Abstract
It has been axiomatic in the field of NF-κB signaling that the formation of a stable complex between NF-κB and the ankyrin repeat protein IκBα precludes the interaction of NF-κB with DNA. Contradicting this assumption, we present stopped-flow fluorescence and NMR experiments that give unequivocal evidence for the presence of a ternary DNA–NF-κB–IκBα complex in solution. Stepwise addition of a DNA fragment containing the κB binding sequence to the IκBα–NF-κB complex results in changes in the IκBα NMR spectrum that are consistent with dissociation of the region rich in proline, glutamate, serine, and threonine (PEST) and C-terminal ankyrin repeat sequences of IκBα from the complex. However, even at high concentrations of DNA, IκBα remains associated with NF-κB, indicated by the absence of resonances of the free N-terminal ankyrin repeats of IκBα. The IκBα-mediated release of NF-κB from its DNA-bound state may be envisioned as the reverse of this process. The initial step would consist of the coupled folding and binding of the intrinsically disordered nuclear localization sequence of the p65 subunit of NF-κB to the well-structured N-terminal ankyrin repeats of IκBα. Subsequently the poorly folded C-terminal ankyrin repeats of IκBα would fold upon binding to the p50 and p65 dimerization domains of NF-κB, permitting the negatively charged C-terminal PEST sequence of IκBα to displace the bound DNA through a process of local mass action.
Keywords: signal transduction, transcription factor
The NF-κB transcriptional factors regulate cell growth, immune responses, inflammatory viral responses, and apoptotic death by binding to thousands of DNA κB sites distributed in genome (1–5). NF-κBs consist of a family of transcriptional activators, made up of either homodimers or heterodimers of RelA (p65), p50, p52, c-Rel, and RelB (4). The heterodimer p65/p50 was the first discovered (6) and is the most abundant form of NF-κB (7, 8). The p50/p65 heterodimer is predominantly regulated by IκBα and IκBβ (1, 7) In resting cells, IκB associates with NF-κB to form an inactive complex, which is sequestered in the cytoplasm (7). Once the pathway is activated, NF-κB is released from the IκB complex and translocates into the nucleus where it binds consensus DNA κB sites (7) and activates downstream genes including the gene coding for IκBα itself, leading to postinduction repression (9).
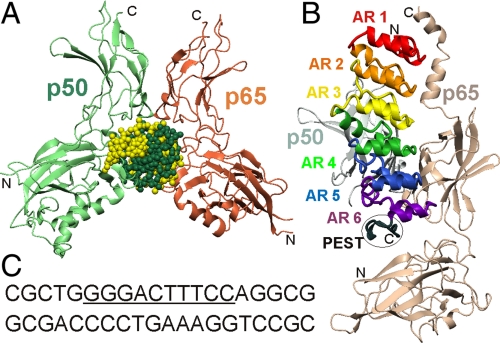
Each NF-κB monomer contains a conserved Rel homology region (RHR) (4, 7, 8) consisting of an N-terminal domain and a C-terminal dimerization domain (10, 11). The two RHRs in a functional NF-κB homo- or heterodimer use various loops from the edges of the N- and C-terminal domains to mediate DNA contacts (Fig. 1A) (11), whereas the ankyrin repeat domain and region rich in proline, glutamate, serine, and threonine (PEST) sequence of IκBα primarily interacts with the C-terminal dimerization domains (Fig. 1B) (12, 13). Interaction of the nuclear localization segment (NLS) at the C terminus of the p65 dimerization domain with the first three ankyrin repeats (ARs) of IκBα and interaction of the N-terminal domain with the IκBα PEST are both critical for establishing the 40 pM binding affinity of the NF-κB–IκBα complex (14, 15). The poor definition of the PEST sequence in the crystal structures (12, 13) is consistent with considerable flexibility in this segment within the complex.
Fig. 1.
Structures of NF-κB complexes. Ribbon representations of the backbones of NF-κB p50 (light green) and p65 (coral) in complex with (A) the immunoglobulin κB DNA sequence (11) (green and yellow spheres) [Protein Data Bank (PDB) ID 1VKX]; (B) the ankyrin repeat domain of IκBα (12) (PDB 1NFI). The ankyrin repeats 1–6 are colored red, orange, yellow, green, blue, and purple, respectively, with the PEST sequence (black) circled. (C) nucleotide sequence of the DNA duplex used in the NMR experiments. The κB recognition sequence is underlined.
We recently showed that IκBα dramatically increases the dissociation rate of NF-κB from the DNA, thus effectively removing NF-κB from transcription sites (16). The weakly folded fifth and sixth ARs of IκBα (17–20) were shown to be essential for this phenomenon, because mutations designed to stabilize AR 5–6 resulted in reduced ability to “strip” NF-κB from the DNA (16, 21). This region undergoes a structural and dynamic transition as it folds upon binding to NF-κB (18, 19), and the center of IκBα, principally AR 3, becomes more flexible in the complex than in the free state (19, 20). In order to probe the role of this localized flexibility of IκBα in its primary function in removal of NF-κB from its cognate DNA complex, we addressed the reverse process, competition by the κB DNA sequence for NF-κB in its complex with IκBα. The effects of the addition of κB DNA on the NMR spectrum of IκBα in complex with the p50/p65 NF-κB heterodimer were assessed by observing the areas of the spectrum where the cross-peaks characteristic of the complex disappeared and were replaced by signals characteristic of the free protein. Somewhat unexpectedly, we observed signals characteristic of the IκBα–NF-κB complex even at high DNA concentrations. We interpret this observation to indicate the presence of a weak ternary complex in solution between NF-κB, IκBα, and DNA. Stopped-flow fluorescence experiments confirm the formation of a ternary complex and show that this complex has lower stability than either the NF-κB–DNA or the NF-κB–IκBα complexes.
Results
Addition of DNA to the NF-κB–IκBα Complex.
A 20-mer DNA duplex containing a κB sequence (Fig. 1C) was prepared and added stepwise in small aliquots to the IκBα–NF-κB complex. During the titration, the one-dimensional spectrum of the imino protons of the DNA are significantly broadened (Fig. S1), providing evidence that the DNA becomes associated with a high-molecular-weight complex. At DNA concentrations lower than that of the IκBα–NF-κB complex, the imino proton resonances of DNA could barely be detected. At concentration ratios > 1∶1, signals corresponding to free DNA are observed, consistent with a stoichiometry of 1∶1 for the complex.
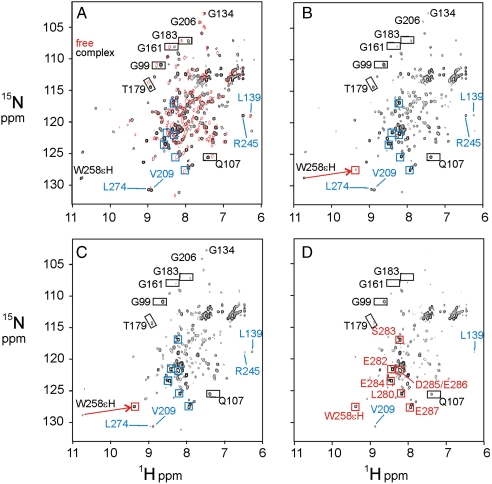
The transverse relaxation optimized heteronuclear single-quantum coherence (TROSY-HSQC) spectrum of the complex between 15N-labeled IκBα(67–287) and the heterodimer of the complete RHRs of p50 and p65 (RelA) is well dispersed, with uniform cross-peak intensity, whereas the spectrum of free IκBα(67–287) shows well-resolved cross-peaks corresponding only to the first four ARs; the cross-peaks for the fifth and sixth ARs are broadened or missing (19) (Fig. 2A). Importantly, some well-separated cross-peaks in the first four ARs have different chemical shifts in the free IκBα as compared to the bound providing a definitive indication of whether ARs 1–4 remain bound (Fig. 2A).
Fig. 2.
Effects of DNA addition on the IκBα–NF-κB complex. (A) Black contours show the 900 MHz 1H-15N TROSY-HSQC spectrum of [2H,15N]-labeled IκBα(67–287) in complex with [2H]-labeled NF-κB p50(39–350)/p65(19–321). Red contours show the 800 MHz 1H-15N TROSY-HSQC spectrum of free [2H,15N]-labeled IκBα (67–287) in the absence of NF-κB or DNA. Blue squares show the positions of cross-peaks of the PEST sequence in free IκBα. (B–D) 1H-15N TROSY-HSQC spectrum at 900 MHz of [2H,15N]-labeled IκBα (67–287) in complex with [2H]-labeled NF-κB p50(39–350)/p65(19–321) in the presence of added DNA in molar ratios (B) 1∶0.5, (C) 1∶1, and (D) 1∶4. The IκBα–NF-κB complex concentration was 0.12 mM in NMR buffer. Representative resonances are labeled. Emerging resonances of the free IκBα PEST sequence are indicated by blue squares in B and labeled red squares in D.
The influence of the addition of DNA on the IκBα in complex with NF-κB was assessed using 1H-15N TROSY-HSQC spectra (Fig. 2). Addition of DNA to the complex in concentration ratios of 1∶0 (Fig. 2A), 1∶0.5 (Fig. 2B), 1∶1 (Fig. 2C), and 1∶4 (Fig. 2D) resulted in a lowered intensity for most of the cross-peaks. However, with the exception of those of the C-terminal PEST sequence, the cross-peaks corresponding to the free form of IκBα do not appear as expected if the IκBα were to dissociate, even at a sixfold molar excess of DNA over IκBα–NF-κB.
The lowered intensity of the IκBα 1H-15N HSQC spectrum in the presence of DNA is consistent with the formation of a high-molecular-weight complex. The fact that the resonances remain in the same positions as for the binary IκBα–NF-κB complex indicates that the DNA has not caused the dissociation of the IκBα from the binary complex. The presence of a high-molecular-weight complex containing DNA is indicated by the broadening of the 1D imino proton spectrum (Fig. S1).
Detection of the Ternary Complex by Fluorescence.
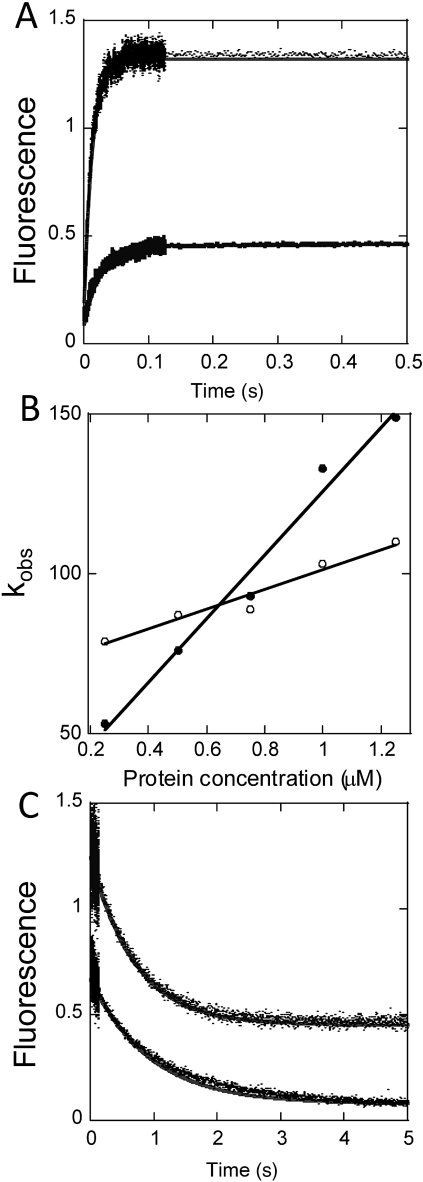
To obtain an independent experimental verification of the formation of the NF-κB–IκBα–DNA ternary complex, we performed stopped-flow fluorescence experiments in which a pyrene-labeled DNA hairpin was rapidly mixed with either NF-κB alone or NF-κB in complex with IκBα, and binding was detected by the change in fluorescence intensity of the pyrene label. Binding of the DNA to NF-κB alone was rapid, with an association rate of 1.05 × 108 M-1 s-1 (Fig. 3 A and B) and a dissociation rate of 0.57 s-1 (Fig. 3C), yielding a calculated binding affinity of 5.5 nM (16). Remarkably, the DNA also associated with the NF-κB–IκBα complex although the association rate was slower, 2.5 × 107 M-1 s-1 (Fig. 3 A and B) and the dissociation rate was faster, 1.4 s-1 (Fig. 3C), giving an overall binding affinity of 56 nM. Importantly, the binding affinity of the ternary complex is tight enough that at the concentrations of the NMR experiments the complex should be 100% formed even at a DNA∶NF-κB–IκBα ratio of 1∶1. All previous reports, particularly of experiments in vivo, have assumed that the binding of DNA and IκBα is mutually exclusive; no stable ternary complex has ever been observed. Our results suggest that the most likely reason that the ternary complex has not been previously observed, for example in gel-shift assays, is due to the presence of a competitive interaction with a higher affinity (formation of the binary complex, with 6 nM affinity, compared to formation of the ternary complex with 56 nM affinity). Nevertheless, at the concentrations used in the NMR experiments (0.12 mM for the IκBα–NF-κB complex), the formation of a 1∶1 complex would be expected if the binding affinity is tighter than 120 nM. Our observations of such a complex yield important insights into the steps that take place during competition between DNA and IκBα for NF-κB.
Fig. 3.
Stopped-flow fluorescence measurements of binary and ternary complex formation. A pyrene-labeled DNA hairpin was combined with either NF-κB or the NF-κB–IκBα complex to compare binding association and dissociation rates for the binary and ternary complexes. (A) Representative binding association curves for pyrene-DNA (0.25 μM) to NF-κB (upper trace) or NF-κB-IκBα (lower trace), with the fits shown as solid lines. (B) Plot of binding rate constant as a function of protein concentration, used to obtain the association rate constant ka for the binary (●) or ternary (○) complex. (C) Dissociation curves obtained from mixing 10-fold excess unlabeled DNA with the NF-κB–DNA (lower trace) or NF-κB–IκBα–DNA (upper trace) complexes, to obtain the dissociation rate constants. Additional experiments with 50-fold and 100-fold excess of unlabeled DNA gave similar results.
Stepwise Effect of DNA Addition on the NF-κB/IκBα Complex.
The presence of an apparent ternary interaction between DNA, NF-κB, and IκBα under NMR conditions allows us to identify the sites on NF-κB-bound IκBα that are preferentially affected by DNA binding. The series of spectra shown in Fig. 2 demonstrate that different areas of IκBα are affected as the DNA concentration is increased. At low DNA concentrations (protein complex∶DNA = 1∶0.5) (Fig. 2B), the most significant change is seen for the signals from C-terminal PEST sequence. A new set of PEST signals emerges, corresponding to the chemical shifts for the PEST sequence in free IκBα (Fig. 2B, blue boxes). This observation suggests that the PEST portion of IκBα is preferentially affected by DNA addition, and that it is dissociated from the complex to form a flexible disordered structure, as observed in the free state.
One particular site, the side chain of the tryptophan residue at position 258, gives a unique site-specific insight into the events that occur as DNA is added to the NF-κB–IκBα complex. In the NF-κB–IκBα complex, the W258(ϵH) is hydrogen bonded to the backbone CO of Q278 (22), immobilizing the PEST sequence on the surface of AR 6 (13). The downfield shift of the W258(ϵH) resonance is the diagnostic feature that identifies this hydrogen bond (22). Addition of DNA at the 1∶0.5 level causes a dramatic upfield shift of this resonance (Fig. 2B, red arrow), which is virtually complete at a molar ratio of 1∶1 (Fig. 2C). The position of the W258(ϵH) cross-peak in Fig. 2C corresponds to that seen when the hydrogen bond is not present (22). These observations are consistent with the dissociation of the PEST sequence from the NF-κB–IκBα complex upon addition of DNA.
Unlike those of the PEST sequence, the remainder of the IκBα resonances do not show evidence of dissociation from the complex. Instead, lowered intensities are observed for resonances corresponding to sites throughout the IκBα molecule, and the intensities do not decrease uniformly. The comparison between V209 (AR 4) and L274 (AR 6) provides an example. The two cross-peaks are of similar intensity in the absence of DNA (Fig. 2A) but the L274 cross-peak is more highly attenuated at a DNA concentration of 1∶1 (Fig. 2C). Similarly, the cross-peak of R245 (AR 5) becomes much weaker than that of L139 (AR 3) at a DNA concentration of 1∶1 (Fig. 2C), although it was stronger before the addition of DNA (Fig. 2A). In general, the intensities of cross-peaks corresponding to residues in AR 5 and 6 are more significantly attenuated than those of residues in other repeats. When excess DNA is added (molar ratio 1∶4.0; Fig. 2D), the intensities of cross-peaks for residues in AR 1–4 also decrease, the resonances of AR 5 and 6 residues become too weak to be clearly detected, the signals corresponding to the bound PEST disappear, and the cross-peaks corresponding to the free PEST residues increase to full intensity (Fig. 2D, red boxes). However, even at a high molar excess of DNA, we did not observe any emerging resonances corresponding to the first four ankyrin repeats of free IκBα.
Localization of DNA Binding from the Spectrum of IκBα.
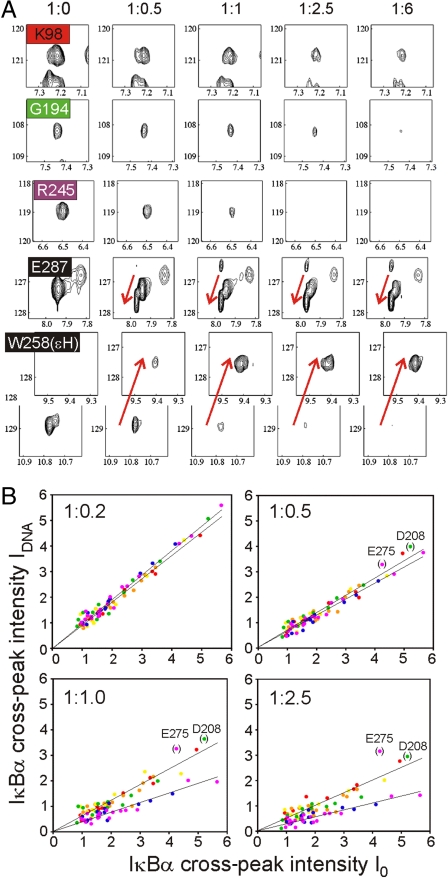
The different local effects observed on the NMR spectra of IκBα in complex with NF-κB as DNA is added are consistent with the structure of the complexes of NF-κB with DNA and IκBα. Fig. 4 shows a comparison of the behavior of representative resonances selected from different ankyrin repeats and from the PEST sequence (a more comprehensive series of resonances is shown in Fig. S2). The sensitivity of the IκBα resonances to the addition of DNA increases from the N terminus to the C terminus of IκBα: The resonances of AR 1–4 are the least sensitive, and most resonances are still observed upon addition of excess DNA. The spectra of AR 1–3 appear to be unchanged between molar ratios 1∶2.5 and 1∶6 (Fig. 4A). For AR 5–6, peak attenuation is more significant, and occurs at lower molar ratios of DNA. As shown in Fig. 2, the PEST sequence shows the most sensitivity to the addition of DNA, with a qualitative difference from the rest of IκBα: The cross-peaks for this region are shifted to their positions in the spectrum of the free protein.
Fig. 4.
Effects of DNA binding for individual ankyrin repeats. (A) Representative cross-peaks from the 900 MHz 1H-15N TROSY-HSQC spectra of NF-κB-bound IκBα are color coded as in Fig. 1B, and shown (plotted at the same contour levels in each panel) at different molar ratios of protein complex to DNA. A more comprehensive collection of similar panels is shown in Fig. S2. (B) Correlation of IDNA [the intensity of an IκBα resonance in complex with NF-κB p50(39–350)/p65(19–321) following addition of a 1∶1 molar ratio of DNA] and I0 [the intensity of the same IκBα resonance in complex with NF-κB p50(39–350)/p65(19–321) in the absence of DNA] at various DNA molar ratios. Each residue is depicted in a colored dot and its position on the plot represents the values of IDNA and I0 associated with the residue at that particular mole ratio of DNA∶IκBα-NF-κB complex. Dots are colored according to the ankyrin repeat, using the same colors as in Fig. 1B. Molar ratios of [NF-κB-IκBα complex] to DNA are shown in each panel. Separate linear fits have been applied to residues of AR 1–4 and AR 5–6. Data for residues D208 and E275 were excluded from the fitting.
Regional differences in the effects of added DNA are plotted quantitatively in Fig. S3, as the experimental intensity ratios (IDNA/I0) of TROSY-HSQC cross-peaks. IDNA represents the cross-peak intensity for NF-κB-bound IκBα in the presence of a 1∶1 molar ratio of DNA and I0 is the intensity of NF-κB-bound IκBα without DNA. The IDNA/I0 ratio gives a measure of the population of IκBα remaining in the initial NF-κB-bound state; residues that are sensitive to DNA have lower values of IDNA/I0. Generally higher values of IDNA/I0 (0.5–0.7) are seen for AR 1–4 and lower values (0.3–0.5) for AR 5–6. A monotonic decrease is observed between L205 and C215, which corresponds to the connecting loop region between AR 4 and AR 5.
This difference between regions of IκBα is found at all molar ratios. Fig. 4B shows the correlation between IDNA and I0 for individual residues under different molar ratios. At low molar ratios (1∶0.2 and 1∶0.5), a uniform linear dependence is seen for all residues. At higher DNA concentrations, the correlation between I0 and IDNA is still linear, but it is divided according to the location of the residue that corresponds to the cross-peak: The linear dependence is shallower for residues in AR 5–6 (blue and purple dots) compared to that for residues in AR 1–4 (red, orange, yellow, and green dots). These results imply that DNA perturbs residues in AR 1–4 and AR 5–6 differently. The reason why a few residues, such as D208 and E275, cannot be fit by either linear correlation is unclear, but might correlate to local dynamics at these particular positions. The data from these cross-peaks were excluded from the fitting procedure.
A plot of the fitted slopes from Fig. S3 as a function of DNA molar ratio for the three portions of IκBα: AR 1–4, AR 5–6, and PEST sequence is shown in Fig. S4. Values represent the fraction remaining in the binary IκBα–NF-κB complex. The three portions demonstrate distinct profiles: Signal decay for the (bound) PEST sequence is the most significant, but AR 5–6 decays more completely than AR 1–4. Each decay fits well to a single exponential curve and the corresponding rate constants are similar, as demonstrated by the similarity of the profiles of curves normalized to the final states (Fig. S4, Inset). These results suggest that the DNA interacts with the NF-κB–IκBα complex in a simple two-state process. However, the regions corresponding to AR 1–4, AR 5–6, and the PEST sequence behave as individual units showing distinct responses to DNA binding during the titration.
Discussion
Addition of DNA to the NF-κB–IκBα complex results in changes to the NMR spectrum of IκBα. The effect of DNA addition differs between three regions of IκBα, AR 1–4, AR 5–6, and PEST, as shown by the separate linear correlations for AR 1–4 and AR 5–6 in Fig. 4B and by the single exponential fits for the three regions in Fig. S4. Upon addition of even small amounts of DNA, the PEST sequence is released from the NF-κB, indicated by the observation of resonances of free PEST in the NMR spectrum and the disappearance of the resonances of NF-κB-bound PEST. The observation of two distinct sets of resonances for each residue indicates slow exchange of IκBα between the binary and ternary complexes, consistent with the nanomolar affinity of the ternary complex. The dissociation of the PEST sequence from the complex is consistent with our understanding of the structures of the NF-κB–IκBα (12, 13) and NF-κB–DNA (11) complexes: The PEST interacts directly with the side chains of residues in the DNA-contact loop of p65 (22).
The observed effect on AR 5–6 must be interpreted differently. AR 5–6 forms a flexible molten globule-like structure in free IκBα (18, 19). The NMR spectroscopic signature of this structure is difficult to distinguish, because most of the cross-peaks for AR 5–6 in the free protein are severely broadened or missing (19). We therefore cannot be completely sure about the state of this region of the protein during the DNA titration. Some insight is obtained from the behavior of the resonances of this region in the spectrum of complexed IκBα. Preferential attenuation of the cross-peaks corresponding to AR 5–6 of the bound IκBα is observed at low-to-medium concentrations of added DNA, indicated by the comparisons in Figs. S2–S4, indicating that it is likely that AR 5–6 are dissociated from the ternary complex. Because the concentration dependence of the peak intensities determined for the PEST and AR 5–6 regions are extremely similar (Fig. S4, Inset), it is likely that AR 5–6 dissociates from NF-κB in a similar slow exchange process to that of the PEST. However, we cannot rule out the participation of an intermediate exchange process where AR 5–6 is weakly associated with NF-κB in the ternary complex. Either mechanism (or a combination of the two) can be invoked to explain the peak attenuation in AR 5–6; both are consistent with the disruption of the coupled folding and binding of AR 5–6 in the binary complex upon addition of DNA.
Our results provide evidence that part of IκBα remains associated when DNA is titrated into the NF-κB-IκBα complex. If IκBα was completely dissociated from the complex by excess DNA, we would expect to see the appearance of resonances belonging to free IκBα. Because the fifth and sixth ARs are weakly folded in free IκBα, we might not expect to see clear evidence for the formation of the free state of this region. However, the NMR spectrum of IκBα(67–206) comprising the first four ARs is well resolved (19, 20). None of the resonances of AR 1–4 of free IκBα(67–206) appear, even at [NF-κB–IκBα complex]∶DNA molar ratios of 1∶6, whereas the cross-peaks characteristic of the NF-κB-bound IκBα remain at roughly the same intensities. We conclude that the IκBα remains associated with the NF-κB–DNA complex in solution under these conditions, forming a ternary complex. The lowered intensity of the cross-peaks corresponding to ARs 1–4, without the appearance of resonances of the free form or any detectable change in the position of the cross-peaks, is most likely due to the formation of a higher-molecular-weight complex, perhaps with a contribution from anisotropic tumbling and an increase in the effective radius due to the loosened structure of the dissociated AR 5 and 6 of IκBα. Given that the binding affinity of IκBα for full-length NF-κB is 40 pM at 37 °C and too tight to measure at 25 °C (14), it is actually not too surprising that the IκBα does not fully dissociate upon DNA binding. We previously showed that, although the interface of the IκBα–NF-κB complex spans over 4,000 Å2, only a few residues at either end are critical for establishing the binding energy, and the interaction between the first AR of IκBα and the NLS polypeptide of NF-κB(p65) alone accounts for approximately half of the binding affinity (14).
These data lend support to a model in which the first four ARs of IκBα remain bound in a ternary complex, where AR 1–4 remain bound to the RelA (p65) NLS polypeptide and parts of the C-terminal dimerization domains of NF-κB p50 and p65, whereas the N-terminal domains of p50 and p65 are associated with the DNA. The intrinsically disordered PEST sequence and the incompletely folded molten globule-like AR 5–6 that are present in free IκBα (19) might well be primed to dissociate readily from NF-κB upon addition of a competing ligand. We may speculate that the lower energy required to dissociate the flexible PEST sequence may predispose this region to dissociate first upon addition of DNA, but it is clear that either the PEST and the AR 5–6 regions dissociate together or AR 5–6 dissociation follows PEST dissociation upon DNA titration. Thus, the DNA-binding induced effects can be integrated as shown in Fig. 5, where the stippled region in the ternary complex represents a conformational ensemble that may include forms where AR 5–6 are free (and molten globule-like, losing their NMR resonances) or bound in the ternary complex (and therefore subject to resonance broadening due to the high molecular weight).
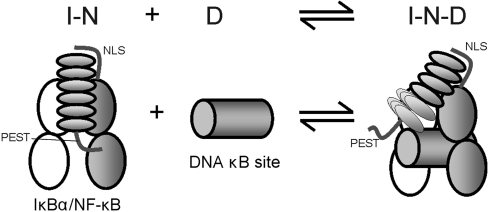
Fig. 5.
Schematic diagram showing the binding model for ternary complex formation. DNA (D) is added to the IκBα-NF-κB complex (I-N). IκBα is represented by a set of six ellipses with the attached PEST sequence, p65 RHR as two gray ellipses with the attached NLS sequence, p50 RHR as two white ellipses, and the DNA κB site as a gray cylinder. Addition of DNA causes the dissociation of the PEST sequence of IκBα from the DNA-binding site of NF-κB and attenuates NMR resonances in AR 5–6, most likely by dissociating AR 5–6 to form the molten globule-like structure characteristic of free IκBα. However, even at high DNA concentrations, where the DNA-binding site is fully occupied and the PEST sequence and AR 5–6 are dissociated from NF-κB, the NMR signals of free IκBα do not appear, indicating that IκBα remains associated, forming a ternary complex (I-N-D).
The observation of ternary complex formation as DNA is added to the NF-κB–IκBα complex can inform on the likely mechanism of the reverse reaction, the enhanced dissociation of NF-κB from DNA mediated by newly synthesized IκBα when the signal is to be turned off (16). When IκBα approaches the DNA-bound NF-κB, the first two ARs, which are stably folded in the free protein, are immediately capable of binding to the intrinsically unstructured NF-κB(p65) NLS segment. This recognition event is completely independent of the recognition of DNA by the NF-κB DNA-binding domains, because a mutant with a deletion of residues 305–325 in the p65 NLS sequence still binds to DNA with the same affinity as the wild-type protein (14). We suggest that these two independent interactions at opposite ends of the NF-κB heterodimer allow the formation of the ternary complex. The remaining ankyrin repeats of IκBα are now in sufficiently close proximity to their binding site on NF-κB for them to gradually form the bound structure, as a result of local mass action. Once the ternary complex is formed, the fifth and sixth ARs can undergo coupled folding and binding in a manner similar to that proposed to occur during the formation of the NF-κB–IκBα complex (19). This process can be likened to the “snap-lock” mechanism of binding of zinc fingers to DNA (23). The final step, binding of the C-terminal PEST sequence, is critical for the dissociation of NF-κB from the DNA: The intrinsically disordered PEST sequence competes successfully for binding to the DNA-contact loop in the NF-κB DNA-binding domain (22), and electrostatic repulsion between the negatively charged PEST sequence and DNA likely contributes to the dissociation of the NF-κB–IκBα complex from the DNA (16).
Materials and Methods
Protein Preparation.
The specific IκBα–NF-κB complex of labeled IκBα with the deuterated heterodimer of p50(39–350) with p65(19–321) (RelA) was prepared by the reported coexpression method (19, 24). Both p50(39–350) and p65(19–321) include their respective N-terminal dimerization and C-terminal DNA-binding domains; p65(19–321) further includes an NLS segment at the C terminus and p50(39–350) incorporates a hexahistidine tag at the N terminus (19). The high molecular size of the complex (∼94 kDa) necessitated the use of extensive deuterium labeling to compensate for rapid NMR relaxation. The [2H]-NF-κB subunits were coexpressed in Escherichia coli cultured in D2O M9 minimal medium. The [2H,15N]-labeled IκBα(67–287) was prepared in the same medium with deuterium-labeled glucose and 15N ammonium sulfate. Protein complexes were purified using the streamlined method described previously (19), where cell lysates derived from the two cultures were mixed and the complex subsequently purified by a nickel affinity chromatography followed by gel filtration. Complexes purified to homogeneity were exchanged into NMR buffer (25 mM Tris pH 7.5/50 mM NaCl/1 mM EDTA/1 mM DTT in 90% H2O/10% D2O). The purities of the protein products were judged to be higher than 95% by SDS-PAGE. The sample 2H enrichment is estimated to be > 90% in IκBα and > 70% in NF-κB, adjudged by 1D NMR spectra. A final sample concentration of 0.12 mM for the complex was used in the titration experiment.
DNA Preparation.
For the NMR experiments, two complementary DNA oligonucleotides (Fig. 1C) were chemically synthesized. The sequence contains the 10-base-pair HIV-κB/Igκ site (underlined) (11, 16) specific for binding of the p50/p65 NF-κB heterodimer. Duplex DNA was prepared by annealing the two strands by heating to 95 °C followed by cooling slowly to room temperature in DNA buffer (25 mM Tris pH 7.5/50 mM NaCl). Duplex DNA was separated from unannealed single-stranded DNA by an FPLC Sephacryl S-100 size exclusion column in NMR buffer. The purified DNA product was concentrated to ∼1 mM stock for the titration experiment. The concentration was estimated measuring the absorbance of the solution at 260 nm (OD260), considering the molar extinction coefficient ε260 to be 244.76 mM-1 cm-1. The purity was judged by OD280/OD260 ratio of ∼1.9. For the stopped-flow experiments, a hairpin DNA containing the IFN-κB recognition sequence (16), GGGAAATTCCTCCCCCAGGAATTTCCC, labeled at the 5′ end with a six carbon amine linker was labeled with pyrene succinimide using established procedures (25).
NMR Titration Experiments.
The prepared DNA duplex was titrated stepwise in small aliquots to the desired concentration into IκBα–NF-κB complex. Only the IκBα component is labeled with 15N, and it is therefore the only component detectable in 1H-15N NMR spectra. TROSY-type HSQC spectra (26) were acquired, with data size 2048 × 128 complex points, 256 scans, and a 1.5-s delay time between each scan. The spectra were acquired at 28 °C on a Bruker Avance900 spectrometer. Data were processed using NMRpipe (27) with linear prediction to 2048 × 256 complex points and were analyzed using NMRView (28) to estimate the peak intensities. Only resonances with adequately resolved peaks in 1H-15N TROSY-HSQC spectra were analyzed: A total of 83 resonances were used in the final analysis, with 12, 14, 11, 13, 12, and 21 resonances from AR 1 to 6, respectively.
Stopped-Flow Fluorescence.
The kinetic experiments were performed on an Applied Photophysics PiStar stopped-flow fluorimeter at 25 °C 0.25 μM starting labeled DNA with 2.5 mL syringe volumes and a 200 μL mixing volume. The association kinetics for the pyrene-DNA were measured in triplicate at six concentrations of NF-κB heterodimer (0.25, 0.5, 0.75, 1, 1.25, 1.5 μM) maintaining a constant concentration of pyrene-DNA (0.25 μM before mixing). In the first 0.125 s, 5,000 data points were collected and another 5,000 for the remainder of the experiment. The dead time of the instrument is approximately 30 ms. The association rate constant for each trace was obtained from single exponential fits (ProFit software). The dissociation rate constant of the NF-κB•pyrene-DNA complex was measured at a fixed concentration of the NF-κB•pyrene-DNA by adding an excess of unlabeled pyrene-DNA (1∶10, 1∶50, and 1∶100, NF-κB∶pyrene-DNA). The experiments to measure the binding affinities for the ternary complex were performed in an identical manner except that IκBα was included at a 1∶1 ratio with the NF-κB.
Supplementary Material
Acknowledgments.
We thank Maria Martinez-Yamout and Peter Wright for valuable discussions, Euvel Manlapaz for technical assistance, and Gerard Kroon for help with NMR experiments. This work was supported by Grant GM71862 from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014323108/-/DCSupplemental.
References
- 1.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 3.Martone R, et al. Distribution of NF-kappaB-binding sites across human chromosome 22. Proc Natl Acad Sci USA. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-κB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 5.Schreiber J, et al. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc Natl Acad Sci USA. 2006;103:5899–5904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen R, Baltimore D. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 7.Gilmore TD. Introduction to NF-κB: Players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin AS. The NF-κB and I-κB proteins: New discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 9.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh G, Van Duyne G, Ghosh S, Sigler PB. Structure of NF-κB p50 homodimer bound to a κB site. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 11.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs MD, Harrison SC. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 13.Huxford T, Huang DB, Malek S, Ghosh G. The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 14.Bergqvist S, et al. Thermodynamics reveal that helix four in the NLS of NF-κB p65 anchors IκBα, forming a very stable complex. J Mol Biol. 2006;360:421–434. doi: 10.1016/j.jmb.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergqvist S, Ghosh G, Komives EA. The IκBα/NF-κB complex has two hot spots, one at either end of the interface. Protein Sci. 2008;17:2051–2058. doi: 10.1110/ps.037481.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergqvist S, et al. Kinetic enhancement of NF-κB•DNA dissociation by IκBα. Proc Natl Acad Sci USA. 2009;106:19328–19333. doi: 10.1073/pnas.0908797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Biophysical characterization of the free IκBα ankyrin repeat domain in solution. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Truhlar SM, Torpey JW, Komives EA. Regions of IκBα that are critical for its inhibition of NF-κB. DNA interaction fold upon binding to NF-κB. Proc Natl Acad Sci USA. 2006;103:18951–18956. doi: 10.1073/pnas.0605794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sue SC, Cervantes C, Komives EA, Dyson HJ. Transfer of flexibility between ankyrin repeats in IκBα upon formation of the NF-κB complex. J Mol Biol. 2008;380:917–931. doi: 10.1016/j.jmb.2008.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cervantes CF, et al. Functional dynamics of the folded ankyrin repeats of IκBα revealed by nuclear magnetic resonance. Biochemistry. 2009;48:8023–8031. doi: 10.1021/bi900712r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreiro DU, et al. Stabilizing IκBα by “consensus” design. J Mol Biol. 2007;365:1201–1216. doi: 10.1016/j.jmb.2006.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sue SC, Dyson HJ. Interaction of the IκBα C-terminal PEST sequence with NF-κB: Insights into the inhibition of NF-κB DNA binding by IκBα. J Mol Biol. 2009;388:824–838. doi: 10.1016/j.jmb.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laity JH, Dyson HJ, Wright PE. DNA-induced α-helix capping in conserved linker sequences is a determinant of binding affinity in Cys2-His2 zinc fingers. J Mol Biol. 2000;295:719–727. doi: 10.1006/jmbi.1999.3406. [DOI] [PubMed] [Google Scholar]
- 24.Chen FE, Kempiak S, Huang DB, Phelps C, Ghosh G. Construction, expression, purification and functional analysis of recombinant NFκB p50/p65 heterodimer. Protein Eng. 1999;12:423–428. doi: 10.1093/protein/12.5.423. [DOI] [PubMed] [Google Scholar]
- 25.Studer SM, Joseph S. Binding of mRNA to the bacterial translation initiation complex. Methods Enzymol. 2007;430:31–44. doi: 10.1016/S0076-6879(07)30002-5. [DOI] [PubMed] [Google Scholar]
- 26.Schulte-Herbruggen T, Sørensen OW. Clean TROSY: Compensation for relaxation-induced artifacts. J Magn Reson. 2000;144:123–128. doi: 10.1006/jmre.2000.2020. [DOI] [PubMed] [Google Scholar]
- 27.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.