Summary of Recent Advances
An estimated 100 trillion microbes colonize human beings, with the majority of organisms residing in the intestines. This microbiota impacts host nutrition, protection, and gut development. Alterations in microbiota composition are associated with susceptibility to various infectious and inflammatory gut diseases. The mucosal surface is not a static barrier that simply prevents microbial invasion but a critical interface for microbiota-immune system interactions. Recent work suggests that dynamic interactions between microbes and the host immune system at the mucosal surface inform immune responses both locally and systemically. This review focuses on intestinal microbiota-immune interactions leading to intestinal homeostasis, and evidence that these interactions at the GI mucosal surface are critical for driving both protective and pathological immune responses systemically.
Introduction
Mammalian mucosal surfaces are host to vast complex microbial ecosystems with distinct compositions dependent on body site [1]. The majority of colonizing bacteria are located in the lower GI tract, where they outnumber human host cells by 10-fold [2]. How do we live in the context of such an enormous microbial burden? In part, the relationship between a host and its microbiota is symbiotic. The host provides space and nutrients, while the microbiota contributes to host nutrition, physiology, and protection [3,4]. A recent metagenomic analysis of the human gut microbiome by the MetaHIT consortium revealed that the gene set obtained from the human microbiome is approximately 150 fold larger than that of a human [5]. This large gene pool likely provides benefit to the host. However, the interplay is complex, and beneficial coexistence comes via many layers of checks and balances.
In germ-free mice, the absence of intestinal microbial colonization leads to stunted epithelial maturation, accumulation of intestinal mucus, reduced expression of specific antimicrobial proteins, and immature development of the mucosal-associated lymphoid tissue associated with reduced populations of T cells, B cells, and sIgA [6]. Reconstitution of mucosal immune development can occur in response to monoassociation with specific bacteria [7,8]. The host responds to colonization by developing several layers of protection, which prevent microbial invasion and contain the microbiota at the mucosal surface.
The germ-free approach has been complemented by the use of genetic mouse models that have targeted specific mucosal barrier components, immune effectors, and cellular pathways involved in host innate and acquired immunity all resulting in disruptions in intestinal homeostasis, and frequently in colitis. Together, the findings from numerous studies highlight the combined importance of the innate and acquired immune systems to respond to the constant stream of changing nutrients, bacteria, and environmental antigens encountered by each individual, while limiting inflammation to avoid interference with normal intestinal function.
This review will focus on recent studies of microbiota-host mucosal immune interactions that contribute to our understanding of intestinal homeostasis, and discuss evidence that suggests that microbiota-host interactions in the gut drive both local and systemic beneficial and pathogenic immune responses. Thus, factors that modulate the biome, such as host genetics, nutrition, and antibiotics, may have a profound impact on host immune response, with far-reaching implications for human health and disease.
The intestinal microbiota
We have gained enormous insight into the composition and community structure of this complex ecosystem by using culture-independent molecular approaches, including the sequencing of 16S rRNA genes and high throughput metagenomic sequencing. The majority of bacterial species in the mammalian gut have been categorized into two phyla by 16S analysis, the gram-negative Bacteroidetes and gram-positive Firmicutes [9]. The dominant subclasses of Firmicutes in the human gut include Clostridiae, Erysipelotrychi, and Bacilli. Other phyla represented at lower abundance include Proteobacteria, Tenericutes, Verrucomicrobia, and Fusobacteria [9-11]. The MetaHIT Consortium recently published a metagenomic sequencing analysis of the human gut microbiome from fecal samples of 124 adult individuals [5**]. 99% of the genes identified were bacterial, and many of the bacterial genes and species were shared between individuals, although there was high interpersonal variability in species abundance. This study defined a minimal gut genome and its associated metabolic activity, and found gut specific gene expression, involving adhesion to host secreted proteins, and digestion of complex sugars such as those found in mucus. Because the microbiota is predominantly bacterial, this review will focus on bacterial-immune interactions.
The Innate Mucosal Barrier
The innate barrier is the primary site of interaction between the microbiota and the host. Defined anatomically by a single epithelial cell layer, a secondary physical barrier is formed by epithelial cell secretion of mucus. While mucus provides a substrate for microbial attachment and nutrition, it also prevents close contact between the microbiota and the epithelial surface. There are two mucus layers in the colon organized around the MUC2 mucin; an inner dense and firmly adherent layer that prevents bacterial penetration, and a looser outer layer where bacterial interaction occurs [12,13]. Absence of MUC2 results in disruption of the mucus layer organization, allowing bacterial contact with the epithelium and colitis [13-15]. In addition to forming a physical barrier and nutrition source for the microbiota, the mucus retains high concentrations of other protective effectors including secreted antimicrobial peptides (AMP) [16] and IgA [17], which localize to the mucus surface layer. Both AMPs and IgA are essential effectors in mediating microbiota-host interactions. Induction of IgA expression and secretion, and some AMP expression require host immune sensing of bacteria. Therefore, despite the physical barriers, the host detects and responds to commensal colonization.
How the host senses bacteria
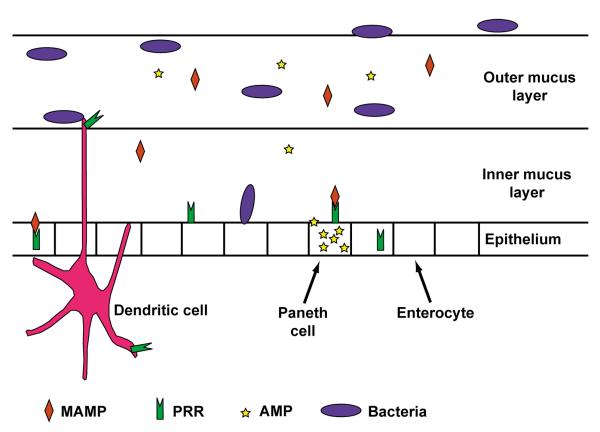
There are several pathways for commensal-immune interaction in a physically and immunologically intact host (Figure 1). Some bacterial species penetrate the mucus and directly contact the epithelial cell layer [18,19], while others are taken up by dendritic cells (DC) that extend dendrites into the intestinal lumen, or by sampling antigen ingested by M cells overlaying Peyer’s patches [20]. Recent work delineated specific subsets of lamina propria (LP) DCs, which respond differently to the intestinal microbiota [21], driving either inflammatory [22] or regulatory responses [21]. In addition to live bacteria, bacterial components such as lipopolysaccharide (LPS), flagellin, CPG DNA, and muramyl dipeptide (MDP) can be sensed by pattern recognition receptors such as Toll-like receptors (TLRs) found on epithelial and immune cell surfaces [23], or nuclear oligomerization domain 1 (NOD1) and 2 (NOD2) receptors found intracellularly. In most instances, signaling through these pattern recognition receptors drives local responses to microbial products, but new evidence highlights that systemic responses to biota-derived peptidoglycan leads to systemic modulation of neutrophil function [24**]. Furthermore, the host is able to sense short chain fatty acids such as butyrate and proprionate, produced as byproducts of bacterial fermentation by the microbiota to determine shifts in bacterial composition [25**,26] and respond.
Figure 1. Mechanisms of host-bacterial sensing at the intestinal mucosal surface.
The host uses several mechanisms to sense and respond to the microbiota. Anatomically, a single epithelial layer separated the host from the intestinal lumen contents, which include food, microbes and microbial antigens. The epithelium secretes a protective mucus layer. The dense inner layer excludes most bacteria, but specific species may penetrate this protection and directly contact the epithelium. Dendritic cells underlay the epithelial surface and can sample antigen delivered by M-cells (not shown), or extend their dendrites into the lumen for direct antigen sampling. Dendritic cells, Paneth cells, and enterocytes express pattern recognition receptors on their surfaces and intracellularly, to detect microbial associated molecular patterns such as LPS, MDP, and CpG DNA.
B cells and IgA-an ever shifting balance
Mucosal B cells necessary for induction of IgA responses are localized to mucosal lymphoid aggregates and Peyer’s patches. The microbiota stimulates mucosal isolated lymphoid follicle (ILF) development by signaling through the NOD1 and CCR6 receptors on intestinal epithelium. Maturation of ILFs into large B-cell clusters requires ensuing bacterial-TLR signaling [27]. Specific protective IgA is induced in mucosal B cells through interactions with intestinal dendritic cells bearing live commensals sampled from the intestinal lumen [28]. Induction occurs in mucosal Peyer’s patches, resulting in the generation of IgA secreting plasma cells that traffic to the mesenteric lymph nodes and ultimately home to the LP. In the LP plasma cells secrete IgA that is transcytosed into the intestinal lumen. IgA is concentrated in the intestinal mucus, where it binds to luminal bacteria and limits access to the epithelium. IgA acts to contain the microbiota, prevent bacterial translocation, and regulate microbiota composition [29,30]. Recently the use of a reversible germ-free colonization system in mice allowed close examination IgA induction and immune memory in response to commensal colonization. After repeated bacterial stimulation, mucosal IgA responses were additive, unlike the synergistic response seen with systemic challenge. Investigation of the IgA repertoire revealed the dynamics of this system. The repertoire represented the current dominant species in the gut, continuously replacing that induced by previous commensal populations [31*]. Thus, unlike the memory Ig responses induced systemically for neutralization and elimination of pathogens, the intestinal IgA response is constantly acclimatizing to the changing microbial environment of the gut.
AMPs and emerging paradigms for intestinal homeostasis
AMPs are at the front lines of interaction between host and commensals. They represent a mechanism of innate host defense conserved throughout the plant and animal kingdom [32]. These membrane active proteins directly kill bacteria, fungi, and enveloped viruses. The classes of AMP in mammals include defensins, cathelicidins, and C-type lectins. In humans, AMP are expressed and secreted by circulating immune cells and epithelial cells at all mucosal surfaces.
Paneth cells (PC), highly specialized secretory cells that inhabit the small intestinal crypts of Lieberkuhn, express and secrete the majority of AMPs and host defense proteins in the small intestine, including α-defensins, secretory phospholipase A2, α1antitrypsin, lysozyme, RegIIIγ, and angiogenins [33]. PCs derive from crypt progenitor cells and express lineage specific markers in response to Wnt signaling, including β-catenin, T-cell factor-4 (TCF-4) which mediate transcription of α-defensins [33]. Therefore, PC development and α-defensin expression and processing are independent of microbial colonization of the gut, although Nod2 expression has been associated with the regulation of specific PC α-defensins [34]. Several other PC effectors including the C-type lectins RegIIIγ, RegIIIβ, and RELMβ are induced via PC intrinsic TLR activation [35,36] in response to intestinal [37,38] and systemic [39] stimulation by bacteria and bacterial products such as LPS and flagellin.
The role of PC AMPs in intestinal homeostasis, and their ability to regulate the composition and abundance of the intestinal microbiota has been a subject of great interest, because of the association between PC abnormalities and Crohn’s disease, a chronic inflammatory bowel disease in humans. PC dysfunction has been associated with a variety of gene defects including Nod2 [40], Atg16l1deficiency [41], Xbp1 deficiency [42], deletion of protein disulfide isomerase, anterior gradient 2 (Agr2−/−)[43], and CD1d deficiency [44], many resulting in loss of intestinal homeostasis and colitis in mouse models. Several of these gene defects in humans have been associated with susceptibility to Crohn’s disease.
The role of PCs in intestinal homeostasis and microbiota regulation has been shown in several distinct mouse models. In one model, PC ablation resulted in increased bacterial translocation to mesenteric lymph nodes, providing support for the significance of PC AMPs in containing the microbiota within the gut [35]. The ability of bacteria to induce PC AMP that subsequently prevent bacterial translocation is one of the direct bacterial-host feedback loops involved in establishing intestinal homeostasis (Figure 2a). However, loss of PC’s did not alter total bacterial numbers in the small intestine [35]. Other evidence supporting the role of PC AMPs in homeostasis comes from studies of Nod2 deficient mice, which have reduced expression of a subset of PC α-defensins among other defects. Nod2−/− mice have abnormal bacterial growth in the terminal ileum [45], reduced crypt antimicrobial activity and severe Th1 dependent granulomatous response to Helicobacter hepaticus infection, which was corrected by PC specific α-defensin transgenic expression [46]. In mice deficient in CD1d, an MHC class I-like molecule, abnormalities of PC degranulation are noted, causing a presumptive deficiency in PC AMPs. As in other PC deficiency models, this resulted in increased bacterial translocation and small intestinal colonization in response to bacterial challenge, as well as alterations in ileal microbiota composition [44].
Figure 2. Bacteria-AMP feedback loops maintain intestinal homeostasis.
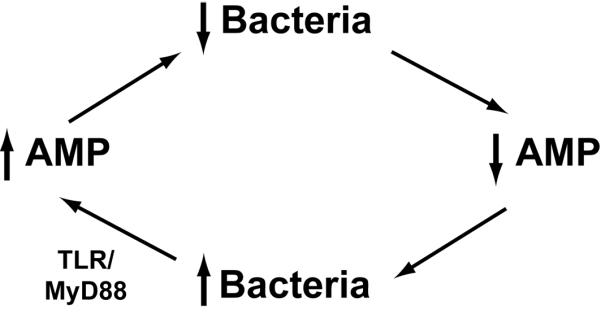
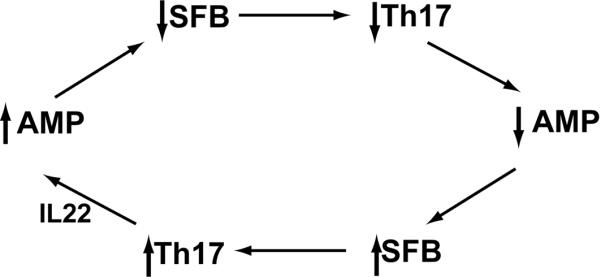
Several bacterial-host feedback loops function at mucosal surfaces to maintain homeostasis. A. Bacterial colonization of the intestinal tract stimulates pattern recognition receptors on intestinal epithelial cells, including Paneth cells. Stimulation of MyD88 dependent TLR pathways results in induction of epithelial AMPs, which then act to prevent bacterial translocation. B. Paneth cell AMPs regulate SFB (and other bacterial species) abundance. SFB colonization triggers DC-dependent Th17 differentiation. IL22, a Th17 cytokine, acts on intestinal epithelial cells and Paneth cells to increase expression of AMPs. AMPs then act to modulate abundance of SFB. Imbalance in this regulatory loop can skew the regulatory/inflammatory balance, resulting in loss of homeostasis, and increased pro-inflammatory responsiveness both locally and systemically.
A specific examination of the role of PC α-defensins in regulating the microbiota was done, using complementary models of defensin-deficiency and defensin excess. Alteration of PC defensin abundance or composition did not change total bacterial numbers [47*] in the intestine, confirming the findings of the PC ablation study. However, alteration of PC defensin composition resulted in significant shifts in small intestinal bacterial colonization. In addition, defensin excess by the transgenic addition of one human PC defensin, DEFA5, resulted in the loss of one specific bacterial group that is found in direct contact with the intestinal epithelial cells, segmented filamentous bacteria (SFB). Interestingly, the Cd1d deficient mice, presumably deficient in PC AMPs, showed an increased abundance of ileal SFB, supporting a role for PC defensins in controlling abundance of this organism [44].
SFB, formally known as Candidatus arthromitis, is an uncultivable spore-forming member of the Firmicutes phylum, Clostridiales class. SFB has been identified in the intestinal tracts of birds, mice, and rabbits, but is not commonly found in humans [48]. It directly contacts the small intestinal epithelium, and can drive complete maturation of host mucosal immune responses [8**,18**].
Commensal bacteria induce epithelial AMPs through induction of mucosal Th17 T cells
SFB has been specifically implicated in inducing mucosal T cell responses. DEFA5 transgenic mice, secondary to their loss of SFB, showed skewed mucosal T cell development, lacking LP Th17 differentiation [47]. Germ-free mice lack LP Th17 T cell development, but reconstitution with SFB alone is capable of inducing Th17 differentiation [8,18], while reconstitution with a complex microbiota lacking SFB, or human microbiota does not [8]. Monoassociation of germ-free mice with SFB stimulates expression of the acute phase reactant serum amyloid A, which induces Th17 differentiation via DC activation [18]. Th17 cells secrete IL17 and IL22, which in turn are responsible for stimulating expression of epithelial AMPs, specifically RegIIIγ [8,18,39,49]. RegIIIγ, as previously noted, can also be induced directly in the intestinal lumen via bacteria-TLR interaction. Its induction via Th17 cytokines can be achieved by luminal triggers, as with SFB [8,18], or by systemic delivery of flagellin [39]. In the intestine, RegIIIγ provides resistance to enteric infection and colonization with organisms such as Citrobacter rodentium [18] and Enterococcus faecalis [38]. This provides an example of a second essential AMP feedback loop, critical for maintaining intestinal homeostasis (Figure 2b).
LP Th17 cells are not the only source of IL17 and IL22 in the intestinal tract. Intraepithelial γδ T cells also express IL17, in response to stimulation by the microbiota [50,51]. In addition, a newly identified population of mucosal IL22 producing cells (RORgt+Nkp46+) has been described, whose differentiation is dependent on interaction with the microbiota [52-54]. IL22 secretion by this cell population also induces epithelial AMP expression [52] and enhances resistance to intestinal infection by Citrobacter rodentium [53].
Commensal bacteria may either induce or suppress mucosal Treg cell differentiation and response
Commensal bacteria also induce regulatory T cells, essential for limiting inflammatory immune responses at mucosal surfaces. The Treg populations that are prominent in the GI tract LP are CD4+FOXP3+ Treg cells and CD4+FOXP-IL10+ Treg cells. The critical role of these cells in host-microbiota homeostasis was made evident by the increased susceptibility to colitis in response to intestinal microbial colonization in animals lacking these functional T cell subclasses [55]. Specific bacterial species, such as Bifidobacterium infantis have been shown to increase the numbers of FOXP3+ Treg cells, and control intestinal inflammation induced by enteric Salmonella infection [56]. Suppression of Th17 production [57*] and induction of Foxp3+ Treg cells [58*] can be accomplished by a single bacterial molecule of a common intestinal commensal, the polysaccharide A (PSA) of Bacteroides fragilis. PSA acts through TLR2 to induce IL10 producing Tregs, and is able to prevent inflammation associated with experimental infectious and inflammatory colitis [57,58]. Faecalibacterium prausnitzii, a member of the Firmicutes phylum and a component of the human intestinal microbiota, also has immunomodulatory activity, inducing IL10 secretion and reducing colitis severity in mouse models. Reduced abundance of this bacterium in patients with Crohn’s disease is associated with greater risk of post-surgical ileal disease recurrence, again showing the ability of the commensal microbiota to enhance regulatory immune responses and reduce inflammation [59].
Conversely, CpG DNA from the microbiota may act through engagement of the TLR9 receptor, to limit Treg differentiation, and enhance effector T cell numbers. In the absence of Tlr9 or reduction of the microbiota by antibiotics, mice showed reduced immune response to oral vaccination or oral infection [60]. Therefore, the ability of the microbiota to control regulatory/effector intestinal immune responses involves a careful balancing act, and how that is achieved is not yet understood.
Implications of homeostatic dysregulation and disease susceptibility
Maintaining a balance between pro and anti-inflammatory processes is essential for normal physiologic function of the GI tract. The specific composition of the microbiota is critical in establishing and maintaining equilibrium at the mucosal surface. In mouse models, disruption of intestinal microbial-immune homeostasis often results in colitis. Interestingly, several of the genetic defects and deficiencies associated with Crohn’s disease, a chronic inflammatory bowel disease in humans, result in disruption of PC function and loss of PC defensins and other effectors, now identified as essential regulators of intestinal microbial colonization. This supports the critical role of the AMP-microbiota-T cell-AMP feedback loop (Figure 2b) in maintaining intestinal homeostasis. However, the importance of intestinal homeostasis extends beyond the GI tract. The microbiota influences systemic innate immune response, by providing a source for peptidoglycan, which primes systemic neutrophil responses via Nod1 signaling [24]. In the face of microbiota depletion, neutrophils are impaired in their ability to kill pathogenic bacteria. Neutrophil activity is restored by administration of Nod1 ligands.
A role for the intestinal microbiota has been suggested in both protection from and provocation of extraintestinal infectious and autoimmune diseases [61-64]. Studies of non-obese diabetic mice demonstrated the critical importance of microbiota-TLR interactions in the incidence of spontaneous development of diabetes in these mice, emphasizing the protective nature of the correct microbial composition in preventing T cell mediated pancreatic islet destruction and diabetes [63] in susceptible hosts. Experimental models for autoimmune arthritis and experimental autoimmune encephalitis (EAE) have confirmed the importance of two specific bacterial species previously identified in regulation of T cell responses. SFB has been shown to drive both autoimmune arthritis [62*] and EAE [61*] through Th17 mechanisms, while both disease processes are attenuated in germ-free animals. Conversely, B. fragilis colonization enhances Foxp3+ Treg induction and protects against the development of EAE [64,65], dependent on PSA expression.
It is evident that the factors that control the biome have profound and far-reaching effects on the immune system. Intestinal effector molecules such as AMPs and IgA, and environmental exposures that modify intestinal bacterial composition, including antibiotic use, diet, and probiotics, are likely capable of modulating both intestinal and systemic immunity. It is likely that the interplay of these factors may underlie, in part, the pathogenesis of a multitude of metabolic, inflammatory, and immune disorders. As but one example, the dramatic increases in autoimmune disease in Westernized countries over the last 50+ years parallels widespread antibiotic use and adoption of the highly processed Western diet, both capable of causing pervasive changes in the microbiota. Continued study of microbiota composition and immune interaction at the intestinal mucosal surface should reveal a greater understanding of essential molecular mechanisms of homeostatic balance, allowing opportunity for both prevention and treatment of mucosal and systemic disease.
Acknowledgments
The author thanks Dr. Calvin Williams and Dr. Charles Bevins for critical reading of this manuscript. The work in Dr. Salzman’s laboratory is supported by NIH grant AI057757.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Reports. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooper LV, Midtvedt T, Gordon JI. How Host-Microbial Interactions Shape the Nutrient Environment of the Mammalian Intestine. Annu. Rev. Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- **5.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. Metagenomic deep sequencing of fecal samples from 124 individuals was used to generate a human intestinal microbial gene catalogue and define a minimal functional gut genome.
- 6.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- **8.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. Along with [18], the authors demonstrated that colonization with a single bacterial species, SFB, drove differentiation of Th17 cells in the intestinal lamina propria, inducing intestinal epithelial AMP expression.
- 9.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Reviews. Microbiology. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. USA. 2010 doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Buller HA, Dekker J, Van Seuningen I, Renes IB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 15.Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, Thornton DJ, Png CW, Crockford TL, Cornall RJ, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Medicine / Public Library of Science. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, Andersson M. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764–771. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- 17.Macpherson AJ, Slack E. The functional interactions of commensal bacteria with intestinal secretory IgA. Current Opinion in Gastroenterology. 2007;23:673–678. doi: 10.1097/MOG.0b013e3282f0d012. [DOI] [PubMed] [Google Scholar]
- **18.Ivanov, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. Along with [8] authors showed that SFB induced intestinal mucosal proinflammatory T cell responses, resulting in induction of epithelial AMPs and resistance to enteric Citrobacter rodentium infection.
- 19.Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host & Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strober W. The multifaceted influence of the mucosal microflora on mucosal dendritic cell responses. Immunity. 2009;31:377–388. doi: 10.1016/j.immuni.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, Luche H, Fehling HJ, Hardt WD, Shakhar G, Jung S. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Niess JH, Adler G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. Journal of Immunology. 2010;184:2026–2037. doi: 10.4049/jimmunol.0901936. [DOI] [PubMed] [Google Scholar]
- 23.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nature Reviews. Immunology. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- **24.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature Medicine. 2010;16:228–231. doi: 10.1038/nm.2087. This study showed that intestinal microbiota-derived peptidoglycan is responsible for priming systemic neutrophil response and enhances the ability of neutrophils to kill pathogenic bacteria.
- **25.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. The authors identify a pathway by which microbiota-derived short chain fatty acids modulate intestinal inflammation by binding and signaling through the GPR43 receptor. This study shows a novel mechanism by which microbiota metabolism in conjunction with diet may regulate mucosal immune response.
- 26.Singh N, Thangaraju M, Prasad PD, Martin PM, Lambert NA, Boettger T, Offermanns S, Ganapathy V. Blockade of dendritic cell development by bacterial fermentation products butyrate and proprionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem. 2010;285:27601–27608. doi: 10.1074/jbc.M110.102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 28.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host & Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- *31.Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. This study describes a novel system of reversible bacterial colonization of mice, used to investigate the details of IgA priming, response, and memory.
- 32.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 33.Ouellette AJ. Paneth cells and innate mucosal immunity. Curr Opin Gastroenterol. 2010 doi: 10.1097/MOG.0b013e32833dccde. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 35.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. Journal of Experimental Medicine. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. Journal of Infectious Diseases. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lesage S, Zouali H, Cezard JP, Colombel JF, Belaiche J, Almer S, Tysk C, O’Morain C, Gassull M, Binder V, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cadwell K, Liu JY, Brown SL, Miyoshi H, J. L, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16L1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [see comment] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao F, Edwards R, Dizon D, Afrasiabi K, Mastroianni JR, Geyfman M, Ouellette AJ, Andersen B, Lipkin SM. Disruption of Paneth and goblet cell homeostasis and increased endoplasmic reticulum stress in Agr2−/− mice. Developmental Biology. 2010;338:270–279. doi: 10.1016/j.ydbio.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nieuwenhuis EE, Matsumoto T, Lindenbergh D, Willemsen R, Kaser A, Simons-Oosterhuis Y, Brugman S, Yamaguchi K, Ishikawa H, Aiba Y, et al. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. Journal of Clinical Investigation. 2009;119:1241–1250. doi: 10.1172/JCI36509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS. Nod2 is required for the regulation of commensal microbiota in the intestine. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biswas A, Liu YJ, Mizoguchi A, Salzman NH, Bevins CL, Kobayashi KS. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc. Natl. Acad. Sci. USA. 2010;107:14739–14744. doi: 10.1073/pnas.1003363107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–82. doi: 10.1038/ni.1825. This study demonstrates that Paneth cell α-defensins modulate the composition of the small intestinal microbiota, which subsequently regulates mucosal T cell differentiation (as shown in [8,18].
- 48.Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, Collins MD. Comparison of 16S rRNA Sequences of Segmented Filamentous Bacteria Isolated from Mice, Rats, and Chickens and Proposal of “Candidatus Arthromitus”. Int J Syst Bacteriol. 1995;45:780–782. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- 49.Ismail AS, Behrendt CL, Hooper LV. Reciprocal interactions between commensal bacteria and gamma delta intraepithelial lymphocytes during mucosal injury. Journal of Immunology. 2009;182:3047–3054. doi: 10.4049/jimmunol.0802705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duan J, Chung H, Troy E, Kasper DL. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host & Microbe. 2010;7:140–150. doi: 10.1016/j.chom.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31:321–330. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 52.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nature Immunology. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, Mills JC, Colonna M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vivier E, Spits H, Cupedo T. Interleukin-22-producing innate immune cells: new players in mucosal immunity and tissue repair? Nature Reviews. Immunology. 2009;9:229–234. doi: 10.1038/nri2522. [DOI] [PubMed] [Google Scholar]
- 55.Chung H, Kasper DL. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr Opin Immunol. 2010;22:455–460. doi: 10.1016/j.coi.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 56.O’Mahony C, Scully P, O’Mahony D, Murphy S, O’Brien F, Lyons A, Sherlock G, MacSharry J, Kiely B, Shanahan F, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathogens. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *57.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- *58.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. These references show that a single bacterial polysaccharide from a common intestinal commensal is able to induce lamina propria Treg development and ameliorate colitis.
- 59.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *61.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA. 2010 doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *62.Wu HJ, Ivanov, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. These references demonstrate that lamina propria Th17 development triggered by intestinal SFB colonization drives systemic autoimmune diseases. These studies demonstrate the far-reaching impact of colonization by specific intestinal microbes on systemic immunity.
- 63.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, Kasper LH. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. Journal of Immunology. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- 65.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010 doi: 10.4049/jimmunol.1001443. in press. [DOI] [PubMed] [Google Scholar]