Abstract
In the peripheral nervous system, target tissues control the final size of innervating neuronal populations by producing limited amounts of survival-promoting neurotrophic factors during development. However, it remains largely unknown if the same principle works to regulate the size of neuronal populations in the developing brain. Here we show that neurotrophin signaling mediated by the TrkB receptor controls striatal size by promoting the survival of developing medium-sized spiny neurons (MSNs). Selective deletion of the gene for the TrkB receptor in striatal progenitors, using the Dlx5/6-Cre transgene, led to a hindpaw-clasping phenotype and a 50% loss of MSNs without affecting striatal interneurons. This loss resulted mainly from increased apoptosis of newborn MSNs within their birthplace, the lateral ganglionic eminence. Among MSNs, those expressing the dopamine receptor D2 (DRD2) were most affected, as indicated by a drastic loss of these neurons and specific down-regulation of the DRD2 and enkephalin. This specific phenotype of mutant animals is likely due to preferential TrkB expression in DRD2 MSNs. These findings suggest that neurotrophins can control the size of neuronal populations in the brain by promoting the survival of newborn neurons before they migrate to their final destinations.
Keywords: brain-derived neurotrophic factor, cell death, striatum, dopamine receptor D1a
A neural circuit consists of several connected nodes that are composed of neurons or nonneuronal cells, and its proper function requires a correct number of cells at each node. Thus, it is important to understand the mechanism by which the size of neuronal populations is determined during development. In the peripheral nervous system (PNS), it has been well documented that developing neurons have to compete for a limited amount of neurotrophic factors produced by their target tissues, and that neurons unable to obtain sufficient amounts of trophic factors die through programmed cell death (1). In this way a peripheral target can control the final size of the innervating neuronal populations through neurotrophic factors. One important family of neurotrophic factors is called neurotrophins, which include nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin 3 (NT3), and neurotrophin 4/5 (NT4/5). Neurotrophins exert many biological effects by binding and activating specific Trk receptor tyrosine kinases: NGF activates TrkA; BDNF and NT4/5 activate TrkB; and NT3 activates TrkC (2). Deletion of neurotrophin genes causes a severe loss of sensory and sympathetic neurons (3). Although ablation of neurotrophic signaling increases programmed cell death in hippocampal dentate gyrus and cerebellar granular layer during the first two postnatal weeks (4, 5), no studies have identified a large population of developing brain neurons that are dependent on a single neurotrophic factor for survival. Therefore, it remains unknown whether neurotrophic factors can coordinate the size of two connected brain regions in the developing brain.
The striatum is the largest component of the basal ganglia. It is responsible for movement control and is associated with addictive behaviors. Its dysfunction is the main cause for the motor disorders associated with Huntington disease (HD) and Parkinson disease (6, 7). Approximately 95% of striatal neurons are medium-sized spiny neurons (MSNs) with the rest being interneurons (8). MSNs, which use γ-aminobutyric acid (GABA) as a transmitter, are born in the lateral ganglionic eminence (LGE) and migrate to the striatum during embryogenesis (9). They are divided into two populations based on their projection sites. MSNs at the origin of the direct pathway directly project to the output nuclei of the basal ganglia, such as the internal segment of the globus pallidus, the substantia nigra pars reticulata, and the ventral pallidum. The other population of MSNs sits at the origin of the indirect pathway and projects to the output nuclei of the basal ganglia via the external segment of the globus pallidus and the subthalamic nucleus (10). Interestingly, MSNs of the two pathways differ in the expression of peptide transmitters and dopamine receptors, such that neurons of the direct pathway express substance P (SP) and the dopamine receptor D1a (DRD1a), whereas neurons of the indirect pathway express enkephalin (Enk) and the dopamine receptor D2 (DRD2) (8). Recent studies using bacterial artificial chromosome (BAC) transgenic mice expressing fluorescent proteins under the control of the promoter for either DRD1a or DRD2 confirm these distinct coexpression patterns in MSNs of the two pathways (11, 12).
In this study we examined the role of TrkB signaling in striatal development by using mouse mutants in which TrkB signaling was abolished in the LGE during early embryogenesis. We found that in these mutant mice the striatal volume was halved and the majority of newly born neurons destined to be DRD2-expressing MSNs died before they could migrate from the LGE to the striatum. These findings demonstrate a critical role for TrkB signaling in striatal development.
Results
Generation of a Striatum-Specific TrkB Mutant.
To selectively delete the TrkB gene in striatal neurons, we used a Cre transgene under the control of the Dlx5/6 enhancer element (Dlx5/6-Cre) (13). At embryonic day 12.5 (E12.5), Dlx5 and Dlx6 are expressed throughout the lateral and medial ganglionic eminences (LGE and MGE) (14), the brain regions that give rise to striatal projection neurons and interneurons, respectively (15–17). To determine in which populations of striatal neurons the Dlx5/6-Cre transgene is expressed, we introduced this transgene into Rosa26 reporter mice, in which the Rosa26 locus expresses β-galactosidase once Cre-mediated recombination has occurred. We found that in the striatum the Dlx5/6-Cre transgene mediated recombination in the vast majority of MSNs expressing the 32-kDa dopamine- and cAMP-regulated phosphoprotein (DARPP-32; Fig. 1A) as well as in interneurons expressing neuropeptide Y (NPY), choline acetyltransferase (ChAT), or parvalbumin (Fig. S1). Dlx5/6-Cre mice were crossed to mice harboring a floxed TrkB allele termed fB (Fig. S2) to generate conditional TrkB knockouts (Dlx5/6-Cre;fB/fB, termed TrkBDlx) or control mice (fB/fB). To determine if TrkB expression was abolished in the striatum of TrkBDlx mice, we analyzed protein extracts prepared from striatal and cortical tissues by immunoblotting. Compared with control mice, levels of both TrkB-F and TrkB-T in the cerebral cortex were slightly reduced in TrkBDlx mice (Fig. 1B), which is consistent with the observation that Dlx5/6-Cre was also expressed in cortical interneurons (13). However, TrkB-F expression in the striatum appeared to be completely abolished in TrkBDlx mice (Fig. 1B). We were still able to detect a small amount of TrkB-T (Fig. 1B), likely due to expression of TrkB-T in glial cells. Thus, we generated a TrkB mutant that lacks TrkB signaling in striatal neurons.
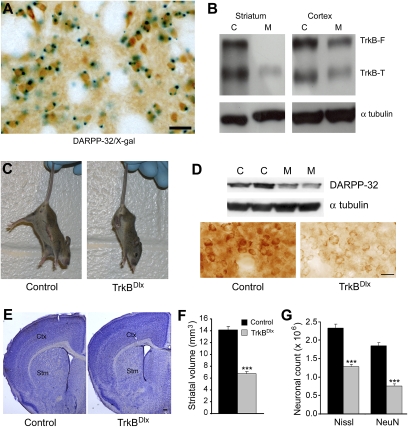
Fig. 1.
Deletion of the TrkB gene in the striatum leads to a large loss of striatal neurons. (A) Dlx5/6-Cre-mediated recombination pattern in the striatum, as revealed by X-gal staining for β-galactosidase (blue) and DARPP-32 immunohistochemistry (brown) in Rosa26/+;Dlx5/6-Cre mice. (Scale bar, 50 μm.) (B) Western blot shows the complete absence of the full-length TrkB receptor (TrkB-F) and a large reduction in the truncated TrkB receptor (TrkB-T) in the striatum of a mutant mouse. Protein extracts were prepared from striata and cortices of Dlx5/6-Cre-mediated TrkB conditional mutant (M) and fB/fB control (C) mice at 3 wk of age and probed with antibodies against the TrkB extracellular domain and α-tubulin. (C) The TrkB mutant (TrkBDlx) at P18 exhibited abnormal hindpaw-clasping phenotype when suspended by its tail. (D) DARPP-32 levels in control (C) and TrkBDlx (M) mice, as revealed by Western blot and immunohistochemistry. (Scale bar, 25 μm.) (E) Representative images of Nissl-stained coronal brain sections from control and TrkBDlx mice. Stm, striatum; Ctx, cerebral cortex. (Scale bar, 500 μm.) (F) Striatal volumes of TrkBDlx and control mice at P21. Note the striatal volume in TrkBDlx mice was reduced by 53% compared with control mice (n = 4 mice each). Error bars represent SEM. Student's t test: ***P < 0.001. (G) Counts of striatal neurons in TrkBDlx and control mice at P21, obtained from Nissl- or NeuN-stained sections. The striatal neuron counts were reduced by 45% (Nissl) and by 58% (NeuN) compared with control mice (n = 4 mice each). Error bars represent SEM. Student's t test: ***P < 0.001.
Deletion of TrkB Leads to a Large Neuronal Loss in the Striatum at Postnatal Day 21.
TrkBDlx mice survived for about 3 wk and displayed a hindpaw-clasping phenotype when suspended by their tails (Fig. 1C). Hindpaw clasping has been associated with motor dysfunction observed in a number of mouse models with neurological disorders (18, 19). To determine whether this abnormal behavior corresponded to biochemical changes occurring in the striatum of the TrkBDlx mice, we measured levels of DARPP-32, which is expressed by all MSNs and is important for proper striatal function (20). There was a large reduction in DARPP-32 levels in TrkBDlx mice compared with control littermates (Fig. 1D). Using an unbiased stereological method to measure striatal volume and count striatal neurons on Nissl-stained coronal sections, we then determined if loss of striatal neurons also contributed to motor dysfunction in TrkBDlx mice (Fig. 1E). The volume and neuronal number of the striatum were reduced by ∼50% in TrkBDlx mice compared with control animals (Fig. 1 F and G). In addition, a specific neuronal marker (NeuN) was used to make sure we did not miss Nissl-stained neurons that might be smaller in size due to cell atrophy in TrkBDlx mice. A similar reduction in NeuN-positive cell counts confirmed the large striatal neuronal loss in TrkBDlx mice (Fig. 1G). Taken together, these results indicate that TrkB signaling is necessary for striatal formation and/or maintenance.
Differential Effect of the TrkB Deletion on DRD2- and Enkephalin-Expressing MSNs Is Due to Preferential Expression of TrkB in These Neurons.
To determine whether TrkB deletion has a differential effect on the two distinct populations of striatal projection neurons, we examined expression of SP and Enk, the markers for MSNs in the direct and indirect pathways, respectively. In TrkBDlx mice there was a marked decrease in Enk immunoreactivity in the external segment of the globus pallidus (Fig. 2A), a projection site of MSNs in the indirect pathway, whereas SP levels were not apparently changed in the substantia nigra pars reticulata (Fig. 2B), a projection site of MSNs in the direct pathway. It was not clear, however, whether the difference in peptide immunoreactivity reflected selective changes in expression of individual genes or in the number of MSNs in each of the two pathways. To address this issue, we measured levels of mRNAs for DRD1a, DRD2, Enk, and SP in the striatum using in situ hybridization. Levels of mRNAs for DRD2 and Enk, expressed by the MSNs in the indirect pathway, were greatly reduced in the striatum of TrkBDlx mice (Fig. 2 D and E). In contrast, levels of mRNAs for SP and DRD1a, expressed by the MSNs in the direct pathway, were not significantly changed (Fig. 2 D and E). Using immunohistochemistry , we further confirmed that DRD1a expression in striatal neurons of TrkBDlx mice was not significantly altered (Fig. 2C). These findings show that TrkB ablation mostly affects MSNs of the indirect pathway.
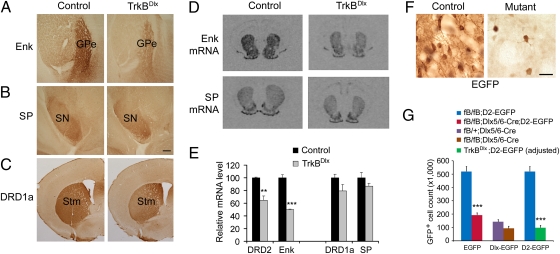
Fig. 2.
Differential effects of TrkB deletion on DRD2- and enkephalin-expressing MSNs at P21. (A) Reduced Enk immunoreactivity in the external segment of the globus pallidus (GPe) of TrkBDlx mice. (B) Normal SP immunoreactivity in the substantia nigra (SN) of TrkBDlx mice. (Scale bar, 250 μm.) (C) Normal expression of DRD1a in the striatum (Stm) of TrkBDlx mice. (D) In situ hybridization showing reduced Enk mRNA levels but normal SP mRNA levels in the striatum of TrkBDlx mice. (E) Quantification of in situ hybridization signals for Drd2, Enk, Drd1a, and SP mRNAs. Note that levels of mRNAs for DRD2 and Enk but not for DRD1a and SP were significantly reduced in TrkBDlx mice (n = 3 mice each). Error bars represent SEM. Student's t test: **P < 0.01; ***P < 0.001. (F) Representative high-magnification images showing fewer EGFP-expressing striatal neurons in fB/fB;Dlx5/6-Cre;D2-EGFP mice (mutant) compared with fB/fB;D2-EGFP mice (control). (Scale bar, 25 μm.) (G) Counts of EGFP-expressing neurons in the striatum. EGFP-expressing striatal neurons were counted in several genotypes of mice to reveal a severe loss of DRD2-expressing MSNs in TrkB mutant mice (n = 4 mice for each genotype). EGFP could be expressed from the Dlx5/6-Cre transgene (Dlx-EGFP) and/or the D2-EGFP transgene. Error bars represent SEM. Student's t test: ***P < 0.001.
The reduced levels of mRNAs for Enk and DRD2 in the TrkB mutant might result from a selective loss of the MSNs in the indirect pathway. To investigate the effect of TrkB deletion on the survival of DRD2-expressing MSNs, we introduced a BAC transgene expressing green fluorescent protein under the control of the Drd2 promoter (D2-EGFP) (21) into TrkBDlx mice and control mice. High-magnification examination of EGFP+ cells demonstrated that the reduced level of Drd2 mRNA in the striatum of TrkBDlx mice resulted mainly from a severe loss of DRD2-expressing neurons (Fig. 2F). The number of EGFP-expressing striatal neurons was reduced by 63% in TrkB mutant mice (fB/fB;Dlx5/6-Cre;D2-EGFP) at 3 wk of age compared with age-matched control mice (fB/fB;D2-EGFP; Fig. 2 F and G). Because the Dlx5/6-Cre transgene also expressed EGFP due to the inclusion of an IRES2-EGFP sequence downstream of the Cre coding region (13), some EGFP-positive neurons in fB/fB;Dlx5/6-Cre;D2-EGFP mice might not be DRD2 neurons, thus causing an underestimation of the loss of DRD2 neurons in TrkBDlx mice. To estimate the number of striatal neurons expressing both EGFP and DRD1a in the TrkB mutant, we counted EGFP-expressing striatal neurons in TrkBDlx (fB/fB;Dlx5/6-Cre) and control (fB/+;Dlx5/6-Cre) mice at postnatal day 21 (P21). The number of neurons labeled with EGFP by the Dlx5/6-Cre transgene in the control mice was small (143,400 ± 16,100) compared with the number of neurons labeled with EGFP by the D2-EGFP transgene (520,300 ± 37,200) at P21 (Fig. 2G), indicating that the expression of Dlx5/6-Cre was turned off in many striatal neurons by P21. Assuming that all EGFP-expressing neurons in fB/fB;Dlx5/6-Cre mice are DRD1a cells, we subtracted this neuronal count from the total number of EGFP+ striatal neurons in fB/fB;Dlx5/6-Cre;D2-EGFP mice to estimate the size of the D2-EGFP-expressing neuronal population in the mutant. The adjusted number revealed that up to 80% of DRD2-expressing striatal neurons might be lost in TrkBDlx mice (Fig. 2G). Therefore, deletion of the TrkB gene in the LGE leads to a 63–80% loss of DRD2 neurons in the striatum.
We next asked if any MSNs of the direct pathway were also lost in TrkBDlx mice by generating fB/fB;Dlx5/6-Cre;Drd1a-tdTomato (mutant) and fB/fB;Drd1a-tdTomato (control) mice. In Drd1a-tdTomato BAC transgenic mice a red fluorescent protein marks MSNs of the direct pathway (12). Counts of tdTomato-expressing cells revealed a 22% loss of DRD1a MSNs in mutant mice at P21 compared with control mice (Fig. S3 A–C). By contrast, no significant changes were found in the numbers of ChAT-, parvalbumin-, or NPY-expressing striatal interneurons (Fig. S3D), although TrkB was expressed in some interneurons expressing either parvalbumin or NPY (Fig. S4). Taken together, these observations indicate that deletion of the TrkB gene in the striatum leads to a severe and relatively selective loss of DRD2-expressing MSNs in the striatum.
The relatively specific effect of TrkB deletion on DRD2/Enk MSNs led to the hypothesis that TrkB expression was restricted to this striatal population. To test this hypothesis, we generated D2-EGFP;TrkBLacZ/+ mice by crossing D2-EGFP transgenic mice to TrkBLaz/+ mice in which expression of β-galactosidase is under the control of the TrkB promoter and thus recapitulates the expression pattern of the TrkB gene (22). Fluorescent immunohistochemistry indicated that β-galactosidase was localized in the cytoplasm as well as in neuropils (Fig. 3). We considered those cells with higher cytoplasmic staining than the surrounding neuropil staining as TrkB-expressing neurons. Using this criterion, we found that 75% (91/122) of TrkB-expressing neurons were positive for EGFP and 43% (91/211) of EGFP+ neurons expressed TrkB in the striatum of adult D2-EGFP;TrkBLacZ/+ mice (Fig. 3 A–C). In concordance with this observation, TrkB was detected in 37% (34/92) of DARPP-32-expressing neurons (Fig. 3 D–F), approximately half of which are DRD2-expressing MSNs (23, 24). These findings may explain the preferential effect of TrkB deletion on DRD2 MSNs, but do not explain the loss of up to 80% of DRD2 neurons in the striatum of the TrkB mutant at P21 (Fig. 2G). To investigate this discrepancy, we examined the TrkB expression pattern in the striatum of young D2-EGFP;TrkBLacZ/+ mice. At P10, TrkB was expressed in many more neurons, so that 98% of EGFP+ striatal neurons (657/670) expressed TrkB (Fig. 3 G–I). To examine TrkB expression in MSNs of the direct pathway, we crossed TrkBLacZ/+ mice to Drd1a-tdTomato mice. We found that 18% (125/690) of DRD1a neurons expressed TrkB in Drd1a-tdTomato;TrkBLacZ/+ mice at P10 (Fig. 3 J–L). Thus, the relative selective loss of DRD2 neurons in TrkBDlx mice results from the preferential expression of TrkB in MSNs of the indirect pathway.
Fig. 3.
TrkB is preferentially expressed in DRD2-positive MSNs. (A–C) Colocalization of TrkB with DRD2 in the striatum of adult TrkBLacZ/+;D2-EGFP mice in which β-galactosidase and EGFP serve as indicators for expression of TrkB and DRD2, respectively. Fluorescent immunohistochemistry with antibodies to β-galactosidase and EGFP shows that the majority of TrkB-expressing neurons also express DRD2 in the adult striatum. (D–F) TrkB expression in striatal MSNs. Antibodies to DARPP-32 and β-galactosidase were used to reveal MSNs and TrkB-expressing cells in the striatum of adult TrkBLacZ/+ mice. (G–I) High coexpression of TrkB and DRD2 in the young striatum. Nearly all EGFP+ cells expressed β-galactosidase in the striatum of TrkBLacZ/+;D2-EGFP mice at P10. (J–L) Low coexpression of TrkB and DRD1a in the striatum of TrkBLacZ/+;Drd1a-tdTomato mice at P10. (Scale bar, 50 μm.)
Increased Apoptosis in Developing Striatal Neurons of TrkBDlx Mice.
The severe neuronal loss observed in the striatum of TrkBDlx mice at P21 could be due to increased cell death during the postnatal period or impaired striatal development during embryogenesis. We first asked if striatal neurons die during the first three postnatal weeks in the absence of the TrkB receptor. We measured striatal volume and counted striatal neurons in TrkBDlx and control mice at P10 and P0. We found that the magnitudes of the reductions in striatal volume and number of striatal neurons in TrkBDlx mice at these two ages were similar to those observed at P21 (Fig. 4 A and B). These findings ruled out the possibility that the loss of striatal neurons in TrkBDlx mice results from increased cell death during the first three postnatal weeks.
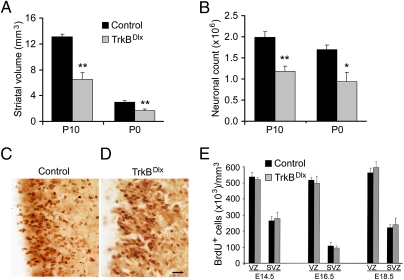
Fig. 4.
Loss of striatal neurons in TrkBDlx mice is not due to impaired striatal neurogenesis. (A) Striatal volumes of TrkBDlx and control mice at P10 and P0. Striatal volume in TrkBDlx mice was reduced by 50% at P10 and 44% at P0 compared with control mice (n = 4 mice each). Error bars represent SEM. Student's t test: **P < 0.01. (B) Striatal neuron counts of TrkBDlx and control mice at P10 and P0. Note that the striatal neuron counts in TrkBDlx mice was reduced by 40% at P10 and 44% at P0 compared with control mice (n = 4 mice each). Error bars represent SEM. Student's t test: *P < 0.05; **P < 0.01. (C and D) Representative images of BrdU-labeled cells in the VZ and SVZ of the LGE at E16.5. (Scale bar, 25 μm.) (E) Quantification of BrdU-positive cells in the VZ and SVZ of the LGE shows no significant difference between control and TrkBDlx embryos at E14.5, E16.5, and E18.5. Error bars represent SEM.
The observation that the loss of striatal neurons in TrkBDlx mice was nearly completed by P0 suggests that TrkB signaling is required for striatal neurogenesis and/or survival of developing striatal neurons. Some studies indicate that TrkB signaling regulates adult neurogenesis in the subventricular zone, affecting production of olfactory bulb interneurons and striatal neurons (25–27). Furthermore, TrkB signaling has been shown to promote proliferation and differentiation of neuronal precursors in the mouse cerebral cortex (28). To examine the effect of TrkB deletion on striatal neurogenesis, which extends from embryonic day 12 (E12) to P2 (29), we used BrdU to label newborn cells. At E14.5, E16.5, and E18.5, pregnant female mice were injected with BrdU (50 mg/kg), and 2 h later embryos were removed to achieve short-term labeling. Numerous BrdU-positive cells were detected in the ventricular and subventricular zones (VZ and SVZ) of the LGE in control and TrkBDlx embryos (Fig. 4 C and D). Quantification revealed no difference in the density of BrdU-labeled cells in the VZ/SVZ between control and TrkBDlx embryos at all three embryonic stages (Fig. 4E). These findings suggest that deletion of the TrkB gene in the LGE does not affect striatal neurogenesis.
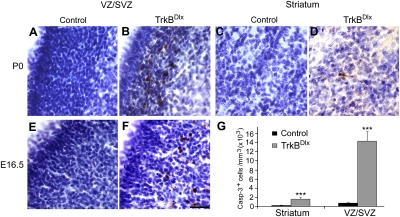
We next asked whether ablation of TrkB signaling leads to increased cell death of striatal neurons during the period of striatal neurogenesis. To this aim, we used immunostaining for activated caspase-3 to detect cells undergoing apoptosis. We observed a great increase in caspase-3-positive cells in the striatal VZ/SVZ of TrkBDlx mice at P0 and E16.5 (Fig. 5 B and F) compared with littermate controls (Fig. 5 A and E). In addition, some caspase-3-positive cells were observed in the TrkBDlx striatum at P0 (Fig. 5D) but not in the control striatum (Fig. 5C). We quantified the number of cells containing the activated caspase-3 at P0 and found that apoptosis increased by 20-fold in the VZ/SVZ and sevenfold in the striatum in TrkBDlx mice compared with littermate controls (Fig. 5G). In agreement with this apoptosis phenotype, the TrkB receptor was expressed in the striatal VZ/SVZ at P0 and E16.5 (Fig. S5). These findings suggest that TrkB signaling is essential for the survival of developing striatal neurons, especially those newborn neurons that still reside in the LGE.
Fig. 5.
Striatal neuronal loss is due to increased apoptosis in the striatal proliferative zone. (A–D) Immunohistochemistry against activated caspase-3 in the VZ/SVZ and striatum at P0. Numerous apoptotic cells were observed in the LGE VZ/SVZ and striatum of TrkBDlx mice. Sections were counterstained with Nissl. (E and F) Immunohistochemistry against activated caspase-3 in the LGE VZ/SVZ of embryos at E16.5. (Scale bar, 25 μm.) (G) Density of cells containing activated caspase-3 in the LGE VZ/SVZ and the striatum of control and TrkBDlx mice at P0. Error bars represent SEM. Student's t test: ***P < 0.001.
Discussion
The vital role of neurotrophins in the survival of developing neurons in the PNS has been well established. For example, deletion of the gene for NGF or its TrkA receptor leads to over 70% neuronal loss in the trigeminal and dorsal root sensory ganglia, and over 95% neuronal loss in the superior cervical sympathetic ganglia (30, 31). Similarly, Bdnf knockout mice exhibit severe neuronal loss in several sensory ganglia (32, 33). Although it has been shown that neurotrophins participate in the maintenance of adult neuronal populations in the brain (34, 35), the role of neurotrophins in the survival of developing neurons in the central nervous system remains largely unknown. A modest increase in postnatal apoptosis was observed in hippocampal and cerebellar granule cells of TrkB and TrkC knockout mice; however, these deletions do not appear to affect the size of these two neuronal populations (4, 5). The redundancy of neurotrophin-mediated signaling pathways in brain regions where more than one Trk receptor is present in a specific neuronal population can provide an explanation for the rather minor effect when one receptor or its ligands are removed. However, the present study supports the idea that a single neurotrophin might be sufficient and necessary to support the survival of some neuronal populations in the brain, because we show here that TrkB signaling is necessary for the survival of immature striatal neurons that will become MSNs of the indirect pathway. Furthermore, this study shows that a large population of developing neurons in the brain is dependent on ligands of a single neurotrophin receptor for survival. Systemic and stereological analysis is necessary to determine whether other populations of neurons in the brain are also dependent on neurotrophic factors for survival during development.
Our results suggest that neurotrophins promote neuronal survival via different modes in the PNS and the striatum. In the PNS, developing neurons depend on neurotrophins for survival after they innervate target tissues (3). However, our findings show that the majority of cell death occurs within the LGE in the absence of TrkB signaling, indicating that developing MSNs need trophic support before they migrate to the striatum and send out axons. These observations suggest that the timing of neurotrophic dependence is different for PNS neurons and CNS neurons. Furthermore, because BDNF, the main ligand for TrkB, is expressed in the substantia nigra but not in the striatum and the cerebral cortex of newborn mice (35, 36), and because nigrostriatal projections have been formed by E16.5 (37, 38), it is possible that BDNF transported anterogradely from the substantia nigra supports the survival of newborn MSNs within the LGE (i.e., innervating neurons controlling the size of their targets). This trophic action is different from the one commonly observed in the PNS, i.e., target tissues controlling the size of innervating neuronal populations. In addition to promoting the survival of developing MSNs during embryogenesis, BDNF-to-TrkB signaling also plays a key role in postnatal dendritic growth of striatal neurons (35, 39).
Our findings show that TrkB is preferentially expressed in DRD2 MSNs at P10 and that the loss of these neurons in TrkBDlx mice is mainly attributable to increased apoptosis within the LGE. These observations suggest that TrkB should be preferentially expressed in newborn neurons destined to be DRD2 MSNs in the LGE; however, it is difficult to demonstrate directly this prediction because the majority of cells in the LGE have not started to express dopamine receptors yet. The observed preferential TrkB expression in DRD2-expressing MSNs of the mature brain may provide some insights into the selectivity of degeneration associated with HD. HD, a dominantly inherited neurodegenerative disorder characterized by abnormalities of movement and cognition along with changes in psychiatric symptoms, is caused by expansion of a polyglutamine tract at the N terminus of huntingtin (htt). The clinical signs and symptoms result from relatively selective degeneration of striatal neurons (6); however, striatal neurons are not uniformly affected in HD. Immunohistochemical studies in patients with HD show a greater decrease in the number of neurons coexpressing DRD2 and Enk (40). These neurons act to terminate movement associated with the basal ganglia or suppress unwanted sequences of movements (10). Hence, the loss of the indirect pathway neurons leads to disinhibition of the thalamus and increased facilitation of the motor cortex, producing hyperkinesias in HD patients (41). Alternatively, direct-pathway neurons coexpressing DRD1a and SP are less affected, and striatal interneurons are mostly spared in patients with HD (42, 43). The CAG expansion in the HD gene has been shown to inhibit Bdnf gene expression and axonal transport of the BDNF protein, leading to a reduction in striatal BDNF levels (44, 45). The deficiency in BDNF may selectively affect DRD2-expressing MSNs due to preferential TrkB expression in these neurons. However, it remains to be determined whether deletion of the TrkB gene in the adult brain causes degeneration of DRD2-expressing MSNs.
Materials and Methods
Animals.
Mice were maintained on a 12-h/12-h light/dark cycle with food and water ad libitum. The Georgetown University Animal Care and Use Committee approved all animal procedures.
Stereology.
We used Stereo Investigator software (MicroBrightField Inc.) to calculate striatal volume and neuronal number as described previously (46).
Additional methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Kenneth Campbell for Dlx5/6-Cre mice, Stefano Vicini for D2-EGFP mice, Nicole Calakos for Drd1a-tdTomato mice, Kevin Jones for Bdnf lox mice, and Filip Vanevski and Emily Waterhouse for critical reading of this manuscript. This work was supported by National Institutes of Health Grants R01 NS050596 (to B.X.), P01 NS016033 (to L.F.R.), and F31 NS060523 (to M.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004744108/-/DCSupplemental.
References
- 1.Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nat Rev Neurosci. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
- 2.Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang EJ, Reichardt LF. Neurotrophins: Roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minichiello L, Klein R. TrkB and TrkC neurotrophin receptors cooperate in promoting survival of hippocampal and cerebellar granule neurons. Genes Dev. 1996;10:2849–2858. doi: 10.1101/gad.10.22.2849. [DOI] [PubMed] [Google Scholar]
- 5.Alcántara S, et al. TrkB signaling is required for postnatal survival of CNS neurons and protects hippocampal and motor neurons from axotomy-induced cell death. J Neurosci. 1997;17:3623–3633. doi: 10.1523/JNEUROSCI.17-10-03623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Dauer W, Przedborski S. Parkinson's disease: Mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 8.Kawaguchi Y. Neostriatal cell subtypes and their functional roles. Neurosci Res. 1997;27:1–8. doi: 10.1016/s0168-0102(96)01134-0. [DOI] [PubMed] [Google Scholar]
- 9.Hamasaki T, Goto S, Nishikawa S, Ushio Y. Neuronal cell migration for the developmental formation of the mammalian striatum. Brain Res Brain Res Rev. 2003;41:1–12. doi: 10.1016/s0165-0173(02)00216-3. [DOI] [PubMed] [Google Scholar]
- 10.Bolam JP, Hanley JJ, Booth PA, Bevan MD. Synaptic organisation of the basal ganglia. J Anat. 2000;196:527–542. doi: 10.1046/j.1469-7580.2000.19640527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day M, et al. Selective elimination of glutamatergic synapses on striatopallidal neurons in Parkinson disease models. Nat Neurosci. 2006;9:251–259. doi: 10.1038/nn1632. [DOI] [PubMed] [Google Scholar]
- 12.Shuen JA, Chen M, Gloss B, Calakos N. Drd1a-tdTomato BAC transgenic mice for simultaneous visualization of medium spiny neurons in the direct and indirect pathways of the basal ganglia. J Neurosci. 2008;28:2681–2685. doi: 10.1523/JNEUROSCI.5492-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: Implications for striatal and olfactory bulb neurogenesis. J Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson SA, et al. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- 15.Olsson M, Campbell K, Wictorin K, Björklund A. Projection neurons in fetal striatal transplants are predominantly derived from the lateral ganglionic eminence. Neuroscience. 1995;69:1169–1182. doi: 10.1016/0306-4522(95)00325-d. [DOI] [PubMed] [Google Scholar]
- 16.Olsson M, Björklund A, Campbell K. Early specification of striatal projection neurons and interneuronal subtypes in the lateral and medial ganglionic eminence. Neuroscience. 1998;84:867–876. doi: 10.1016/s0306-4522(97)00532-0. [DOI] [PubMed] [Google Scholar]
- 17.Anderson SA, Marín O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- 18.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 19.Mantamadiotis T, et al. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 20.Svenningsson P, et al. DARPP-32: An integrator of neurotransmission. Annu Rev Pharmacol Toxicol. 2004;44:269–296. doi: 10.1146/annurev.pharmtox.44.101802.121415. [DOI] [PubMed] [Google Scholar]
- 21.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 22.Xu B, et al. The role of brain-derived neurotrophic factor receptors in the mature hippocampus: Modulation of long-term potentiation through a presynaptic mechanism involving TrkB. J Neurosci. 2000;20:6888–6897. doi: 10.1523/JNEUROSCI.20-18-06888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson KD, Reiner A. Immunohistochemical localization of DARPP-32 in striatal projection neurons and striatal interneurons: Implications for the localization of D1-like dopamine receptors on different types of striatal neurons. Brain Res. 1991;568:235–243. doi: 10.1016/0006-8993(91)91403-n. [DOI] [PubMed] [Google Scholar]
- 24.Ouimet CC, Greengard P. Distribution of DARPP-32 in the basal ganglia: An electron microscopic study. J Neurocytol. 1990;19:39–52. doi: 10.1007/BF01188438. [DOI] [PubMed] [Google Scholar]
- 25.Zigova T, Pencea V, Wiegand SJ, Luskin MB. Intraventricular administration of BDNF increases the number of newly generated neurons in the adult olfactory bulb. Mol Cell Neurosci. 1998;11:234–245. doi: 10.1006/mcne.1998.0684. [DOI] [PubMed] [Google Scholar]
- 26.Pencea V, Bingaman KD, Wiegand SJ, Luskin MB. Infusion of brain-derived neurotrophic factor into the lateral ventricle of the adult rat leads to new neurons in the parenchyma of the striatum, septum, thalamus, and hypothalamus. J Neurosci. 2001;21:6706–6717. doi: 10.1523/JNEUROSCI.21-17-06706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chmielnicki E, Benraiss A, Economides AN, Goldman SA. Adenovirally expressed noggin and brain-derived neurotrophic factor cooperate to induce new medium spiny neurons from resident progenitor cells in the adult striatal ventricular zone. J Neurosci. 2004;24:2133–2142. doi: 10.1523/JNEUROSCI.1554-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartkowska K, Paquin A, Gauthier AS, Kaplan DR, Miller FD. Trk signaling regulates neural precursor cell proliferation and differentiation during cortical development. Development. 2007;134:4369–4380. doi: 10.1242/dev.008227. [DOI] [PubMed] [Google Scholar]
- 29.Marchand R, Lajoie L. Histogenesis of the striopallidal system in the rat. Neurogenesis of its neurons. Neuroscience. 1986;17:573–590. doi: 10.1016/0306-4522(86)90031-x. [DOI] [PubMed] [Google Scholar]
- 30.Smeyne RJ, et al. Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature. 1994;368:246–249. doi: 10.1038/368246a0. [DOI] [PubMed] [Google Scholar]
- 31.Crowley C, et al. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- 32.Jones KR, Fariñas I, Backus C, Reichardt LF. Targeted disruption of the BDNF gene perturbs brain and sensory neuron development but not motor neuron development. Cell. 1994;76:989–999. doi: 10.1016/0092-8674(94)90377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ernfors P, Lee KF, Jaenisch R. Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature. 1994;368:147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- 34.Xu B, et al. Cortical degeneration in the absence of neurotrophin signaling: Dendritic retraction and neuronal loss after removal of the receptor TrkB. Neuron. 2000;26:233–245. doi: 10.1016/s0896-6273(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 35.Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J Neurosci. 2004;24:4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baquet ZC, Bickford PC, Jones KR. Brain-derived neurotrophic factor is required for the establishment of the proper number of dopaminergic neurons in the substantia nigra pars compacta. J Neurosci. 2005;25:6251–6259. doi: 10.1523/JNEUROSCI.4601-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voorn P, Kalsbeek A, Jorritsma-Byham B, Groenewegen HJ. The pre- and postnatal development of the dopaminergic cell groups in the ventral mesencephalon and the dopaminergic innervation of the striatum of the rat. Neuroscience. 1988;25:857–887. doi: 10.1016/0306-4522(88)90041-3. [DOI] [PubMed] [Google Scholar]
- 38.Hu Z, Cooper M, Crockett DP, Zhou R. Differentiation of the midbrain dopaminergic pathways during mouse development. J Comp Neurol. 2004;476:301–311. doi: 10.1002/cne.20230. [DOI] [PubMed] [Google Scholar]
- 39.Rauskolb S, et al. Global deprivation of brain-derived neurotrophic factor in the CNS reveals an area-specific requirement for dendritic growth. J Neurosci. 2010;30:1739–1749. doi: 10.1523/JNEUROSCI.5100-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiner A, et al. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci USA. 1988;85:5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calabresi P, et al. Synaptic transmission in the striatum: From plasticity to neurodegeneration. Prog Neurobiol. 2000;61:231–265. doi: 10.1016/s0301-0082(99)00030-1. [DOI] [PubMed] [Google Scholar]
- 42.Ferrante RJ, Beal MF, Kowall NW, Richardson EP, Jr., Martin JB. Sparing of acetylcholinesterase-containing striatal neurons in Huntington's disease. Brain Res. 1987;411:162–166. doi: 10.1016/0006-8993(87)90694-9. [DOI] [PubMed] [Google Scholar]
- 43.Ferrante RJ, et al. Morphologic and histochemical characteristics of a spared subset of striatal neurons in Huntington's disease. J Neuropathol Exp Neurol. 1987;46:12–27. doi: 10.1097/00005072-198701000-00002. [DOI] [PubMed] [Google Scholar]
- 44.Zuccato C, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 45.Gauthier LR, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 46.Xie Y, Hayden MR, Xu B. BDNF overexpression in the forebrain rescues Huntington's disease phenotypes in YAC128 mice. J Neurosci. 2010;30:14708–14718. doi: 10.1523/JNEUROSCI.1637-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.