Abstract
The 10S self-inhibited monomeric conformation of myosin II has been characterized extensively in vitro. Based upon its structural and functional characteristics, it has been proposed to be an assembly-competent myosin pool in equilibrium with filaments in cells. It is known that myosin filaments can assemble and disassemble in nonmuscle cells, and in some smooth muscle cells, but whether or not the disassembled pool contains functional 10S myosin has not been determined. Here we address this question using human airway smooth muscle cells (hASMCs). Using two antibodies against different epitopes on smooth muscle myosin II (SMM), two distinct pools of SMM, diffuse, and stress-fiber–associated, were visualized by immunocytochemical staining. The two SMM pools were functional in that they could be interconverted in two ways: (i) by exposure to 10S- versus filament-promoting buffer conditions, and (ii) by exposure to a peptide that shifts the filament-10S equilibrium toward filaments in vitro by a known mechanism that requires the presence of the 10S conformation. The effect of the peptide was not due to a trivial increase in SMM phosphorylation, and its specificity was demonstrated by use of a scrambled peptide, which had no effect. Based upon these data, we conclude that hASMCs contain a significant pool of functional SMM in the 10S conformation that can assemble into filaments upon changing cellular conditions. This study provides unique direct evidence for the presence of a significant pool of functional myosin in the 10S conformation in cells.
Keywords: filament assembly, myosin phosphorylation, nuclear myosin II, stress fibers, cytoskeleton
The ability to adopt a self-inhibited conformation appears to be a common function of motor proteins. The structural mechanisms for inhibition are varied but in general involve the intramolecular interaction of the noncatalytic tail domains with the catalytic or regulatory domains. It is generally assumed that these inhibited monomeric forms, which have been characterized in vitro, also exist in cells but this has been difficult to demonstrate directly.
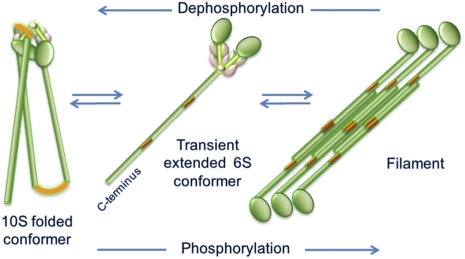
The smooth muscle isoform, smooth muscle myosin II (SMM), like many other myosin II isoforms such as nonmuscle myosin II (NMM), forms a self-inhibited conformer that has been extensively studied in vitro ([reviewed by Cremo and Hartshorne (1)]. This self-inhibited conformer (10S, the conformation of myosin II that sediments at 10 Svedbergs) has a more compact shape than the extended conformer (6S, the conformation of myosin II that sediments at 6 Svedbergs) (2, 3). Fig. 1 illustrates the equilibrium between 10S and filaments, which most likely proceeds through the transient 6S intermediate (4). The position of the equilibrium depends upon the solvent conditions (5, 6). At physiological ionic strength in the presence of ATP, myosin II is most stable in the 10S conformation if the regulatory light chains (RLC) are unphosphorylated. Myosin filament assembly is promoted by phosphorylation of the RLCs at Ser-19 by myosin light chain kinase. This filamentous phosphorylated state is responsible for force generation in smooth muscle.
Fig. 1.
Schematic of conformational states of SMM derived from in vitro studies. Green, tail with red bending regions and globular heads; brown and pink represent ELC, and RLC, respectively (Fig. 4 gives a more detailed description of 10S structure). Equilibration between filaments and 10S likely proceeds through the transient extended monomeric 6S conformer. Phosphorylation at S19 on RLC promotes filament assembly and dephosphorylation promotes disassembly of filaments to 10S. Only one head is shown on the filament for clarity. In both the filament and 6S schematics, the heads are drawn to the same scale as in the 10S schematic, but the tails are drawn to a smaller scale, making them about three times shorter than they actually are.
The relative concentration of the 10S conformation in solution in vitro also depends upon the total myosin concentration, with filament assembly promoted at higher concentration (4, 7). The interconversion between filaments and 10S in the cell, if it occurs, may be highly regulated reflecting the need for precise spatiotemporal control of myosin II-containing structures. If so, the amount of 10S could be higher than predicted from simple mass action even at the high myosin concentrations typical of muscle cells, for example.
The compact shape of the 10S conformer is due to the tail bending twice to allow one of the hairpin bends to specifically interact with the head region containing the active site (8) (Fig. 1A). Although the head-tail interaction is not required for low ATPase activity, it further reduces the ATPase by factor of ~10. The 10S conformer binds weakly to actin. Based upon these structural and functional characteristics, 10S has been proposed to be an assembly-competent myosin pool in equilibrium with filaments in cells, which would consume ATP in limited amounts and would not compete with activated myosin for actin binding.
We are interested in whether or not the 10S conformer of myosin II actually exists in cells and if it is present, whether or not it is in a functional equilibrium with myosin filaments. If 10S myosin is present, its distinct structural features could provide binding sites for signaling and regulatory proteins that mediate changes in contractility and motility. Because the 10S conformer is essentially switched off with respect to actin-activated ATPase activity, its presence would limit the total turnover of ATP by myosin, thus impacting the cellular energetics. Its ability to access cellular contractile structures could also limit the rate of myosin assembly in a precise spatiotemporal manner.
Although the questions posed above have not been answered for any cell type we have chosen to address them by investigating SMM in human airway smooth muscle cells (hASMCs). The significant role of SMM in smooth muscle contraction in general is obvious but the airway in particular is interesting because several studies have suggested that myosin filaments can undergo reversible filament assembly in intact airway muscle (9). This raises the possibility that a significant amount of 10S myosin may exist in this muscle type. The cultured hASMCs used here (10–19) are defined as smooth muscle cells because (i) they express smooth muscle restricted genes, i.e., SMM, calponin, smooth muscle tropomyosin, α and γ smooth muscle actins, SM22, and smoothelin; (ii) growth arrest of proliferating cells by serum deprivation in the presence of TGF-β further up-regulates expression of these genes in the so-called “contractile” phenotype; (iii) they express many different functionally coupled receptors and activation of these receptors causes changes in cell Ca2+ and RLC phosphorylation as in intact airway muscle; (iv) they contract when embedded in a 3D collagen matrix and relax in response to agents that increase cAMP; and (v) they synthesize the same cytokines and chemokines as primary airway smooth muscle cells. These molecular and functional characteristics are all shared with intact smooth muscle tissue.
We show that a peptide that specifically inhibits the intramolecular interaction between the hairpin bend in the tail and the heads causes a clearly discernable diffuse SMM pool to incorporate into a stress-fiber-associated pool. Because this peptide requires the presence of 10S myosin to exert its effects, these data strongly suggest that at least some of the SMM in the diffuse pool is in the 10S conformation. This study provides unique direct evidence for the presence of a significant pool of functional myosin in the 10S conformation in any cell type.
Results
Two Antibodies Detect SMM Not NMM in hASMCs.
The specificities of the two different anti-myosin (HC) antibodies used in this study toward smooth and nonmuscle myosin are documented in Fig. S1. The polyclonal anti-SMM antibody recognized chicken gizzard SMM with 47 ± sixfold greater affinity than platelet nonmuscle myosin (NMMIIA) (Fig. S1A). The previously described anti-gizzard SMM monoclonal antibody MM19 (20), raised against the assembly-competent domain at the tip of the tail (Fig. 1), recognized gizzard SMM as expected but not NMMIIA. The predominant nonmuscle isoform in hASMCs is NMMIIB, rather than NMMIIA, but it is unlikely that MM19 recognizes NMMIIB due to 9 nonidentical amino acids in the epitope (seven nonidentical for IIA). Fig. S1B shows the effect of preabsorbing the MM19 antibody with gizzard SMM before staining proliferating confluent hASMCs. Loss of staining suggests that MM19 detects primarily SMM. Western blots of both proliferative and contractile phenotype whole-cell lysates showed a single band comigrating with the myosin HC when probed with the anti-SMM antibody (Fig. S1C), suggesting that it specifically recognizes SMM in hASMCs. Both antibodies primarily recognize SMM, not NMM or other proteins in hASMCs.
Two Antibodies Detect Different SMM Pools.
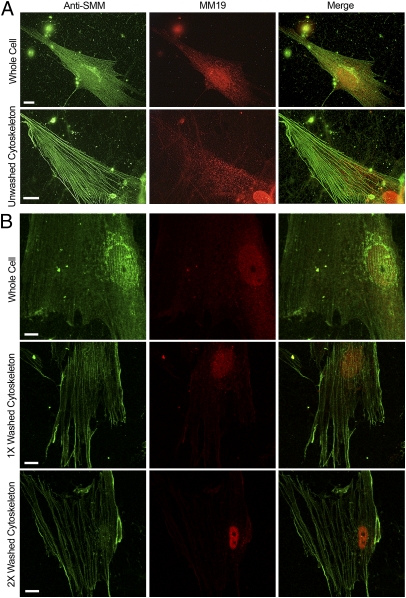
We reasoned that the two antibodies described above might detect different pools of SMM in hASMCs. The epitope for the MM19 antibody (the assembly-competent domain) is likely to be less exposed in filaments and more exposed in 10S, as previously found for a similar antibody (21). Because binding of MM19 to SMM could cause filament disassembly, it is important to note that cells were fixed before exposure to the antibody in all experiments. Fig. 2A compares staining of a whole cell versus an unwashed cytoskeleton prepared by Triton skinning. In the whole cell (top row), anti-SMM stained prominent stress-fibers along with a more diffusely distributed pool interspersed between the stress-fibers. In contrast, MM19 preferentially stained the diffuse pool in the cytoplasm with little staining of the stress fibers. Both antibodies showed perinuclear staining, most likely indicating the Golgi apparatus. The distinction between the two pools was accentuated in the unwashed cytoskeletons (Fig. 2A, second row). The diffuse pool was clearly localized between stress fibers and stress-fibers were preferentially visualized with anti-SMM. Myosin staining can be seen in wisps outside the cell. Fig. 2B shows confocal images of whole cells versus cytoskeletons that have been gently washed one or two times with PBS buffer lacking ATP (to avoid stress fiber disassembly). Note that this procedure depleted MM19 but not anti-SMM staining further (compare with Fig. 2A), suggesting that most of the diffuse pool is not stably associated with stress-fibers as predicted for 10S myosin. Because MM19 stains the diffuse pool, the SMM here may have a more exposed assembly-competent domain at the tip of the tail, which is also a characteristic of 10S SMM.
Fig. 2.
Visualization of SMM pools in contractile phenotype cells. (A) Untreated whole (63×) versus Triton-permeabilized cells (cytoskeletons; 100×) that were not washed. Anti-total SMM stain (green), MM19 stain (red), and merged images (Right). Controls without primary antibodies were black. (B) Confocal images (100×) showing the effect of washing the cytoskeletons with PBS without ATP. (Top Row) Whole cell, middle cytoskeleton prepared as in Materials and Methods but further washed with PBS one time (Middle Row) and two times (Bottom Row). (All scale bars, 10 μm.)
The MM19 antibody also appeared to strongly stain the nuclei. Washing of the cytoskeletons (Fig. 2B) did not deplete this pool. Interestingly, smooth and nonmuscle myosin II have recently been identified in the nuclei of rat colonic smooth muscle cells where they interact with chromatin (22).
SMM Pools Can Be Partially Interconverted in α-Toxin–Permeabilized hASMCs.
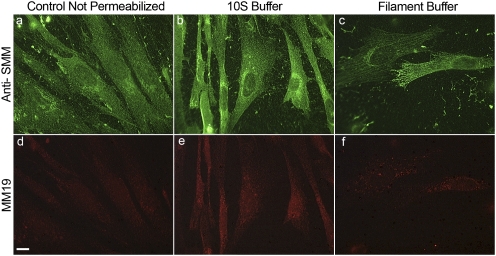
To investigate the relationship between the stress fiber and diffuse pool, we studied proliferating confluent cells that were mildly permeabilized with α-toxin to allow control of the intracellular milieu with two different buffer conditions. 10S-promoting buffer contains MgATP but no Ca2+ that favors dephosphorylated myosin, whereas filament-promoting buffer favors myosin phosphorylation by endogenous myosin light chain kinase/calmodulin (Ca2+ and phosphatase inhibitor). α-Toxin creates 1.4-nm holes in the membrane (23) that are too small to allow filamentous or monomeric SMM to exit the cell (21). We used proliferating confluent cells because they were more stable to permeabilization and changes in buffer conditions than cells that were differentiated to the contractile phenotype. Fig. 3 A and D show untreated cells (not permeabilized with α-toxin). These cells showed similar diffuse staining for the anti-SMM (a, green) and MM19 (d, red) with few stress fibers compared with cells differentiated to the contractile phenotype (compare with Fig. 2A). Permeabilization with α-toxin and gentle exposure to 10S-promoting buffer induced little change (Fig. 3 B and E). In contrast, exposure to filament-promoting buffer (Fig. 3 C and F) accentuated the stress-fibers visualized with the anti-SMM antibody. At the same time, MM19 staining was significantly reduced to a residual punctate pattern. These data suggest that SMM can be interconverted between stress fiber and diffuse pools.
Fig. 3.
Effect of buffer conditions on SMM pools in α-toxin–permeabilized confluent proliferating cells. Green represents anti-SMM; red, MM19 antibody. Control not permeabilized (A, D); α-toxin permeabilized and exposed to 10S buffer (B, E) and α-toxin permeabilized exposed to filament buffer (C, F). (Scale bar, 10 μm.) Images D, E, and F were despeckled using National Institutes Health ImageJ software to reduce camera noise.
To quantify the interconversion between SMM pools observed in Fig. 3, we compared the amount of SMM in α-toxin–permeabilized cells that were similarly exposed to 10S or filament buffer, either with or without a subsequent Triton extraction (whole cell versus cytoskeleton/nuclear) using the protocol for Fig. 2. Table 1 shows that before permeabilization with α-toxin, ~60% of the SMM was in the cytoskeleton/nuclear fraction (protein not extracted by Triton), which represents the stress fiber and nuclear pool from Fig. 2. After α-toxin permeabilization and exposure to filament buffer, SMM in the cytoskeleton/nuclear fraction increased from ~60 to 75%, suggesting that ~15% of the total SMM assembled from the soluble pool into the cytoskeleton, consistent with Fig. 3. Permeabilized cells treated with 10S buffer had less (~40%) SMM in the cytoskeleton/nuclear fraction than unpermeabilized cells as expected, suggesting that about ~20% of the SMM disassembled from stress fibers to the soluble cytosolic pool. These data suggest that the buffer-induced changes in anti-SMM and MM19 staining in Fig. 3 represent about 15–20% of the total SMM. The changes in the pools did not appear to be due to changes in the actin cytoskeleton because there were no detectable differences in actin staining under the same conditions. This was expected because neither the filament- nor the 10S-promoting buffers promote actin depolymerization in vitro.
Table 1.
Quantification of SMM in cytoskeletal/nuclear versus soluble cytosolic pools
| Treatment | SMM in cytoskeletal/nuclear pool (% of total)* |
| PBS control, no permeabilization | 57 ± 10 |
| Permeabilized + filament buffer | 74 ± 4† |
| Permeabilized + 10S buffer | 41 ± 6‡ |
*For each treatment, cells were either skinned or not skinned with Triton to prepare cytoskeletons as described in Materials and Methods. To quantify the SMM, unskinned cells or cytoskeletons were solubilized with M-Per Extraction Buffer including HALT protease inhibitor. The SMM content was determined by slot blot using the SMM antibody with respect to a standard curve prepared with purified SMM. Data are from three determinations, each from an independent experiment with proliferating confluent cells. The total amount of SMM varied between treatments by a maximum of 16%. Means and SDs are indicated.
†Mean is significantly different from control as determined by t test, P = 0.05.
‡Mean is significantly different from control as determined by t test, P = 0.08.
Peptide A Shifts the Equilibrium from 10S to Filaments in Vitro.
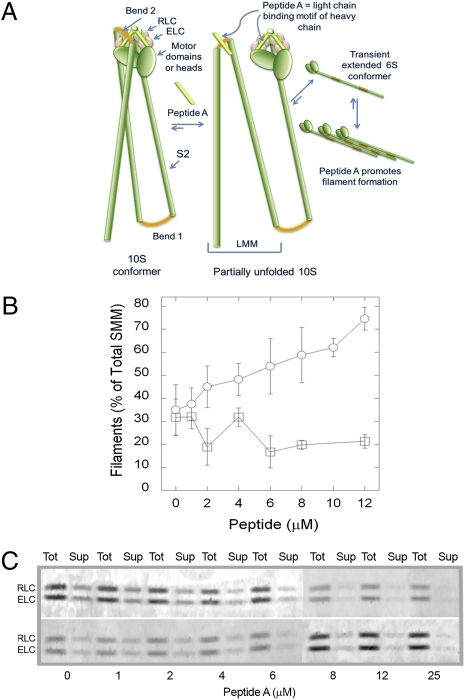
The key question is whether or not the diffuse pool contains 10S SMM. It is possible that this pool contains only small filaments that have an exposed C-terminal filament-assembly domain. Our approach was to examine the effects of a reagent that is known to specifically interfere or compete with intramolecular head-tail interactions in the 10S conformation. Such a reagent should have no effect upon small SMM filaments, but should promote assembly of existing 10S SMM into filaments. Therefore we investigated the effects of a peptide (Peptide A; Fig. 4A) derived from the sequence of the SMM HC immediately N-terminal of the head–tail junction (position of the invariant proline) including the RLC binding motif. We have previously shown that in the 10S conformation both the N- and C-terminal lobes of the RLC interact with Bend 2 in the light meromyosin (LMM) region of the SMM tail (24, 25) (Fig. 4A). This places Bend 2 proximal to the Peptide A region of the HC (RLC binding motif). The LMM portion of the tail has been shown to interact with a peptide similar to Peptide A (26). The predicted effect of the peptide is to compete with the intramolecular head–tail interaction by binding to Bend 2 in the LMM region. Fig. 4B shows that Peptide A shifted the normal filament-10S equilibrium toward filaments in vitro in a concentration-dependent manner, as expected (26), whereas a scrambled version of peptide A had no effect. If the cellular diffuse pool contains 10S myosin, it should be driven toward the stress-fiber pool by Peptide A but not by the scrambled peptide (see Peptide A Promotes a Redistribution of the Diffuse to the Stress-Fiber–Associated SMM Pools).
Fig. 4.
Peptide A mechanism and effects on filament-monomer equilibrium in vitro. (A) Schematic showing mechanism of inhibition of SMM hairpin bend (Bend 2) interaction with the light chain region by Peptide A. Green indicates HC including the globular motor domains, the S2 descending region of the tail, and the light meromyosin (LMM) region of the tail; red, bends in HC; pink, ELC; and brown, RLC. The 10S conformer (left) is fully bent with Bend 2 in LMM interacting with heads. Cartoon is partly based upon structural data from Seow (8) and Salzameda et al. (24). Peptide A, which is identical in sequence to the light chain binding motif of the HC (light green), will compete with that region for interaction with Bend 2, stabilizing the 6S conformer and promoting filaments. (B) Effects of synthetic peptides on equilibrium between filaments and soluble SMM in vitro. Chicken gizzard SMM (1 mg/mL) with Peptide A (○) or scrambled peptide (□) in 1 mM MgCl2, 150 mM NaCl, 10 mM Imidazole (pH 7.0), 0.1 mM EGTA, 0.1 mM DTT, and 1 mM ATP. The filaments were pelleted by ultracentrifugation, and the amount of soluble and total SMM was determined by a Bradford assay. Data represent the results from three independent experiments on two different SMM preparations. Error bars represent SEM. (C) Images of the light chain region from Coomassie-stained SDS gels of samples from two different (Upper and Lower) in vitro Peptide A titrations using equal loading volumes of the sample before (Tot) and after (Sup) pelleting filaments using the same protocol as in B.
We have considered an alternative mechanism for the effect of Peptide A. It could compete with the HC for RLC binding, thus generating RLC-deficient myosin, which cannot adopt the 10S conformation (27). If this mechanism is significant, the prespin samples from Fig. 4B should have less RLC than the postspin supernatants. Fig. 4C shows the light chain region from gels of 2 Peptide A titrations before (Tot) and after (Sup) pelleting filaments. Analysis of the RLC/ELC density ratios using a paired one-tailed t test showed that there was no significant difference between the totals and supernatants (P = 0.04). These data provide strong evidence that Peptide A is not generating a measurable amount of RLC-deficient myosin at least in vitro. These data are also in accord with the fact that Mg2+ ions, a constituent of all buffers, stabilize RLC-HC binding. EDTA and prolonged elevated temperatures are required to efficiently remove the RLC from the HC (27). Finally, it is known that RLC-deficient myosin aggregates in a nonnative manner through intermolecular interaction of the exposed HC IQ regions (27). Such aggregated myosin would not be competent to assemble into filaments. Peptide A appears to redistribute myosin from the soluble to the stress fiber pool (Fig. 5). This functional property of the redistributing myosin strongly suggests that it is competent to assemble into filaments, inconsistent with RLC-deficient myosin. For these reasons, and with consideration of our data in Fig. 4C, we favor the mechanism described in Fig. 4A.
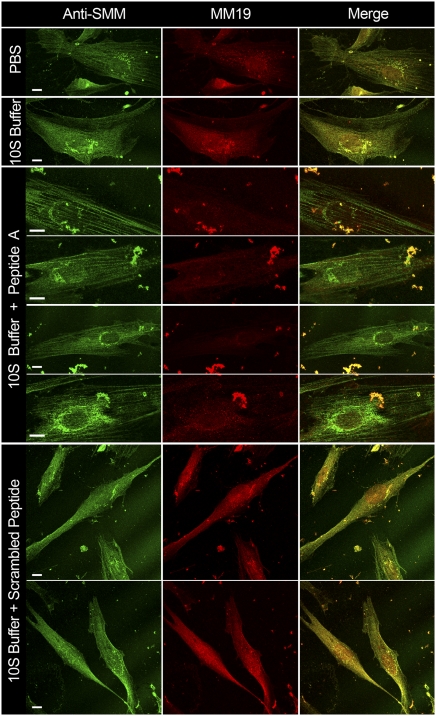
Fig. 5.
Effect of synthetic peptides on nonconfluent proliferating hASMCs. First column, hASMCs stained with anti-SMM; second column, stained with MM19; third column, merged. Row labels indicate treatments. PBS, control not permeabilized, exposed to PBS; 10S buffer, α-toxin–permeabilized exposed to 10S buffer; 10S Buffer + Peptide A or scrambled peptide, exposed to 30 μM Peptide A or scrambled peptide, in 10S buffer. Bright particles are cell debris. All images were taken on a confocal microscope, 63× or 100×. (All scale bars, 10 μm.)
Peptide A Promotes a Redistribution of the Diffuse to the Stress-Fiber–Associated SMM Pools.
Fig. 5 shows nonconfluent proliferating hASMCs cells dual-labeled with the anti-SMM (green) and MM19 (red) antibodies. We used nonconfluent cells for this experiment because they appeared to show more intense MM19 staining than did the confluent cells shown in Fig. 3D. As with the confluent cells, MM19 appeared to preferentially stain a diffusely distributed pool versus a stress fiber-associated pool, whereas anti-SMM appeared to label both pools (Fig. 5). Interestingly, in the control merged image (top right), MM19 staining is often seen also in a punctate pattern decorating the stress fibers. This pattern was not evident in the proliferating confluent cells shown in Fig. 3. The significance of this finding needs further investigation, but it suggests that the stress fibers contain regions in which the extreme C-termini of the SMM tails are exposed to the solvent.
After α-toxin permeabilization and exposure to a 10S-promoting buffer, fewer stress fibers were evident with more colocalization of the two antibody staining patterns. In striking contrast, the inclusion of Peptide A in the 10S-promoting buffer induced a prominent stress-fiber pattern that was strongly labeled with anti-SMM but sparsely labeled with MM19. The MM19 staining was often seen between the prominent stress fibers. Notably, the inclusion of the scrambled peptide in the 10S-promoting buffer had little or no effect with most SMM remaining in a diffuse pattern similar to that seen in 10S buffer without peptide. Because the mechanism for the effects of Peptide A requires the presence of the 10S conformer, these data strongly suggest that at least some of the SMM in the diffuse pool is in the 10S conformation.
The effect of Peptide A in 10S buffer to drive cytosolic myosin into stress fibers (Fig. 5) was similar to (but more pronounced than) the effect of filament buffer in Fig. 3. Both treatments also appeared to deplete nuclear myosin, suggesting that it can exit the nucleus and, like the soluble cytosolic myosin, assemble into the stress fibers under these conditions. The scrambled peptide did not deplete the nuclear myosin pool. These data suggest that at least some of the nuclear myosin is in the 10S conformation.
Due to multiple washing steps on permeabilized cells, cell debris is often observed in Fig. 5. However, we noted that the presence of Peptide A appeared to enhance the cell debris, whereas the scrambled peptide did not. The reason for this is not known but may be related to the dual effect of 10S buffer (depolymerizing conditions) followed by strongly polymerizing conditions induced by Peptide A. This could promote detachment of some of the cells from the coverslip surface.
Effect of Peptide A Was Not Due to Increased SMM Phosphorylation.
It is known that phosphorylation of SMM promotes filament formation and destabilizes the 10S conformation in vitro (Fig. 1). It is possible that the effect of Peptide A in Fig. 5 was simply due to increased SMM phosphorylation. Table 2 shows that the ratio of anti-pRLC to anti-total RLC antibody staining was unchanged by Peptide A, or the scrambled peptide. Therefore we conclude that Peptide A promotes filament assembly by destabilizing existing 10S SMM in the cell, in a manner consistent with the effect in vitro (Fig. 4B).
Table 2.
Quantification of pRLC to RLC ratio in hASMCs
| Treatment | Average ratio pRLC/RLC (% of control)* |
| PBS control, no permeabilization | 100 ± 19 |
| 10S buffer, no peptide | 85 ± 26 |
| 10S buffer + Peptide A | 82 ± 46 |
| 10S buffer + scrambled peptide | 103 ± 31 |
*Cells were dual-labeled with anti-pRLC and anti-total RLC antibodies, and the fluorescence for each secondary antibody was quantified in defined regions of 20 different cells for each condition using ImageJ software (National Institutes of Health). Perinuclear and nuclear areas were avoided. SEM values are indicated. Differences in means were not statistically significant (P = 0.05 using t test).
Discussion
We present several lines of evidence that are consistent with the idea that 10S SMM exists in hASMCs. First, we visualized a diffusely distributed cytosolic pool of myosin that was not tightly associated with filament-containing stress fibers (Fig. 2), consistent with the monomeric structure of 10S (Figs. 1 and 4A). Preferential staining of the diffuse versus stress-fiber pool with the MM19 antibody suggested that the SMM in the diffuse pool has an accessible HC C terminus, which is a property of 10S SMM (Fig. 4A). The behavior of the diffuse pool upon exposing α-toxin permeabilized cells to both filament- and 10S-promoting buffers (Fig. 3) was consistent with the known behavior of 10S SMM in vitro (Fig. 1). SMM appeared to assemble from the diffuse pool into the stress-fibers in filament-promoting conditions. Using a quantitative biochemical approach, we showed that a significant amount (~15–20%) of the total cellular SMM behaved in this manner (Table 1). The SMM pools in unpermeabilized cells appeared to be intermediate between the two extreme buffer conditions, consistent with the idea that 10S SMM also exists in the unpermeabilized cells (Fig. 5).
All evidence above is consistent with the presence of a pool of 10S SMM in hASMCs. This evidence however is insufficient to directly demonstrate that 10S exists, as other forms of SMM, such as small filaments would likely behave in a similar manner. We characterized Peptide A that destabilized the 10S conformation by a mechanism that is consistent with the extensive structural information about the head-tail interaction in 10S SMM (8, 24, 25) (Fig. 4A). Adding Peptide A resulted in more filamentous myosin at the expense of soluble myosin in vitro (Fig. 4B). Exposure of cells to Peptide A caused a striking shift of the diffuse to the filamentous myosin pool even in a 10S-promoting buffer, but a scrambled version of the peptide did not (Fig. 5). The extent of conversion to a stress-fiber pool caused by Peptide A in 10S buffer appeared to be much greater than that caused by filament buffer without Peptide A (Fig. 3 C and F) or in unpermeabilized cells (Figs. 3 A and D and 5, Top Row). This is in accord with the mechanism described in Fig. 4A. If the 10S conformer is prevented from forming because of Peptide A, the monomer-filament equilibrium should be shifted far toward filaments. If there is sufficient peptide to saturate the system, any remaining SMM is presumably either not accessible to the peptide or is not in the 10S conformation. Our data do not suggest that all of the SMM in the diffuse pool is in the 10S conformation. However, we conclude that functional 10S myosin is present because a significant portion of the diffuse pool appeared to be sensitive to Peptide A (Fig. 5).
The extent to which 10S myosin is present in smooth muscles most likely depends upon the state of differentiation, disease, and function of the muscle. For example, there is good evidence that myosin undergoes reversible filament assembly in differentiated airway smooth muscle [(9) and references therein] and in the rat anococcygeus (28–30) upon muscle activation. If a measurable amount of the disassembled pool is in the 10S conformation, addition of Peptide A to these permeabilized muscles may increase the total number and length of filaments. In contrast, using the phasic muscle from chicken gizzards, a previous study (21) showed that 10S myosin was not detectable in relaxed or activated muscle or upon exposure of the permeabilized muscle to a 10S-promoting buffer. Interestingly, these differences in muscle types may be due to filament stabilizing proteins such as telokin (31), as visceral muscles, including gizzard, express higher levels of telokin than vascular (tonic) (32) and airway (33) muscles.
In summary, we conclude that functional 10S SMM exists in hASMCs. This conclusion is based upon the behaviors of two general pools of SMM, diffuse (not stress fiber associated) and stress-fiber–associated, which were distinguished by immunocytochemical dual staining of cultured hASMCs using two anti-SMM specific antibodies. We showed that the diffuse pool has many behaviors consistent with the presence of 10S SMM. However, the key observation is that a peptide that shifts the filament-10S equilibrium toward filaments in vitro, by a known mechanism that requires the presence of 10S, caused a striking shift of the diffuse to the stress-fiber pool. The specificity of the effect was demonstrated by the fact that a scrambled version of the peptide had no effect (Fig. 5) and that neither peptide caused RLC phosphorylation levels to change (Table 2). We conclude that hASMCs contain a significant pool of functional SMM in the 10S conformation that can assemble into filaments upon changing cellular conditions. As this study provides a unique direct demonstration of 10S myosin in cells, our findings should have a broad impact on the understanding of the cellular regulation of myosin II filament assembly and on regulation of other motor proteins that can adopt self-inhibited structures.
Materials and Methods
Materials, Western Blots, Immunocytochemistry and Imaging, and Cells.
Details of standard materials and methods and previously described details of the hASMCs and culture (34) are given in SI Text.
Preparation of α-Toxin–Permeabilized hASMCs.
HASMCs grown on collagen-coated glass coverslips were rinsed three times quickly with relaxing buffer (130 mM KOH, 130 mM proprionic acid, 4 mM MgCl2, 20 mM Tris·maleate, and 2 mM KEGTA, pH 7.0), and treated with α-toxin (100 U/mL in relaxing buffer) for 1 h at room temperature. After rinsing cells once quickly with relaxing buffer, either 10S buffer (150 mM KCl, 10 mM NaPi, 5 mM EGTA, 4 mM MgATP, and 1 mM DTT, pH 7.0), or filament buffer (135 mM KOH, 2.5 mM MgCl2, 2.2 mM CaCl2, 0.5 mM ATP, 2 mM EGTA, 1 mM DTT, and 50 nM calyculin A, pH, adjusted to 6.5 with proprionic acid) was added to cells and incubated for 30 min. Two controls, α-toxin–treated or untreated cells, were treated with relaxing buffer for the duration of the experiment. After buffer removal, cells were immediately fixed and stained.
Preparation of Cytoskeletons.
Cytoskeletons were prepared by rinsing cells on coverslips three times quickly with 1× PBS followed by permeabilization with 0.1% Triton-X-100 in 1× PBS, added gently and directly onto coverslip to avoid cell dissociation, for 5 min at room temperature. PBS contained 137 mM NaCl, 2.7 mM KCl, 4.3 mM NaPO4, and 1.4 mM KPO4, pH 7.4. The Triton solution was removed by capillary action using a tissue, followed by fixation. Control cells were treated in parallel with 1× PBS for 5 min. For the washing experiment in Fig. 2B, cells were skinned and Triton was removed as above, and PBS was similarly added and removed either once or twice before fixation.
Effect of Synthetic Peptides.
Peptide A (QQLTAMKVIQRNC(t-butyl)AAYLKLRNWQWWRLFTKV) and its scrambled version (TQLWRAAFVLLKQYWLINNKKWTMRVRQAC) were purified to >95% by HPLC and verified by MALDI-MS. Peptide A corresponds to residues 817–847 of the chicken SMM HC (myosin-11; NCBI Reference Sequence: NP990605.1). α-Toxin–skinned hASMCs were washed and treated with 10S or filament buffer +/− 30 μM Peptide A or scrambled peptide for 30 min at room temperature. After buffer removal cells were immediately fixed and dual-labeled. The same controls were used as above.
Supplementary Material
Acknowledgments
We thank Dr. Sean Ward (University of Nevada School of Medicine, Reno, NV) for assistance with imaging. This work was supported by National Institutes of Health (NIH) National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant 5R01AR040917-20 (to C.R.C. and K.C.F.) and NIH Heart, Lung, and Blood Institute Grant HL077726 (to W.T.G). NIH National Center for Research Resources Centers of Biomedical Research Excellence Grant 5 P20 RR018751 supported the Imaging and Protein Expression Core Facility. Mass spectrometry was performed at the University of Nevada Proteomics Core Facility, Reno, NV, and supported by NIH Grant P20 RR-016464 from the IDeA Networks of Biomedical Research Excellence Program of the National Center for Research Resources.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011784108/-/DCSupplemental.
References
- 1.Cremo CR, Hartshorne DJ. In: Myosins: A Superfamily of Molecular Motors. Coluccio LM, editor. Vol. 7. Dordrecht, Netherlands: Springer; 2008. pp. 171–222. [Google Scholar]
- 2.Onishi H, Wakabayashi T. Electron microscopic studies of myosin molecules from chicken gizzard muscle I: The formation of the intramolecular loop in the myosin tail. J Biochem. 1982;92:871–879. doi: 10.1093/oxfordjournals.jbchem.a134001. [DOI] [PubMed] [Google Scholar]
- 3.Trybus KM, Huiatt TW, Lowey S. A bent monomeric conformation of myosin from smooth muscle. Proc Natl Acad Sci USA. 1982;79:6151–6155. doi: 10.1073/pnas.79.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kendrick-Jones J, Smith RC, Craig R, Citi S. Polymerization of vertebrate non-muscle and smooth muscle myosins. J Mol Biol. 1987;198:241–252. doi: 10.1016/0022-2836(87)90310-x. [DOI] [PubMed] [Google Scholar]
- 5.Trybus KM, Lowey S. Conformational states of smooth muscle myosin. Effects of light chain phosphorylation and ionic strength. J Biol Chem. 1984;259:8564–8571. [PubMed] [Google Scholar]
- 6.Onishi H, Wakabayashi T, Kamata T, Watanabe S. Electron microscopic studies of myosin molecules from chicken gizzard muscle II: The effect of thiophosphorylation of the 20K-dalton light chain on the ATP-induced change in the conformation of myosin monomers. J Biochem. 1983;94:1147–1154. doi: 10.1093/oxfordjournals.jbchem.a134459. [DOI] [PubMed] [Google Scholar]
- 7.Trybus KM. Assembly of cytoplasmic and smooth muscle myosins. Curr Opin Cell Biol. 1991;3:105–111. doi: 10.1016/0955-0674(91)90172-u. [DOI] [PubMed] [Google Scholar]
- 8.Burgess SA, et al. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Seow CY. Myosin filament assembly in an ever-changing myofilament lattice of smooth muscle. Am J Physiol Cell Physiol. 2005;289:C1363–C1368. doi: 10.1152/ajpcell.00329.2005. [DOI] [PubMed] [Google Scholar]
- 10.Salinthone S, et al. Overexpression of human Hsp27 inhibits serum-induced proliferation in airway smooth muscle myocytes and confers resistance to hydrogen peroxide cytotoxicity. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1194–L1207. doi: 10.1152/ajplung.00453.2006. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe S, et al. Expression of functional leukotriene B4 receptors on human airway smooth muscle cells. J Allergy Clin Immunol. 2009;124:59–65. doi: 10.1016/j.jaci.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma P, et al. Expression of the dystrophin-glycoprotein complex is a marker for human airway smooth muscle phenotype maturation. Am J Physiol Lung Cell Mol Physiol. 2008;294:L57–L68. doi: 10.1152/ajplung.00378.2007. [DOI] [PubMed] [Google Scholar]
- 13.Redhu NS, et al. Proinflammatory and Th2 cytokines regulate the high affinity IgE receptor (FcepsilonRI) and IgE-dependant activation of human airway smooth muscle cells. PLoS ONE. 2009;4:e6153. doi: 10.1371/journal.pone.0006153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran T, et al. Laminin-binding integrin alpha7 is required for contractile phenotype expression by human airway myocytes. Am J Respir Cell Mol Biol. 2007;37:668–680. doi: 10.1165/rcmb.2007-0165OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran T, McNeill KD, Gerthoffer WT, Unruh H, Halayko AJ. Endogenous laminin is required for human airway smooth muscle cell maturation. Respir Res. 2006;7:117–132. doi: 10.1186/1465-9921-7-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia YC, et al. Functional expression of IgG-Fc receptors in human airway smooth muscle cells. Am J Respir Cell Mol Biol. 2010 doi: 10.1165/rcmb.2009-0371OC. 10.1165/rcmb.2009-0371OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long X, Bell RD, Gerthoffer WT, Zlokovic BV, Miano JM. Myocardin is sufficient for a smooth muscle-like contractile phenotype. Arterioscler Thromb Vasc Biol. 2008;28:1505–1510. doi: 10.1161/ATVBAHA.108.166066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosens R, et al. Caveolae facilitate muscarinic receptor-mediated intracellular Ca2+ mobilization and contraction in airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1406–L1418. doi: 10.1152/ajplung.00312.2007. [DOI] [PubMed] [Google Scholar]
- 19.Gosens R, et al. Cooperative regulation of GSK-3 by muscarinic and PDGF receptors is associated with airway myocyte proliferation. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1348–L1358. doi: 10.1152/ajplung.00346.2007. [DOI] [PubMed] [Google Scholar]
- 20.Ikebe M, et al. The tip of the coiled-coil rod determines the filament formation of smooth muscle and nonmuscle myosin. J Biol Chem. 2001;276:30293–30300. doi: 10.1074/jbc.M101969200. [DOI] [PubMed] [Google Scholar]
- 21.Horowitz A, Trybus KM, Bowman DS, Fay FS. Antibodies probe for folded monomeric myosin in relaxed and contracted smooth muscle. J Cell Biol. 1994;126:1195–1200. doi: 10.1083/jcb.126.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Sarna SK. Nuclear myosin II regulates the assembly of preinitiation complex for ICAM-1 gene transcription. Gastroenterology. 2009;137:1051–60. doi: 10.1053/j.gastro.2009.03.040. 1060 e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song L, et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 24.Salzameda B, Facemyer KC, Beck BW, Cremo CR. The N-terminal lobes of both regulatory light chains interact with the tail domain in the 10 S-inhibited conformation of smooth muscle myosin. J Biol Chem. 2006;281:38801–38811. doi: 10.1074/jbc.M606555200. [DOI] [PubMed] [Google Scholar]
- 25.Olney JJ, Sellers JR, Cremo CR. Structure and function of the 10 S conformation of smooth muscle myosin. J Biol Chem. 1996;271:20375–20384. doi: 10.1074/jbc.271.34.20375. [DOI] [PubMed] [Google Scholar]
- 26.Katoh T, Kodama T, Fukushima A, Yazawa M, Morita F. Evidence for involvement of a 12-residue peptide segment of the heavy chain in the neck region of smooth muscle myosin in formation of the 10S conformation. J Biochem. 1995;118:428–434. doi: 10.1093/oxfordjournals.jbchem.a124925. [DOI] [PubMed] [Google Scholar]
- 27.Trybus KM, Lowey S. The regulatory light chain is required for folding of smooth muscle myosin. J Biol Chem. 1988;263:16485–16492. [PubMed] [Google Scholar]
- 28.Xu JQ, Gillis JM, Craig R. Polymerization of myosin on activation of rat anococcygeus smooth muscle. J Muscle Res Cell Motil. 1997;18:381–393. doi: 10.1023/a:1018634412117. [DOI] [PubMed] [Google Scholar]
- 29.Gillis JM, Cao ML, Godfraind-De Becker A. Density of myosin filaments in the rat anococcygeus muscle, at rest and in contraction. II. J Muscle Res Cell Motil. 1988;9:18–29. doi: 10.1007/BF01682145. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe M, Takemori S, Yagi N. X-ray diffraction study on mammalian visceral smooth muscles in resting and activated states. J Muscle Res Cell Motil. 1993;14:469–475. doi: 10.1007/BF00297209. [DOI] [PubMed] [Google Scholar]
- 31.Shirinsky VP, et al. A kinase-related protein stabilizes unphosphorylated smooth muscle myosin minifilaments in the presence of ATP. J Biol Chem. 1993;268:16578–16583. [PubMed] [Google Scholar]
- 32.Gallagher PJ, Herring BP. The carboxyl terminus of the smooth muscle myosin light chain kinase is expressed as an independent protein, telokin. J Biol Chem. 1991;266:23945–23952. [PMC free article] [PubMed] [Google Scholar]
- 33.Herring BP, Smith AF. Telokin expression is mediated by a smooth muscle cell-specific promoter. Am J Physiol. 1996;270:C1656–C1665. doi: 10.1152/ajpcell.1996.270.6.C1656. [DOI] [PubMed] [Google Scholar]
- 34.Gosens R, et al. Role of caveolin-1 in p42/p44 MAP kinase activation and proliferation of human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2006;291:L523–L534. doi: 10.1152/ajplung.00013.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.