Abstract
The use of molecular methods is altering our understanding of the microbial biosphere and the complexity of the tree of life. Here, we report a newly discovered uncultured plastid-bearing eukaryotic lineage named the rappemonads. Phylogenies using near-complete plastid ribosomal DNA (rDNA) operons demonstrate that this group represents an evolutionarily distinct lineage branching with haptophyte and cryptophyte algae. Environmental DNA sequencing revealed extensive diversity at North Atlantic, North Pacific, and European freshwater sites, suggesting a broad ecophysiology and wide habitat distribution. Quantitative PCR analyses demonstrate that the rappemonads are often rare but can form transient blooms in the Sargasso Sea, where high 16S rRNA gene copies mL−1 were detected in late winter. This pattern is consistent with these microbes being a member of the rare biosphere, whose constituents have been proposed to play important roles under ecosystem change. Fluorescence in situ hybridization revealed that cells from this unique lineage were 6.6 ± 1.2 × 5.7 ± 1.0 μm, larger than numerically dominant open-ocean phytoplankton, and appear to contain two to four plastids. The rappemonads are unique, widespread, putatively photosynthetic algae that are absent from present-day ecosystem models and current versions of the tree of life.
Keywords: algal diversity, aquatic photosynthesis, phylogeny, plastid evolution, uncultured eukaryotes
Photosynthetic marine organisms perform roughly half of the primary production on earth (1). In addition to cyanobacteria, phytoplankton taxa distributed across the eukaryotic tree are responsible for photosynthetic CO2 uptake (2, 3). Eukaryotic phytoplankton taxa differ significantly in relative abundances depending on environmental conditions, as do their contributions to primary production, which are poorly quantified (4–6). Analyses of environmental ribosomal RNA (rRNA) genes have transformed our understanding of such microbial eukaryotes. Eukaryotic algae possess both nuclear (18S) and plastid (16S) rRNA genes, and environmental sequence surveys using both markers have revealed that lineages such as the stramenopiles (heterokonts), haptophytes, and alveolates are diverse and complicated groups (7–14). Although environmental sequences from uncultured organisms can be linked to abundance and ecological roles using FISH and quantitative PCR (qPCR), much of the biology of the organisms from which environmental sequences are derived remains unknown.
Newly discovered lineages can help to elucidate fundamental issues in eukaryotic evolution, such as the number and type of endosymbiotic events that gave rise to the current distribution of plastids. Many ecologically relevant eukaryotic groups, including haptophytes, cryptophytes, and stramenopiles, harbor red algal-derived plastids of secondary or tertiary endosymbiotic origin, but how these organisms (and their photosynthetic organelles) are related is unclear (15–18). We present evidence for the existence of a previously unrecognized microalgal lineage, herein referred to as rappemonads. Phylogenetic analyses of near-complete environmental rRNA operon sequences reveal that rappemonads are related to but distinct from known haptophyte and cryptophyte algae and constitute a diverse group distributed across marine and freshwater ecosystems.
Results and Discussion
Rappemonads: A Diverse and Evolutionarily Distinct Microalgal Lineage.
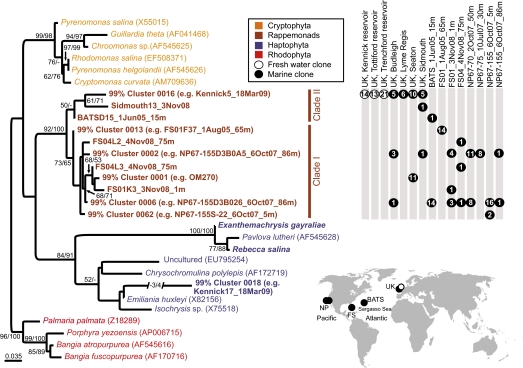
Preliminary phylogenetic analyses of database 16S rDNA sequences identified a unique plastid sequence, OM270, deposited over a decade ago from Atlantic waters near Cape Hatteras, North Carolina (19). OM270 seemed to represent a previously undescribed red algal secondary plastid-bearing organism (19). To determine whether this environmentally derived sequence represents an ecologically significant group, we designed PCR primers to OM270 and a similar recently deposited sequence (MC622-32) (Table S1) (14). Environmental plastid 16S rDNA clone libraries were constructed from samples originating in the Northeast Pacific Ocean, in two subtropical North Atlantic regions, the Florida Straits and the Sargasso Sea, and in coastal and oligotrophic freshwater sites in the United Kingdom (Fig. 1, Inset and Table S2) using these primers. A diverse set of rDNA sequences related to OM270 was recovered (Fig. 1). Furthermore, some UK coastal marine and freshwater sequences fell within the same 99% identity cluster (Fig. 1). This suggests that the physiology of the group may be relatively flexible to different aquatic habitats, although it is still unclear how 16S rDNA sequence variability relates to genome or ecological diversity within eukaryotic algae; this particular 99% cluster may encompass numerous ecotypes. The 16S rDNA analyses demonstrate that this group is distinct from known algae and possesses considerable diversity. Pairwise 16S rDNA identities range down to 94% identity (across a 492-nt fragment) within the group, whereas cryptophytes and haptophytes range down to 96% and 87% identity, respectively. The latter is considered a phylum and includes organisms as disparate as the pavlovales, the soft-bodied picohaptophytes that form a major portion of oceanic picoplankton biomass (6), and coccolithophores, which have ornate calcium carbonate plates (20). The overall level of 16S rDNA divergence within the newly discovered group identified herein is notable, and it presumably results in differing genomic repertoires and physiologies. Contrast, for example, the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum, which share 98% identity over the 492-nt fragment and 96% identity across the entire plastid 16S rRNA gene but whose genomes share only 30–40% of protein-encoding genes (21).
Fig. 1.
Environmental diversity and sample locations. Maximum likelihood phylogenetic tree of environmental 16S rDNA sequences obtained herein (bold) as well as OM270. (Right) Clone library sample sites (Table S2) and the number of sequences obtained are shown, including marine samples from the North Pacific (NP), Florida Straits (FS), and BATS, as well as UK coastal and freshwater samples. The scale bar indicates the inferred number of nucleotide substitutions per site. Bootstrap support values (≥50%) are from RaxML and Log-Det distance analyses, respectively. (Inset) Map shows the approximate positions of the sites sampled.
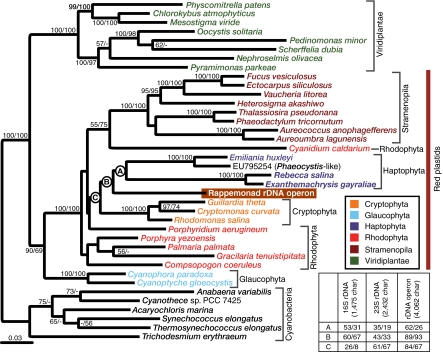
To resolve the phylogenetic position of this distinctive group further, a region spanning the 16S rRNA gene through to the 23S locus (including the intergenic transcribed spacers and two tRNA encoding genes) was amplified and sequenced (Table S3). This allowed us to establish the group's evolutionary position relative to known red algae and algae with red algal-derived secondary plastids (specifically cryptophytes, haptophytes, and stramenopiles). Analyses of these near-complete environmental plastid rDNA operons, together with sequences obtained herein from the haptophytes Rebecca salina and Exanthemachrysis gayraliae, showed that the newly discovered group is a unique lineage that branches deeply within the haptophyte and cryptophyte radiation. The group specifically appears to sister the haptophytes (Fig. 2 and Fig. S1), with alternative topology tests rejecting placement of the unique plastid rDNA operon sequences as a sister to cryptophytes or to the stramenopiles (Fig. S2). This suggests that the unique group is a sister clade to the haptophytes but does not rule out it being sister to the wider haptophyte/cryptophyte clade (Fig. 2 and Fig. S2). Haptophytes are a diverse and anciently diverged lineage (20) of measurable environmental importance (6, 22). Based on bootstrap analyses, it is possible that rappemonads represent a deeply diverged and previously unrecognized haptophyte or haptophyte-like group (Fig. 2). The unique and deep-branching position, based on the rDNA operon analysis, establishes this group as a unique lineage which, in the absence of detailed morphological data, we name the “rappemonads,” in reference to the publication by Rappé et al. (19), which initially reported the OM270 clone sequence.
Fig. 2.
Maximum likelihood (ML) phylogenetic tree of plastid 16S-Ile tRNA-Ala tRNA-23S rDNA sequences rooted with select cyanobacteria. Sequences in bold were generated as part of this study, including newly obtained Pavlovophyceae sequences. The scale bar indicates the inferred number of nucleotide substitutions per site. Bootstrap support values (≥50%) are from ML and Log-Det distance analyses, respectively. (Inset) Bootstrap support for nodes labeled A, B, and C using 16S rDNA, 23S rDNA, and near-complete rDNA operon alignments (representative 16S and 23S rDNA tree topologies are shown in Fig. S1).
Environmental Distribution and Cellular Morphology of the Rappemonads.
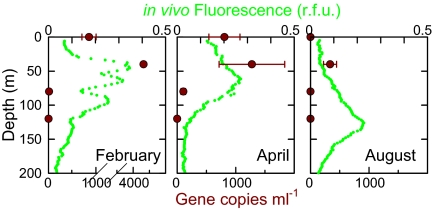
To investigate distributions in the environment, a rappemonad-specific qPCR assay was developed (SI Materials and Methods). Sequencing of environmental qPCR products and tests against nontarget and plasmid controls verified rappemonad specificity (SI Materials and Methods). The unique lineage was detected in 23 of 48 marine euphotic-zone samples ranging from 15 ± 14–4,318 ± 38 16S rRNA gene copies mL−1 (Table S4). Sixteen samples in which cells were not detected showed high inhibition, an issue frequently seen with environmental DNA extractions and generally thought to be caused by the presence of unidentified inhibitory substances (23). This required dilution of these samples to levels at which detection limits were poor (in one case, minimum detection was 647 copies mL−1, although detection was typically much better than this; Table S4); these data were not included in additional statistical analyses comparing abundance with environmental parameters. High 16S rRNA gene copies mL−1 were detected in what appeared to be a late-winter bloom in sub surface waters at the Bermuda Atlantic Time-series site (BATS) (Fig. 3). The water column at this time showed several fluorescence maxima, and rappemonads were concentrated at the shallowest of these, indicating that deeper maxima were composed of other taxa. Very few or no rappemonads were detected in stratified summertime conditions, when there was a pronounced deep chlorophyll maximum (Fig. 3). In addition, 11 of 12 samples from a North Pacific anticyclonic eddy, in which colder and more nutrient-rich waters, akin to the higher nutrient availability in late-winter BATS samples, were brought to the surface, resulting in a shallower mixed layer, had measureable gene copy numbers (averaging 186 ± 78 gene copies mL−1). Other samples from the 500-mile transect in the North Pacific had fewer (Table S4).
Fig. 3.
Rappemonad distributions in the Sargasso Sea in 2003. Seasonal transitions are shown for BATS, as revealed by qPCR assays for the 16S rRNA gene (dark red, gene copies mL−1). In vivo chlorophyll fluorescence is shown in relative fluorescence units (green, r.f.u.). Note differences in x-axis scales.
Although depth, temperature, salinity, phosphate, chlorophyll a, and nitrate plus nitrite were measured, no statistically significant differences were identified between samples where rappemonads were detected and those samples where none were detected (t tests, Mann–Whitney rank sum), considering only those in which nutrient concentrations were above detection limits. The upper range of phosphate concentrations in rappemonad-positive samples was 0.69 μM, lower than for all samples (1.19 μM). In addition, chlorophyll a ranged from 0.07–0.69 μg L−1 for rappemonad-containing samples and from 0.03–2.71 μg L−1 for all samples. Average temperatures of the samples investigated and those that contained rappemonads were identical (17 ± 4 °C). Rappemonad sequences were detected in waters ranging from 11 °C to 24 °C, with sequences also recovered from 26 °C waters (Table S2), although this sample was not screened by qPCR because the DNA was not extracted in a quantitative manner. The temperature range of rappemonad-containing samples again indicates that this lineage may have a broad ecophysiological range.
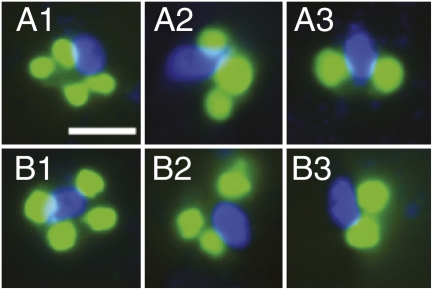
We also characterized rappemonads morphologically. Oligonucleotide probes targeting two different 16S rRNA regions of distinct rappemonad subgroups within clade I (Fig. 1) were designed for use with tyramide signal amplification FISH and verified for specificity on a series of nontarget controls. The probes were applied to two samples for which FISH filters were available and notable gene copies mL−1 were detected by qPCR. Rappemonads (n = 88) measured 5.7 ± 1.0 (SD) μm in width (shortest dimension) and 6.6 ± 1.2 (SD) μm in length (longest dimension). Each cell appeared to contain two, three, or four plastids (Fig. 4 and Fig. S3), with four being the most common [46 (52%) of 88 cells]; it is conceivable that instances of three or four organelles associated with a single nucleus correspond to dividing stages of the cell. Alternatively, plastids can be bilobed, giving the appearance of multiple plastids when only one is present, and cell orientation can bias imaging. By comparison, the plastids of haptophytes mostly occur singly or in pairs and are often bilobed (2). The microscopy analyses also revealed a faint reddish fluorescence colocalized with the hybridized plastid compartments using a DAPI filter set (excitation, G365; emission, LP420), presumably derived from residual chlorophyll pigments.
Fig. 4.
Fluorescence micrographs of rappemonads in the North Pacific. The DAPI-stained nucleus (blue) was often slightly elongated with a tapering end. Two to four plastids appeared to be present per cell (green, tyramide signal amplification FISH-labeled). Cells shown in A1–A3 were detected using the RappeA probe, whereas cells in B1–B3 were detected with the RappeB probe. (Scale bar: 5 μm.)
The average cell biovolume of rappemonads was 112 μm3, and the average carbon content was 27 pg of carbon cell−1, based on cellular dimensions and an established carbon conversion factor (24). This is a significantly greater cellular carbon content than in the picophytoplankton (<2–3 μm in diameter) that dominate such regions, for example, small haptophytes that range from 1–3 pg of carbon cell−1 (6). Large cell size may also be responsible for the rarity of reported rappemonad sequences. The majority of environmental 16S (and 18S) rDNA clone libraries, especially those using primer sets targeting plastid-16S rRNA genes, are constructed from water prefiltered through 3-μm pore-sized filters (25, 26), which would select against these cells. In addition, a high abundance of heterotrophic bacteria (105–106 mL−1) could effectively swamp 16S rDNA libraries constructed using universal primers, such that few plastid-derived sequences are attained.
What Are Rappemonads?
We explored the possibility that rappemonad plastid 16S rDNA sequences are derived from other lineages represented in 18S rDNA phylogenies. One recently reported uncultured group is the biliphytes (or picobiliphytes). This putatively plastid-bearing eukaryotic lineage was previously detected in marine 18S rDNA clone libraries (27), and subsequent phylogenetic analyses indicated that they represented an uncultured lineage possibly related to cryptophytes, although no analysis provided bootstrap support for this relationship in excess of 50% (28, 29). Several lines of evidence suggest that rappemonads do not correspond to a putative plastid associated with biliphytes. First, the latter appear to be smaller than rappemonads, with two biliphyte clades being 3.5 ± 0.9 × 3.0 ± 0.9 and 4.1 ± 1.0 × 3.5 ± 0.8, respectively (29). Biliphytes detected here in the North Pacific, using FISH probes (28, 29), were similar in size to those reported by Cuvelier et al. (29). The highly punctate phycobilin-like (orange) fluorescence reported previously (28, 29) was not seen colocalized with the hybridized (North Pacific) cells, however, and biliphytes were not present at levels significantly above background counts for negative controls. Given that punctate orange fluorescence is detected in some (28, 29) but not all instances, biliphytes are likely not obligate photoautotrophs but rather facultative mixotrophs or phagotrophs, whereby transient detection of orange fluorescence could represent ingested prey items (e.g., the cyanobacterium Synechococcus).
We also applied a two-step group-specific nested 18S rDNA PCR protocol targeting the majority of known biliphyte diversity (SI Materials and Methods) but did not recover any biliphyte sequences from the same freshwater samples for which we had previously recovered rappemonad plastid 16S rDNA sequences. Although reaction-specific PCR biases cannot be completely ruled out, this is consistent with the idea that the biliphyte 18S rDNA and rappemonad plastid 16S rDNA do not share the same environmental distribution, and thus correspond to two separate groups. In addition, although rappemonads and biliphytes were recovered in marine 16S and 18S rDNA clone libraries, respectively, from the same depth and site (Table S2, station 67–155) for which 768 clones were sequenced per size fraction and primer pair, rappemonads were only found in the 3- to 20-μm size fraction, whereas biliphyte 18S rDNA sequences were in the 0.8- to 3-μm size fraction. Finally, we used a combination of nested forward primers and general reverse primers to amplify most of the biliphyte nuclear 18S-ITS1-5.8S-ITS2-28S rRNA gene cluster. A multigene phylogeny was then constructed, and the biliphyte branching position within eukaryotes was retested, using more phylogenetic information than previously published (28, 29). Like previous studies, the resulting phylogeny lacked bootstrap support above 50% for placement of biliphytes with respect to known eukaryotic groups. Furthermore, the phylogeny demonstrated that biliphytes are separated from cryptophyte and haptophyte algae by multiple branches resolved with weak bootstrap support, although the maximum likelihood bootstrap analyses demonstrated that glaucophytes formed a moderately supported clade (71%) with cryptophytes and katablepharids, to the exclusion of biliphytes (Fig. S4). This suggests a different branching relationship than that from 18S rDNA analyses alone (28, 29).
Alternative phylogenetic topology tests of the nuclear biliphyte and plastid rappemonad alignments were then used to investigate whether we could reject the hypothesis that the two groups represented equivalent branching positions on the plastid and nuclear rDNA trees. These data support the placement of rappemonad plastid sequences with the wider haptophyte/cryptophyte plastid radiation, whereas placement of the biliphyte lineage remains ambiguous (Fig. S2). The data suggest that the rappemonad plastid and biliphyte nucleus have incongruent ancestries, although this may result from secondary or tertiary endosymbioses or from potentially methodological artifacts. Taken in sum, multiple lines of indirect evidence do not support rappemonads and biliphytes being one and the same cellular entity. One possibility is that rappemonads harbor a plastid derived from a common ancestor it shared with haptophytes, and thus represent a lineage that diverged before the diversification of Pavlovophyceae and Coccolithophyceae (or Prymnesiophyceae), the only two classes of haptophytes known to date. Detection of recently obtained environmental 18S rDNA sequences showing affinity to the host component of haptophytes is consistent with this hypothesis (6, 30, 31).
Conclusion
We have reported the discovery of a putatively photosynthetic diverse plastid-bearing microbial lineage in marine and freshwater environments. Together with definitive proof that rappemonads contain plastids and perform photosynthesis, elucidation of their evolution and functional ecology will help to address how these organisms thrive. For example, should the presence of rappemonads in both marine and freshwater systems indicate relatively flexible halotolerance, this could be advantageous under conditions in which oceans are freshening as a result of ice melt, such as the Arctic (32). Microbes within the rare biosphere are thought to be essential for system stability and important under climate change scenarios (33). Furthermore, future initiatives to identify the specific eukaryotic host lineage corresponding to these plastid 16S rDNA sequences and its specific relationship to known haptophytes will enhance understanding of the origin, spread, and diversification of red algal-derived secondary plastids.
The discovery of unique plastid-bearing lineages, such as the rappemonads described herein, demonstrates that our current understanding of aquatic microbial community structure is far from complete. This has important repercussions for the current inability to model global biogeochemistry under perturbation scenarios, which relies not only on knowledge of “who is there” but on their resilience to change and capacity for both acclimation and adaptation.
Materials and Methods
Detailed descriptions of environmental sampling procedures and locations, DNA extraction protocols, clone library construction, and DNA sequencing are provided in SI Materials and Methods. Identification of chimeric sequences, molecular phylogenetic analyses and alternative topology tests, qPCR procedures and controls, and tyramide signal amplification FISH experiments are described in SI Materials and Methods. Tables S5–S8 present a list of PCR primers for biliphyte 18S rRNA gene amplification (Table S5), primers for biliphyte nucleus-encoded rRNA gene cluster amplification (Table S6), 99% sequence clusters for the phylogeny shown in Fig. 1 (Table S7), and oligonucleotide probes for TSA-FISH experiments (Table S8).
Supplementary Material
Acknowledgments
We thank the captains and crews of the research vessels Oceanus, Walton Smith, and Western Flyer as well as cruise participants, particularly M. Cuvelier, A. Engman, and E. Demir. Some DNA samples were kindly provided by R. Paerl, J. Zehr, S. Giovannoni, and C. Carlson. Rebecca salina materials were provided by R. Kamikawa, T. Matsumoto, and T. Nakayama. We thank G. Weinstock, the Washington University St. Louis Genome sequencing center, J. Vassar, and R. Gausling for clone library assistance. N. Onodera assisted with sequencing haptophyte rDNA operons. J.W.H. was supported by a University of Exeter Studentship, M.D.M.J. was supported by Natural Environment Research Council (London) Grant NE/F011709/1, and T.A.R. thanks the Leverhulme Trust for fellowship support and the Natural Environment Research Council (London) and Biotechnology and Biological Sciences Research Council for funding. At-sea research, qPCR development, and some library construction and sequencing as well as A.Z.W., H.M.W., and S.S. were supported by National Science Foundation Grant OCE-0836721, Moore Foundation Grant MMI-1668, and the David and Lucille Packard Foundation. E.K. and the Archibald Laboratory were supported by Dalhousie University's Centre for Comparative Genomics and Evolutionary Bioinformatics and the Tula Foundation. A.Z.W. and J.M.A. acknowledge support from the Canadian Institute for Advanced Research's Integrated Microbial Biodiversity Program.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The newly obtained sequences reported in this paper were deposited in the GenBank database (accession nos. HM594190–HM594247 and HM595040–HM595172).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013337108/-/DCSupplemental.
References
- 1.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 2.Graham LE, Graham JM, Wilcox LW. Algae. San Francisco, CA: Benjamin Cummings (Pearson); 2009. [Google Scholar]
- 3.Lane CE, Archibald JM. The eukaryotic tree of life: Endosymbiosis takes its TOL. Trends Ecol Evol. 2008;23:268–275. doi: 10.1016/j.tree.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Li WKW. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: Measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. [Google Scholar]
- 5.Bidigare RR, Ondrusek ME. Spatial and temporal variability of phytoplankton pigment distributions in the central equatorial Pacific Ocean. Deep Sea Research Part II-Topical Studies in Oceanography. 1996;43:809–833. [Google Scholar]
- 6.Cuvelier ML, et al. Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. Proc Natl Acad Sci USA. 2010;107:14679–14684. doi: 10.1073/pnas.1001665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon-van der Staay SY, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- 8.López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- 9.Berney C, Fahrni J, Pawlowski J. How many novel eukaryotic ‘kingdoms’? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2004;2:13. doi: 10.1186/1741-7007-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards TA, Vepritskiy AA, Gouliamova DE, Nierzwicki-Bauer SA. The molecular diversity of freshwater picoeukaryotes from an oligotrophic lake reveals diverse, distinctive and globally dispersed lineages. Environ Microbiol. 2005;7:1413–1425. doi: 10.1111/j.1462-2920.2005.00828.x. [DOI] [PubMed] [Google Scholar]
- 11.Guillou L, et al. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata) Environ Microbiol. 2008;10:3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- 12.Massana R, Pedrós-Alió C. Unveiling new microbial eukaryotes in the surface ocean. Curr Opin Microbiol. 2008;11:213–218. doi: 10.1016/j.mib.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Worden AZ, Not F. In: Microbial Ecology of the Ocean. Kirchman DL, editor. New York: Wiley; 2008. pp. 159–196. [Google Scholar]
- 14.McDonald SM, Sarno D, Scanlan DJ, Zingone A. Genetic diversity of eukaryotic ultraphytoplankton in the Gulf of Naples during an annual cycle. Aquat Microb Ecol. 2007;50:75–89. [Google Scholar]
- 15.Reyes-Prieto A, Weber APM, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- 16.Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- 17.Bodył A, Stiller JW, Mackiewicz P. Chromalveolate plastids: Direct descent or multiple endosymbioses? Trends Ecol Evol. 2009;24:119–121. doi: 10.1016/j.tree.2008.11.003. author reply 121–122. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Puerta MV, Delwiche CF. A hypothesis for plastid evolution in chromalveolates. J Phycol. 2008;44:1097–1107. doi: 10.1111/j.1529-8817.2008.00559.x. [DOI] [PubMed] [Google Scholar]
- 19.Rappé MS, Suzuki MT, Vergin KL, Giovannoni SJ. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl Environ Microbiol. 1998;64:294–303. doi: 10.1128/aem.64.1.294-303.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medlin LK, Saez AG, Young JR. A molecular clock for coccolithophores and implications for selectivity of phytoplankton extinctions across the K/T boundary. Marine Micropaleontology. 2008;67:69–86. [Google Scholar]
- 21.Bowler C, Vardi A, Allen AE. Oceanographic and biogeochemical insights from diatom genomes. Annu Rev Mar Sci. 2010;2:333–365. doi: 10.1146/annurev-marine-120308-081051. [DOI] [PubMed] [Google Scholar]
- 22.Liu H, et al. Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans. Proc Natl Acad Sci USA. 2009;106:12803–12808. doi: 10.1073/pnas.0905841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guy RA, Payment P, Krull UJ, Horgen PA. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl Environ Microbiol. 2003;69:5178–5185. doi: 10.1128/AEM.69.9.5178-5185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worden AZ, Nolan JK, Palenik B. Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component. Limnol Oceanogr. 2004;49:168–179. [Google Scholar]
- 25.Fuller NJ, et al. Analysis of photosynthetic picoeukaryote diversity at open ocean sites in the Arabian Sea using a PCR biased towards marine algal plastids. Aquat Microb Ecol. 2006;43:79–93. [Google Scholar]
- 26.Lepère C, Vaulot D, Scanlan DJ. Photosynthetic picoeukaryote community structure in the South East Pacific Ocean encompassing the most oligotrophic waters on Earth. Environ Microbiol. 2009;11:3105–3117. doi: 10.1111/j.1462-2920.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- 27.Romari K, Vaulot D. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol Oceanogr. 2004;49:784–798. [Google Scholar]
- 28.Not F, et al. Picobiliphytes: A marine picoplanktonic algal group with unknown affinities to other eukaryotes. Science. 2007;315:253–255. doi: 10.1126/science.1136264. [DOI] [PubMed] [Google Scholar]
- 29.Cuvelier ML, et al. Widespread distribution of a unique marine protistan lineage. Environ Microbiol. 2008;10:1621–1634. doi: 10.1111/j.1462-2920.2008.01580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slapeta J, Moreira D, López-García P. The extent of protist diversity: Insights from molecular ecology of freshwater eukaryotes. Proc Biol Sci. 2005;272:2073–2081. doi: 10.1098/rspb.2005.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi XL, Marie D, Jardillier L, Scanlan DJ, Vaulot D. Groups without cultured representatives dominate eukaryotic picophytoplankton in the oligotrophic South East Pacific Ocean. PLoS ONE. 2009;4:e7657. doi: 10.1371/journal.pone.0007657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li WKW, McLaughlin FA, Lovejoy C, Carmack EC. Smallest algae thrive as the Arctic Ocean freshens. Science. 2009;326:539. doi: 10.1126/science.1179798. [DOI] [PubMed] [Google Scholar]
- 33.Caron DA, Countway PD. Hypotheses on the role of the protistan rare biosphere in a changing world. Aquat Microb Ecol. 2009;57:227–238. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.