Abstract
Background
DevR (also called as DosR) is a two-domain response regulator of the NarL subfamily that controls dormancy adaptation of Mycobacterium tuberculosis (M. tb). In response to inducing signals such as hypoxia and ascorbic acid, the N-terminal receiver domain of DevR (DevRN) is phosphorylated at Asp54. This results in DevR binding to DNA via its C-terminal domain (DevRC) and subsequent induction of the DevR regulon. The mechanism of phosphorylation-mediated activation is not known. The present study was designed to understand the role of the N- and C-terminal domains of DevR in DevR regulon genes activation.
Methodology/Principal Findings
Towards deciphering the activation mechanism of DevR, we compared the DNA binding properties of DevRC and DevR and correlated the findings with their ability to activate gene expression. We show that isolated DevRC can interact with DNA, but only with the high affinity site of a representative target promoter. Therefore, one role of DevRN is to mask the intrinsic DNA binding function of DevRC. However, unlike phosphorylated DevR, isolated DevRC does not interact with the adjacent low affinity binding site suggesting that a second role of DevRN is in cooperative binding to the secondary site. Transcriptional analysis shows that consistent with unmasking of its DNA binding property, DevRC supports the aerobic induction, albeit feebly, of DevR regulon genes but is unable to sustain gene activation during hypoxia.
Conclusions/Significance
DevR is a unique response regulator that employs a dual activation mechanism including relief of inhibition and cooperative interaction with binding sites. Importantly, both these functions reside outside the C-terminal domain. DevRN is also essential for stabilizing DevR and sustaining autoregulation under hypoxia. Hence, both domains of DevR are required for robust transcription activation.
Introduction
Bacterial persistence is a hallmark of tuberculosis (TB). Following a TB infection, the individual usually mounts an effective immune response that leads to a cessation of disease progression due to the formation of granulomas around infective foci. Clinical studies suggest that the bacilli within these granulomas remain dormant in untreated individuals, causing latent infection that can last a lifetime [1], [2]. Oxygen limitation during granuloma development has been proposed to be one of the main signals that alter the metabolic status of bacteria to a state of dormancy [3]. Two-component systems are majorly involved in sensing and responding to changing environments in bacteria [4]. Numerous studies have demonstrated the relevance of the DevR-DevS two-component system in virulence and adaptation of Mycobacterium tuberculosis (M. tb) to putative granuloma signals including hypoxia, nitric oxide, carbon monoxide and ascorbic acid [5]–[13]. It mediates the induction of ∼48 genes referred to as the DevR regulon [8] and this genetic response is essential for bacterial adaptation and persistence under hypoxia [14], [15].
DevR (Rv3133c, also called DosR) is one of the best characterized transcriptional regulators of M. tb. It is a typical two-domain response regulator of the NarL subfamily [5] and its N-terminal domain that contains a phosphorylation site, Asp54, is connected to the C-terminal DNA binding domain (DevRC) by a linker sequence [16]–[19]. The target genes of the DevR regulon were predicted to contain one, two or more putative DevR binding sites (Dev boxes) in their upstream regions [8]. We have shown the importance of cooperative binding of DevR to two or more sites for the full induction of some of these genes. Close packing of the binding sites and an overlap of the Transcription start point (TSP)-proximal binding site with the -35 promoter element were common features of the target promoters that were analyzed [20]–[22]. While DevRC interaction with DNA oligonucleotides containing two consensus binding sequences was shown by crystal structure analysis [18], phosphorylation of intact DevR at Asp54 was found to be essential for interaction with DNA [20]. The importance of phosphorylation was supported by visualizing extensive interactions between the N- and C-terminal domains in the DevR structure that mask the DNA binding domain. A helix rearrangement mechanism was proposed to alleviate this inhibition [19].
The present study was designed to understand the role of the N- and C-terminal domains in activation of the DevR regulon genes. We compared the DNA binding properties of DevRC and DevR and correlated the findings with their ability to activate gene expression. We show that DevRC activates albeit weakly, the aerobic expression of the DevR regulon. The inability of DevRC to support full induction is attributed, at least in part, to a failure to cooperatively recruit DevR to adjacently-placed secondary binding sites. We also show that devRC transcript and DevRC protein levels are not maintained during hypoxia. The present study reveals the multifunctional role of the DevRN domain. In addition to receiving the phosphosignal at Asp54 from DevS and DosT kinases [16], [17], [23], DevRN suppresses the DNA binding and transcription-activating ability of unphosphorylated DevR under aerobic conditions, sponsors cooperative binding of DevR with secondary binding sites, and it is required for sustaining DevR stability and autoregulation during hypoxia. Thus DevR action is mediated by both its N-terminal and C-terminal domains.
Materials and Methods
Plasmids, bacterial strains, and culture conditions
All plasmids and bacterial strains used in this study are described in Tables 1 and 2, respectively. M. tb strains were cultured at 37°C in Dubos medium containing 0.05% Tween-80 plus 0.5% albumin, 0.75% dextrose and 0.085% NaCl (DTA medium). Escherichia coli (E. coli) strains and culture conditions were as described earlier [20]. Antibiotics were used at the following concentrations: hygromycin at 50 µg/ml for M. tb and 200 µg/ml for E. coli, kanamycin at 20 µg/ml for M. tb and 50 µg/ml for E. coli.
Table 1. Plasmids used in this study.
| Plasmid | Relevant featuresa | Source/Reference |
| pUS-DevRC | pET28a overexpressing DevR C-terminal domain cloned in NdeI site | This study |
| pAV-DevR | pET28a overexpressing full length wild type DevR cloned in NdeI site | [12] |
| pJFR19 | E. coli-Mycobacterium integrating shuttle plasmid with 3-kbacetamidase promoter, Hygr | [43] |
| pFPV27 | E. coli-Mycobacterium shuttle plasmid with promoter less gfp, Kanr | [44] |
| pET28a | E. coli expression vector (with N-terminal His6 tag), Kanr | Novagen |
| pMG86 | pJFR19 containing devR-devS expressed from acetamidase promoter, Hygr | [45] |
| pTGS | pFPV27 containing tgs1 promoter (-143 to +45), Kanr | [22] |
| p3131 | pFPV27 containing Rv3131 promoter (-150 to +48), Kanr | [22] |
| pSD POperon devR | pJFR19 containing devR (cloned between NdeI and XbaI sites), full-length DevR is expressed from Rv3134c-devRS operon promoter (-608 to +998, ref. 20) cloned in NdeI and BstBI sites | S.D.Majumdar and J.S.Tyagi, 2010, unpublished |
| pUS POperon devRC | pJFR19 containing devRC (cloned between NdeI and XbaI sites), DevRC is expressed from Rv3134c-devRS operon promoter cloned in NdeI and BstBI sites | This study |
| pUS PAcet devRC | pJFR19 containing devRC (cloned between NdeI and XbaI sites), DevRC expressed from constitutive acetamidase promoter cloned in NdeI and BstBI sites | This study |
| pUS Phsp60 devRC | pJFR19 containing devRC (cloned between NdeI and XbaI sites), DevRC expressed from constitutive hsp60 promoter cloned in NdeI and BstBI sites | This study |
The coordinates of the promoters (in parentheses) are with reference to the transcription start point (TSP) of tgs1;
Hygr, hygromycin resistance;
Kanr, kanamycin resistance.
Table 2. Strains used in this study.
| M. tb strain | Relevant features | Source/Reference |
| H37Rv | WT laboratory strain of M. tuberculosis (M. tb) | Laboratory collection |
| ΔdevR | 447-bp BalI deletion in M. tb H37Rv devR gene (deletes DevR amino acid residues from position 40 to191) | [46] |
| Comp13 | ΔdevR complemented with plasmid pSD POperon devR, expresses full-length DevR protein | S.D. Majumdar,Ph.D. ThesisSubmitted, 2010 |
| Comp5 | ΔdevR complemented with plasmid pUS POperon devRC, expresses DevRC protein | This study |
| Comp6 | ΔdevR complemented with plasmid pUS PAcet devRC, expresses DevRC protein | This study |
| Comp7 | ΔdevR complemented with plasmid pUS Phsp60 devRC, expresses DevRC protein | This study |
Construction of DevRC over-expressing plasmid and purification of DevRC
The devR C-terminal domain coding sequence (141-217 amino acids of DevR) was amplified from M. tb H37Rv DNA by PCR (Table 3), and cloned into pET28a to generate pUS-DevRC which expresses the C-terminal domain. N-terminal His6-tagged DevRC and full-length DevR proteins (referred to as DevRC and DevR, respectively) were overexpressed in E. coli C43 (DE3) from pUS-DevRC and pAV-DevR, respectively using standard procedures. The recombinant proteins were purified by standard techniques and used in EMSA and DNase I footprinting experiments.
Table 3. Primers used in this study.
| Primer | Sequence (5′→ 3′) | Application |
| devRC NdeI F | CGGACCCATATGCAGGACCCGCTATCAGGC | Cloning of devRC in pJFR19 |
| devRC XbaI R | CCGCTCTAGACCTGTTGTCATGGTCCATCACCGGGTG | |
| hsp60 BstBI F | CCGTTCGAAGGTGACCACAACGACGCGCCCGC | Cloning of hsp60 promoter in pJFR19 |
| hsp60 NdeI R | CCGCATATGTGCGAAGTGATTCCTCCGGATCG | |
| devRC NdeI F | CGGACCCATATGCAGGACCCGCTATCAGGC | Cloning of devRC in pET28a |
| devRC NdeI R | CCGCATATGCTATCATGGTCCATCACCGGGTGG | |
| RT16S F | ATGACGGCCTTCGGGTTGTAA | Real Time RT PCR (ref. 13) |
| RT16S R | CGGCTGCTGGCACGTAGTTG | Real Time RT PCR (ref. 13) |
| RT3134c F | CTGGCTGGGTCGGCCTTAGC | Real Time RT PCR (ref. 13) |
| RT3134c R | TGACCTGGGAGGTTGTCG | Real Time RT PCR (ref. 13) |
| RTdevRC F5 | CGAGGATCCCTGTTGTCATGGTCCAT | Real Time RT PCR |
| RTdevR R | CGCGGCTTGCGTCCGACGTTC | Real Time RT PCR |
| RT devS F | TACTGACCGACCGGGATCGT | Real Time RT PCR (ref. 13) |
| RTdevS R | AGAGCCGCTGGATGACATGG | Real Time RT PCR (ref. 13) |
| RT1738 F | CGACGAACACGAAGGATTGA | Real Time RT PCR |
| RT1738 R | ACACCCACCAATTCCTTTTCC | Real Time RT PCR |
| RT2031c F | CGCACCGAGCAGAAGGA | Real Time RT PCR |
| RT2031c R | ACCGTGCGAACGAAGGAA | Real Time RT PCR |
| RTtgs1 F | CAGTGATTTGCGTCGCTACAG | Real Time RT PCR |
| RTtgs1 R | ACATCATTGATGGTGACGTCG | Real Time RT PCR |
| RT3131 F | CGATCAGGCCGATGTCGCCTT | Real Time RT PCR |
| RT3131 R | TCACCTCCTGGCACCGGCC | Real Time RT PCR |
| LH1 | CGAGTCGACAGAGCACGAAGGCTCGCCAGCGGAGG ACCTTTGGCCCTGCGTCGACCGA | Gel shift assays (P+S box) (ref. 22) |
| LH2 | TCGGTCGACGCAGGGCCAAAGGTCCTCCGCTGGCGA GCCTTCGTGCTCTGGTCGACTCG | Gel shift assays (P+S box) (ref. 22) |
| 3130F | TGGCTGCCGGGCCTTTCCCAT | DNase I footprinting (ref. 22) |
| 3131R | CATGGTCAGCGCCTTCCCCGG | DNase I footprinting (ref. 22) |
NdeI, XbaI, and BstBI restriction enzyme sites are underlined.
Construction of M. tb strains expressing DevRC
The gene sequences encoding the DevR C-terminal domain were amplified from H37Rv DNA by PCR. The amplified DNA was cloned into the integrative plasmid pJFR19 to generate pUS PAcet devRC. For DevRC expression from the native Rv3134c-devRS operon promoter, the operon promoter was excised from plasmid pSD POperon devR and cloned upstream of the DevRC-coding sequence to generate pUS POperon devRC. For expression from the hsp60 promoter, the operon promoter (in pUS POperon devRC) was replaced with the hsp60 promoter to generate plasmid pUS Phsp60 devRC, These integrating plasmids, namely, pUS POperon devRc, pUS PAcet devRC, and pUS Phsp60 devRc were individually electroporated into M. tb ΔdevR bacteria to generate Comp5, Comp6 and Comp7 strains, respectively (Table 2).
Western blotting
Frozen stocks of M. tb strains were revived in DTA medium, subcultured thrice and grown with vigorous shaking (120 ml in a 500-ml flask) in a shaker incubator at 220 rpm till ∼A 595 0.2-0.3, and subsequently processed for immunoblotting and RNA analysis (below). Briefly, a 20 ml aliquot was chilled on ice (‘aerobic’), centrifuged immediately at 5,000 g for 10 min at 4°C and the pellet was stored at −20°C. Sixty ml of the culture was distributed (10 ml aliquots in 50 ml tubes that were tightly closed) and kept standing for 1, 3 and 5 days (‘hypoxic’). The cells were harvested from dedicated culture tubes after appropriate incubation and whole cell lysates were prepared as described [24]. SigA protein was used as internal control. HspX and SigA proteins were detected in the lysates (containing ∼15 µg protein) by western blotting using polyclonal anti-HspX and anti-SigA antibodies as described earlier [25].
RNA isolation
The remaining culture (40 ml from above) was harvested and RNA was isolated. Briefly, a 20 ml aliquot was chilled on ice and centrifuged immediately as described above (‘aerobic’) and the remaining culture was kept standing for 5 days (‘hypoxic’). The harvested cell pellets were each resuspended in 1 ml of TRI reagent (Molecular Research Center, USA), and lysed in a mini bead beater using 0.1 mm zirconium/silica beads (Biospec, USA). RNA was purified as described earlier [20].
Reverse transcription (RT) and Real Time PCR
Two hundred nanograms of DNA-free RNA was reverse transcribed into cDNA using 50 U of Multi Scribe reverse transcriptase and random hexamer primers as per manufacturer's instructions (Applied Biosystems, USA). The cDNA was subjected to real time PCR using gene specific primers (Table 3) and Power SYBR Green PCR Master Mix in a MyiQ thermal cycler (Bio-Rad, USA). The primers were designed using the Primer3 program (http://workbench.sdsc.edu) and gene sequence data obtained from TubercuList (http://genolist.pasteur.fr/TubercuList). Reaction conditions were 94°C (10 min) followed by 40 cycles of 94°C (30 s), 56–65°C (45 s) and 72°C (30 s). A RT-negative (without Reverse Transcriptase) reaction was used to account for residual DNA if any and transcript numbers were normalized to that of 16S rRNA. The normalized copy number values were then used to determine the relative quantities (RQ) of individual gene transcripts. Three independent cultures were each analyzed in duplicate and the results are expressed as Mean ± SD.
EMSA and DNase I footprinting
The binding patterns of full-length DevR and DevRC proteins were compared in EMSA and DNase I footprinting experiments. EMSA assays were performed with purified DevR or DevRC protein and DNA fragments containing double-stranded oligonucleotides corresponding to the P+S binding sites located in the tgs1-Rv3131 intergenic promoter region. When used, full-length DevR was phosphorylated by incubating it with 50 mM acetyl phosphate for 20 min at 25°C in 40 mM Tris-Cl (pH 8.0) and 5 mM MgCl2. EMSA and DNase I footprinting analysis were carried out as described previously [22]. The sequences of the primers used in EMSA and DNase I footprinting are shown in Table 3.
GFP reporter assay
Aerobic GFP reporter assays were conducted in DTA medium as described previously [20]. The promoter activity is expressed in Relative Fluorescence Units (RFU)/OD595 of GFP as Mean values of RFU/OD ± standard deviation of three independent experiments, each in triplicate.
Results
Towards determining the role of the C-terminal domain of DevR in transcriptional regulation, we analyzed the in vitro binding property of its isolated C-terminal domain, DevRC. We also compared DevRC and full-length DevR proteins with respect to their ability to activate the transcription of target genes.
Isolated DevRC domain interacts with DNA but is deficient in cooperative interactions
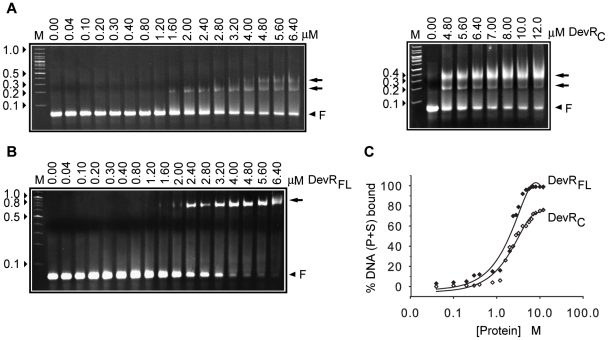
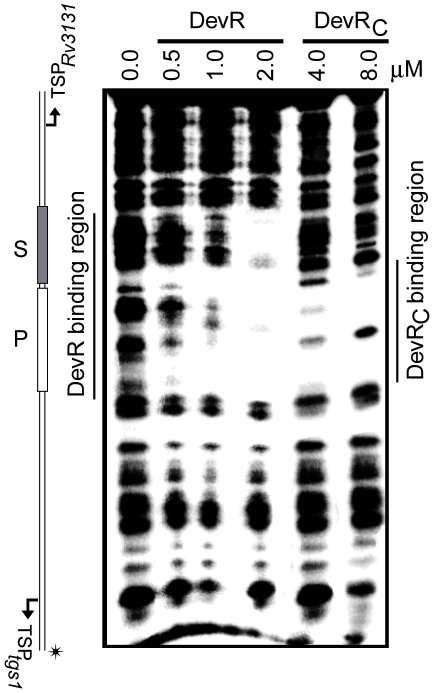
The tgs1-Rv3131 intergenic promoter region was used to assess DevRC interaction with DNA because these divergent promoters are regulated by DevR interaction with two binding sites, P and S [22]. At first, EMSA assays were carried out using double-stranded oligonucleotides containing P and S binding sites (called as P+S). The interaction between DevRC and P+S DNA generated two progressive DevRC-DNA complexes; first, a faster moving species (alongside ∼200 bp DNA marker), was observed and subsequently a slower migrating complex (alongside <400 bp DNA marker), also appeared at higher protein concentrations (Fig. 1A). This was significantly different from the interaction of full-length DevR which produced a single DNA-protein complex of low mobility that migrated alongside ∼700 bp DNA marker without an intermediate species, even at low protein concentrations, suggesting the interaction to be strongly cooperative (Fig. 1B). Another notable difference was that only partial saturation of DNA was observed even at very high concentration of DevRC (Fig. 1A, 1C). DNase I footprinting analysis of DevRC with the tgs1-Rv3131 intergenic region revealed that it binds to the primary site, but fails to cooperatively bind to the adjacent site, unlike full-length DevR which protected both the sites (Fig. 2). The underlying reason for obtaining two bound complexes with DevRC in EMSA is not well understood. Because the S site is not bound to DevRC, the appearance of the slower migrating species at higher protein concentration (>3 µM) is likely to be a result of interactions involving P-site bound DevRC species.
Figure 1. EMSA analysis.
Interaction of DevRC (A) and full-length DevR (B), with tgs1-Rv3131 promoter DNA. Double-stranded oligonucleotides having P+S box sequences belonging to the tgs1-Rv3131 divergent promoters were incubated with increasing concentrations of DevRC or DevR. Arrow, DNA-protein complex; F, free oligonucleotides, arrowheads indicate molecular weight markers in kilobase pairs (lane M), (C) Fraction of bound DNA (from Fig. 1A, B) plotted against protein concentration.
Figure 2. DevRC is defective in cooperative binding to DNA.
DNase I footprinting of DevRC or DevR on tgs1-Rv3131 intergenic DNA containing P and S binding sites. Bent arrows indicate the positions of the TSPs. 32P radiolabeled DNA strand is indicated by an asterisk.
DevRC feebly activates DevR regulon gene expression under aerobic conditions
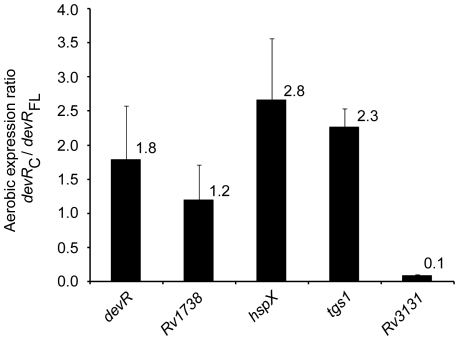
To address whether the DevR N-terminal domain has a regulatory role in suppressing DevR regulon gene expression under aerobic conditions, we asked whether DevRC could activate transcription in the absence of the inducing signal (i.e. under aerobic conditions). For this, we compared the relative quantities of tgs1, Rv3131 and selected DevR regulon transcripts in aerobic M. tb cultures of similar genetic background that produce DevRC (Comp5 strain) or full-length protein (Comp13 strain) from an identical chromosomal location. An ∼2-fold higher level of aerobic devR transcripts was estimated in Comp5 vs. Comp13 bacteria (Fig. 3), demonstrating that DevRC autoregulates transcription in aerobic cultures. This is noteworthy because in WT DevR-expressing cultures, autoregulation is dependent on DevR phosphorylation which occurs under hypoxic and not under aerobic conditions [20]. The expression of a target gene, tgs1, was also elevated > 2 fold in DevRC-expressing aerobic cultures (Fig. 3) and this aerobic overexpression was confirmed by GFP reporter assay using pTGS (mean aerobic GFP fluorescence ∼450 RFU/OD vs. ∼42 RFU/OD in the presence of WT DevR). The expression of various other DevR regulon genes was also induced, albeit modestly, in aerobic Comp5 bacteria (upto ∼3-fold, Fig. 3).
Figure 3. Aerobic expression of selected DevR target genes.
The relative aerobic transcript levels of selected genes were estimated by real time RT-PCR analysis in DevRC-expressing Comp 5 bacteria and expressed in relation to that in aerobic Comp13 cultures (expressing full-length DevR).
DevRC supports the aerobic expression of HspX
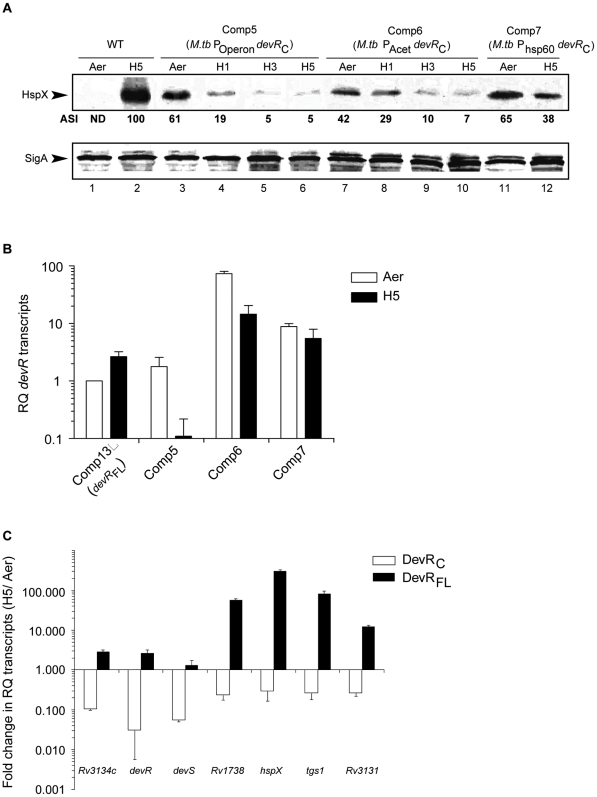
The ability of DevRC to mediate gene induction was confirmed at the protein level. HspX protein was detected in aerobic DevRC-expressing M. tb strains (Comp5), but not in aerobic cultures expressing full-length protein. Because HspX expression is DevR dependent, its expression implies the presence of an adequate amount of DevRC in aerobic Comp cultures (Fig. 4A). However, despite analyzing a large quantity of protein by immunoblotting (∼80 µg), DevRC was undetectable in Comp bacterial lysates (see Discussion). An artefactual increase in HspX expression during centrifugation of DevRC-expressing M. tb was ruled out by the absence of HspX expression in aerobic WT cultures that were processed in parallel. Moreover, activation by phosphorylation (ie. during hypoxia/centrifugation) is not relevant for DevRC because it lacks the phosphorylatable N-terminal domain.
Figure 4. DevR regulon expression in DevRC-expressing cultures declines during hypoxia.
(A) M. tb lysates (15 µg protein) were immunoblotted using rabbit anti-HspX or anti-SigA polyclonal sera and the blots were analyzed using Quantity One software (Bio-Rad, USA). The normalised intensities of the HspX-derived signals (with respect to those of SigA) are denoted as Arbitrary Signal Intensities (ASI) with respect to those obtained in 5 days hypoxic WT cultures (H5). ‘Aer’, aerobic; H1 H3 and H5 refer to 1, 3 and 5 days hypoxic cultures, respectively; ND, not detected. (B) Relative Quantity (RQ) of devRC transcripts in different Comp strains determined by real time RT-PCR analysis. (C) Real time RT-PCR analysis of DevR regulon transcripts. Fold change in the relative quantity of transcripts under ‘hypoxic’ vs. ‘aerobic’ conditions (fold decrease in Comp5 and fold increase in Comp13) is shown.
DevR regulon induction is not sustained by DevRC during hypoxia
Since DevR is physiologically relevant for regulon induction during hypoxia, the ability of DevRC to support hypoxic expression was examined next. The results of qRT-PCR and western blot analysis demonstrate that contrary to wild type DevR-expressing bacteria, gene induction is not sustained during hypoxia in DevRC-expressing Comp 5 bacteria (Fig. 4). An approximately 3- to 18-fold reduction in devR transcripts and regulon transcripts was observed in hypoxic Comp5 cultures in striking contrast to an ∼2- to 300-fold increase in Comp13 bacteria (expressing full-length DevR) under identical conditions (Fig. 4C). These results establish that DevRC-expressing bacteria have an autoregulation defect and an associated defect in regulon induction under hypoxia. The induction defect was also noted at the level of protein expression; HspX protein levels progressively decreased by >10-fold in Comp5 bacteria over 5 days in contrast to the sustained hypoxic induction noted in WT M. tb cultures (Fig. 4A, lanes 5 and 6). The decrease in HspX protein levels in Comp5 bacteria paralleled the decline in hspX transcripts on day 5 (Fig. 4C). As expected, SigA was constitutively expressed in all the strains under aerobic and hypoxic conditions.
Possible reasons for the decline in DevR regulon expression in hypoxic Comp5 cultures are that devRC transcripts are unstable or poorly expressed from the native promoter and therefore unable to maintain DevRC levels at a level adequate for autoregulation and target genes induction during hypoxia. To address these questions, two additional M. tb strains, Comp6 and Comp7, were constructed wherein DevRC is expressed from the constitutive acetamidase and hsp60 promoters, respectively, each with its own translational signals. Note that Comp6 and Comp7 are identical to Comp5 except for the promoter that is used to transcribe devRC. Transcription from the acetamidase and hsp60 promoters (in Comp6 and Comp7 bacteria) did enhance devRC transcript levels; the relative quantity (RQ) of devRC transcripts increased to ∼15 and ∼5, respectively, vs. <0.2 in Comp5 cultures (Fig. 4B). However, in spite of an increase in devRC transcripts during hypoxia, HspX levels were not sustained, particularly in Comp6 cultures (Fig. 4A, lanes 9 and 10), and the expression of other genes of the regulon also declined in these strains during hypoxia (data not shown). Therefore, we infer that DevRC is not stable at the protein level in the absence of DevRN in M. tb cultures. These results are in contrast to WT bacteria wherein DevR regulon products are induced and maintained during the 5-day hypoxia period. We conclude that in addition to the cooperativity defect, likely reasons for the failure of the hypoxic response are the selective instability of truncated DevRC protein and/or inability of DevRC to support its own transcription owing to missing of crucial interactions with the transcription machinery in Comp bacteria.
Discussion
Recently we showed by analyzing some target genes of the DevR regulon that robust induction depends on the binding of native phosphorylated DevR protein to two or more binding sites located in target promoters [20]–[22]. A DevRC-DNA complex was visualized by others from crystal structure analysis [18], and therefore we hypothesized that perhaps DevRC could support robust aerobic expression of DevR regulon genes. To address this possibility, we characterised the isolated C-terminal domain of DevR with respect to its DNA binding properties in vitro and its role in transcriptional activation in vivo. In the present study, expression analysis suggests that DevRC does indeed support aerobic gene expression, but only at a modest level. An analysis of the arrangement of Dev boxes at target promoters and the pattern of their occupancy provides insights into the underlying defect. We show that DevRC does bind to DNA but it is not recruited to the adjacent binding site at a target promoter unlike intact DevR protein. This difference in binding property is crucial because we know that complete occupancy of the binding sites is functionally important for full induction [22]. For example, DevRC does not bind to the S box in the tgs1-Rv3131 intergenic region and this defect is associated with the lack of Rv3131 aerobic expression. Taking together the results of previous and present findings, we attribute the poor aerobic induction of target genes, in fair measure, to the failure of DevRC to mediate cooperative interactions. The target promoters are characterized by an overlap of the TSP-proximal binding site with the -35 promoter element [20]–[22]. Therefore, another possible contributory factor is that interactions between DevRN and RNA polymerase are necessary for transcriptional activation and these are missing in DevRC-expressing bacteria. A consideration of all the results supports masking by DevRN of the intrinsic DNA binding activity of DevRC in the intact protein as a regulatory mechanism to prevent the aerobic induction of regulon genes.
We also compared the mechanism of DevR activation with that proposed for other response regulators, including those belonging to the NarL family. Many of the response regulators are placed in one of two classes with respect to the consequences of phosphorylation and mechanism of activation. In the first class, phosphorylation of the N-terminal domain activates the DNA binding activity of the protein by triggering its oligomerisation as in OmpR, ArcA and NtrC [26]–[30]. In the second class, the regulatory domain is believed to act negatively on the DNA binding function and phosphorylation is thought to relieve this inhibition by triggering a conformational change and/or inducing dimerization or oligomerization as in FixJ, PhoB, StyR, NarL and Spo0A [31]–[35]. Indeed, the isolated C-terminal domains of several response regulators, such as FixJ, PhoB, SsrB, Spo0A, and RhaS bind to DNA and activate transcription [31], [32], [36]–[40]. Since intact DevR binds to DNA only upon phosphorylation [20], and isolated DevRC exhibits DNA binding ability (this study), DevR resembles response regulators of the second class and uses domain separation as a key mechanism of activation. Our findings are substantiated by the proposal of domain rearrangement that was made from structural analysis [19]. However, relief of inhibition is not the only mechanism of activation in some response regulators. The isolated C-terminal domain of NarL, a close homologue of DevR, binds to DNA but does not activate transcription [35], implying a regulatory role for its N-terminal domain. NtrC from Salmonella typhimurium resembles DevRC in that its C-terminal domain is defective in cooperative interaction and its N-terminal domain is required for this function [29]. However, NtrC differs from DevR in that it binds to DNA as an unphosphorylated protein but its binding efficiency is enhanced by phosphorylation [29]. These comparisons highlight the rich diversity in the activation mechanisms employed by various response regulators, including those belonging to the same family. DevR is a unique example of a regulator that exploits an activation mechanism involving both relief of inhibition and cooperative binding to control gene induction. Importantly, both these functions reside in the N-terminal domain and/or linker region of DevR. As we have not examined it, we cannot rule out the effect of phosphorylation on the oligomerization status of DevR. Additionally, since DevR appears to interact with the transcriptional machinery to activate transcription, the role of the individual domains in these interactions remains to be elucidated.
The sequential binding of DevR to high affinity and low affinity sites may constitute a safety mechanism to tightly regulate induction and prevent regulon activation in the absence of the inducing signals. Since DevR plays a key role in M. tb dormancy it is considered to be a novel target for the development of drugs effective against dormant organisms [8], [41]. In principle, DevR-mediated signalling can be intercepted at any of the steps in the signalling cascade, including, signal sensing, DevS/DosT sensor kinase activation, transfer of the phosphosignal to DevR and binding of DevR to target DNA [42]. We recently provided a proof-of-concept for interfering with M. tb dormancy by inhibiting DevR activity through a small molecule [15]. Because the present study shows that cooperative binding is crucial for gene activation, hence blocking of cooperativity offers an additional step at which DevR can be effectively intercepted.
In conclusion, the major findings of this study are (i) the intrinsic DNA binding activity of DevRC and aerobic expression of the DevR regulon is masked by DevRN, (ii) DevRC fails to interact cooperatively with the binding sites at target promoters, and (iii) DevRC activates transcription of the DevR regulon genes under aerobic conditions, but only weakly, and the induction is not sustained during hypoxia. The binding property of DevRC is in striking contrast to intact phosphorylated DevR, which binds to two or more upstream binding sites and in a highly cooperative manner. From these findings we conclude that the determinant(s) of cooperativity are located outside of the C-terminal domain. These determinants are likely to fulfill a very important function in a genomic context wherein DevR binding sites may vary widely in their strengths; and cooperativity would play a key role in recruiting DevR to all the binding sites at target promoters. In addition to cooperativity, these determinants also provide other vital and essential functions that include autoregulation during hypoxia as well as imparting stability to DevR protein and providing surfaces for interacting with the transcriptional machinery. Thus, the activity and function of DevR is determined by both its C-terminal and N-terminal domains.
Acknowledgments
We sincerely thank Dr. Neil G. Stoker, Royal Veterinary College, London, UK, for the generous gift of M. tb ΔdevR (dosR) deletion mutant strain; Dr Malini Rajagopalan, University of Texas Health Center, USA, for the generous gift of the integrating plasmids, pJFR19 and pMG86, and Dr. U. Sharma, AstraZeneca, Bangalore, India, for Anti-SigA antibody.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant from the Department of Biotechnology, Government of India to JST. JST is thankful to the Department of Biotechnology, Government of India for a Tata Innovation Fellowship. USG and SC are thankful to the Council for Scientific and Industrial Research (CSIR) for Senior Research Associateship (Scientist's Pool Scheme) and Research Associateship, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Grange JM. The mystery of the mycobacterial ‘persistor’. Tuber Lung Dis. 1992:73249–73251. doi: 10.1016/0962-8479(92)90128-7. [DOI] [PubMed] [Google Scholar]
- 2.Stead WW. Pathogenesis of a first episode of chronic pulmonary tuberculosis in man: recrudescence of residuals of the primary infection or exogenous reinfection? Am Rev Respir Dis. 1967;95:729–745. doi: 10.1164/arrd.1967.95.5.729. [DOI] [PubMed] [Google Scholar]
- 3.Wayne LG, Sohaskey CD. Nonreplicating persistence of mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 4.Hoch JA, Silhavy TJ. American Society for Microbiology, Washington, D.C.: American Society for Microbiology Press; 1995. Two-component signal transduction. [Google Scholar]
- 5.Dasgupta N, Kapur V, Singh KK, Das TK, Sachdeva S, et al. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber Lung Dis. 2000;80:141–159. doi: 10.1054/tuld.2000.0240. [DOI] [PubMed] [Google Scholar]
- 6.Kumar A, Toledo JC, Patel RP, Lancaster JR, Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhotra V, Sharma D, Ramanathan VD, Shakila H, Saini DK, et al. Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol Lett. 2004;231:237–245. doi: 10.1016/S0378-1097(04)00002-3. [DOI] [PubMed] [Google Scholar]
- 8.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, et al. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci U S A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiloh MU, Manzanillo P, Cox JS. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe. 2008;3:323–330. doi: 10.1016/j.chom.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majumdar SD, Sharma D, Vashist A, Kaur K, Taneja NK, et al. Co-expression of DevR and DevR(N)-Aph proteins is associated with hypoxic adaptation defect and virulence attenuation of Mycobacterium tuberculosis. PLoS One. 2010;5:e9448. doi: 10.1371/journal.pone.0009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taneja NK, Dhingra S, Mittal A, Naresh M, Tyagi JS. Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C. PLoS One. 2010;5:e10860. doi: 10.1371/journal.pone.0010860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boon C, Dick T. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J Bacteriol. 2002;184:6760–6767. doi: 10.1128/JB.184.24.6760-6767.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta RK, Thakur TS, Desiraju GR, Tyagi JS. Structure-based design of DevR inhibitor active against nonreplicating Mycobacterium tuberculosis. J Med Chem. 2009;52:6324–6334. doi: 10.1021/jm900358q. [DOI] [PubMed] [Google Scholar]
- 16.Roberts DM, Liao RP, Wisedchaisri G, Hol WG, Sherman DR. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J Biol Chem. 2004;279:23082–23087. doi: 10.1074/jbc.M401230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini DK, Malhotra V, Dey D, Pant N, Das TK, et al. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology. 2004;150:865–875. doi: 10.1099/mic.0.26218-0. [DOI] [PubMed] [Google Scholar]
- 18.Wisedchaisri G, Wu M, Rice AE, Roberts DM, Sherman DR, et al. Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency. J Mol Biol. 2005;354:630–641. doi: 10.1016/j.jmb.2005.09.048. [DOI] [PubMed] [Google Scholar]
- 19.Wisedchaisri G, Wu M, Sherman DR, Hol WG. Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation. J Mol Biol. 2008;378:227–242. doi: 10.1016/j.jmb.2008.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauhan S, Tyagi JS. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J Bacteriol. 2008;190:4301–4312. doi: 10.1128/JB.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chauhan S, Tyagi JS. Interaction of DevR with multiple binding sites synergistically activates divergent transcription of narK2-Rv1738 genes in Mycobacterium tuberculosis. J Bacteriol. 2008;190:5394–5403. doi: 10.1128/JB.00488-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chauhan S, Tyagi JS. Powerful induction of divergent tgs1-Rv3131 genes in Mycobacterium tuberculosis is mediated by DevR interaction with a high-affinity site and an adjacent cryptic low-affinity site. J Bacteriol. 2009;191:6075–6081. doi: 10.1128/JB.00310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saini DK, Malhotra V, Tyagi JS. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 2004;565:75–80. doi: 10.1016/j.febslet.2004.02.092. [DOI] [PubMed] [Google Scholar]
- 24.Rodrigue S, Brodeur J, Jacques PE, Gervais AL, Brzezinski R, et al. Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. J Bacteriol. 2007;189:1505–1513. doi: 10.1128/JB.01371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagchi G, Chauhan S, Sharma D, Tyagi JS. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology. 2005;151:4045–4053. doi: 10.1099/mic.0.28333-0. [DOI] [PubMed] [Google Scholar]
- 26.Harlocker SL, Bergstrom L, Inouye M. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J Biol Chem. 1995;270:26849–26856. doi: 10.1074/jbc.270.45.26849. [DOI] [PubMed] [Google Scholar]
- 27.Jeon Y, Lee YS, Han JS, Kim JB, Hwang DS. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J Biol Chem. 2001;276:40873–40879. doi: 10.1074/jbc.M104855200. [DOI] [PubMed] [Google Scholar]
- 28.Nakashima K, Kanamaru K, Aiba H, Mizuno T. Signal transduction and osmoregulation in Escherichia coli. A novel type of mutation in the phosphorylation domain of the activator protein, OmpR, results in a defect in its phosphorylation-dependent DNA binding. J Biol Chem. 1991;266:10775–10780. [PubMed] [Google Scholar]
- 29.Porter SC, North AK, Wedel AB, Kustu S. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 30.Weiss V, Claverie-Martin F, Magasanik B. Phosphorylation of nitrogen regulator I of Escherichia coli induces strong cooperative binding to DNA essential for activation of transcription. Proc Natl Acad Sci U S A. 1992;89:5088–5092. doi: 10.1073/pnas.89.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Da Re S, Bertagnoli S, Fourment J, Reyrat JM, Kahn D. Intramolecular signal transduction within the FixJ transcriptional activator: in vitro evidence for the inhibitory effect of the phosphorylatable regulatory domain. Nucleic Acids Res. 1994;22:1555–1561. doi: 10.1093/nar/22.9.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellison DW, McCleary WR. The unphosphorylated receiver domain of PhoB silences the activity of its output domain. J Bacteriol. 2000;182:6592–6597. doi: 10.1128/jb.182.23.6592-6597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kahn D, Ditta G. Modular structure of FixJ: homology of the transcriptional activator domain with the -35 binding domain of sigma factors. Mol Microbiol. 1991;5:987–997. doi: 10.1111/j.1365-2958.1991.tb00774.x. [DOI] [PubMed] [Google Scholar]
- 34.Leoni L, Ascenzi P, Bocedi A, Rampioni G, Castellini L, et al. Styrene-catabolism regulation in Pseudomonas fluorescens ST: phosphorylation of StyR induces dimerization and cooperative DNA-binding. Biochem Biophys Res Commun. 2003;303:926–931. doi: 10.1016/s0006-291x(03)00450-9. [DOI] [PubMed] [Google Scholar]
- 35.Maris AE, Sawaya MR, Kaczor-Grzeskowiak M, Jarvis MR, Bearson SM, et al. Dimerization allows DNA target site recognition by the NarL response regulator. Nat Struct Biol. 2002;9:771–778. doi: 10.1038/nsb845. [DOI] [PubMed] [Google Scholar]
- 36.Feng X, Walthers D, Oropeza R, Kenney LJ. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol. 2004;54:823–835. doi: 10.1111/j.1365-2958.2004.04317.x. [DOI] [PubMed] [Google Scholar]
- 37.Galinier A, Garnerone AM, Reyrat JM, Kahn D, Batut J, et al. Phosphorylation of the Rhizobium meliloti FixJ protein induces its binding to a compound regulatory region at the fixK promoter. J Biol Chem. 1994;269:23784–23789. [PubMed] [Google Scholar]
- 38.Grimsley JK, Tjalkens RB, Strauch MA, Bird TH, Spiegelman GB, et al. Subunit composition and domain structure of the Spo0A sporulation transcription factor of Bacillus subtilis. J Biol Chem. 1994;269:16977–16982. [PubMed] [Google Scholar]
- 39.Ireton K, Rudner DZ, Siranosian KJ, Grossman AD. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 40.Wickstrum JR, Skredenske JM, Kolin A, Jin DJ, Fang J, et al. Transcription activation by the DNA-binding domain of the AraC family protein RhaS in the absence of its effector-binding domain. J Bacteriol. 2007;189:4984–4993. doi: 10.1128/JB.00530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy DJ, Brown JR. Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect Dis. 2007;7:84. doi: 10.1186/1471-2334-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saini DK, Tyagi JS. High-throughput microplate phosphorylation assays based on DevR-DevS/Rv2027c 2-component signal transduction pathway to screen for novel antitubercular compounds. J Biomol Screen. 2005;10:215–224. doi: 10.1177/1087057104272090. [DOI] [PubMed] [Google Scholar]
- 43.Chauhan A, Madiraju MV, Fol M, Lofton H, Maloney E, et al. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J Bacteriol. 2006;188:1856–1865. doi: 10.1128/JB.188.5.1856-1865.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valdivia RH, Hromockyj AE, Monack D, Ramakrishnan L, Falkow S. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene. 1996;173:47–52. doi: 10.1016/0378-1119(95)00706-7. [DOI] [PubMed] [Google Scholar]
- 45.Fol M, Chauhan A, Nair NK, Maloney E, Moomey M, et al. Jagannath C, Madiraju MV, Rajagopalan M: Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol Microbiol. 2006;60:643–657. doi: 10.1111/j.1365-2958.2006.05137.x. [DOI] [PubMed] [Google Scholar]
- 46.Parish T, Smith DA, Kendall S, Casali N, Bancroft GJ, et al. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect Immun. 2003;71:1134–1140. doi: 10.1128/IAI.71.3.1134-1140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]