dr., MR6 Room 1714, Charlottesville, VA 22908, Tel: 434-924-5575, Fax: 434-924-0075, E-mail: mt2f@virginia.edu.
References
- 1.Verweij JJ, Schinkel J, Laeijendecker D, van Rooyen MA, van Lieshout L, Polderman AM. Real-time PCR for the detection of Giardia lamblia. Mol Cell Probes. 2003;17:223–225. doi: 10.1016/s0890-8508(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 2.Webster KA, Smith HV, Giles M, Dawson L, Robertson LJ. Detection of Cryptosporidium parvum oocysts in feces: comparison of conventional coproscopical methods and the polymerase chain reaction. Vet Parasitol. 1996;61:5–13. doi: 10.1016/0304-4017(95)00811-x. [DOI] [PubMed] [Google Scholar]
- 3.Haque R, Roy S, Siddique A, Mondal U, Rahman SM, Mondal D, Houpt E, Petri WA., Jr Multiplex real-time PCR assay for detection of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium spp. Am J Trop Med Hyg. 2007;76:713–717. [PubMed] [Google Scholar]
- 4.Verweij JJ, Brienen EA, Ziem J, Yelifari L, Polderman AM, Van Lieshout L. Simultaneous detection and quantification of Ancylostoma duodenale, Necator americanus, and Oesophagostomum bifurcum in fecal samples using multiplex real-time PCR. Am J Trop Med Hyg. 2007;77:685–690. [PubMed] [Google Scholar]
- 5.Ng CT, Gilchrist CA, Lane A, Roy S, Haque R, Houpt ER. Multiplex real-time PCR assay using Scorpion probes and DNA capture for genotype-specific detection of Giardia lamblia on fecal samples. J Clin Microbiol. 2005;43:1256–1260. doi: 10.1128/JCM.43.3.1256-1260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruijnesteijn van Coppenraet LE, Wallinga JA, Ruijs GJ, Bruins MJ, Verweij JJ. Parasitological diagnosis combining an internally controlled real-time PCR assay for the detection of four protozoa in stool samples with a testing algorithm for microscopy. Clin Microbiol Infect. 2009;15:869–874. doi: 10.1111/j.1469-0691.2009.02894.x. [DOI] [PubMed] [Google Scholar]
- 7.Calderaro A, Gorrini C, Bommezzadri S, Piccolo G, Dettori G, Chezzi C. Entamoeba histolytica and Entamoeba dispar: comparison of two PCR assays for diagnosis in a non-endemic setting. Trans R Soc Trop Med Hyg. 2006;100:450–457. doi: 10.1016/j.trstmh.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 8.Calderaro A, Gorrini C, Montecchini S, Peruzzi S, Piccolo G, Rossi S, Gargiulo F, Manca N, Dettori G, Chezzi C. Evaluation of a real-time polymerase chain reaction assay for the laboratory diagnosis of giardiasis. Diagn Microbiol Infect Dis. 2010;66:261–267. doi: 10.1016/j.diagmicrobio.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 9.ten Hove R, Schuurman T, Kooistra M, Moller L, van Lieshout L, Verweij JJ. Detection of diarrhoea-causing protozoa in general practice patients in The Netherlands by multiplex real-time PCR. Clin Microbiol Infect. 2007;13:1001–1007. doi: 10.1111/j.1469-0691.2007.01788.x. [DOI] [PubMed] [Google Scholar]
- 10.ten Hove RJ, van Esbroeck M, Vervoort T, van den Ende J, van Lieshout L, Verweij JJ. Molecular diagnostics of intestinal parasites in returning travellers. Eur J Clin Microbiol Infect Dis. 2009;28:1045–1053. doi: 10.1007/s10096-009-0745-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visser LG, Verweij JJ, Van Esbroeck M, Edeling WM, Clerinx J, Polderman AM. Diagnostic methods for differentiation of Entamoeba histolytica and Entamoeba dispar in carriers: performance and clinical implications in a non-endemic setting. Int J Med Microbiol. 2006;296:397–403. doi: 10.1016/j.ijmm.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Verweij JJ, Pit DS, van Lieshout L, Baeta SM, Dery GD, Gasser RB, Polderman AM. Determining the prevalence of Oesophagostomum bifurcum and Necator americanus infections using specific PCR amplification of DNA from fecal samples. Trop Med Int Health. 2001;6:726–731. doi: 10.1046/j.1365-3156.2001.00770.x. [DOI] [PubMed] [Google Scholar]
- 13.Guy RA, Payment P, Krull UJ, Horgen PA. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl Environ Microbiol. 2003;69:5178–5185. doi: 10.1128/AEM.69.9.5178-5185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, van Lieshout L, Polderman AM. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol. 2004;42:1220–1223. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiria AE, Prasetyani MA, Hamid F, Wammes LJ, Lell B, Ariawan I, Uh HW, Wibowo H, Djuardi Y, Wahyuni S, Sutanto I, May L, Luty AJ, Verweij JJ, Sartono E, Yazdanbakhsh M, Supali T. Does treatment of intestinal helminth infections influence malaria? Background and methodology of a longitudinal study of clinical, parasitological and immunological parameters in Nangapanda, Flores, Indonesia (ImmunoSPIN Study) BMC Infect Dis. 2010;10:77. doi: 10.1186/1471-2334-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niesters HG. Clinical virology in real time. J Clin Virol. 2002;25((Suppl 3)):S3–S12. doi: 10.1016/s1386-6532(02)00197-x. [DOI] [PubMed] [Google Scholar]
- 17.Verweij JJ, Canales M, Polman K, Ziem J, Brienen EA, Polderman AM, van Lieshout L. Molecular diagnosis of Strongyloides stercoralis in fecal samples using real-time PCR. Trans R Soc Trop Med Hyg. 2009;103:342–346. doi: 10.1016/j.trstmh.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Luminex . Sample Protocol for Oligonucleotide Hybridization. Toronto: 2006. Protocol. [Google Scholar]
- 19.Banoo S, Bell D, Bossuyt P, Herring A, Mabey D, Poole F, Smith PG, Sriram N, Wongsrichanalai C, Linke R, O'Brien R, Perkins M, Cunningham J, Matsoso P, Nathanson CM, Olliaro P, Peeling RW, Ramsay A. Evaluation of diagnostic tests for infectious diseases: general principles. Nat Rev Microbiol. 2006;4:S20–S32. doi: 10.1038/nrmicro1570. [DOI] [PubMed] [Google Scholar]
- 20.Haque R, Roy S, Kabir M, Stroup SE, Mondal D, Houpt ER. Giardia assemblage A infection and diarrhea in Bangladesh. J Infect Dis. 2005;192:2171–2173. doi: 10.1086/498169. [DOI] [PubMed] [Google Scholar]


