Abstract
Nucleosome deposition occurs on newly synthesized DNA during DNA replication and on transcriptionally active genes via nucleosome-remodeling complexes recruited by activator proteins and elongating RNA polymerase II. It has been long believed that histone deposition involves stable H3–H4 tetramers, such that newly deposited nucleosomes do not contain H3 and H4 molecules with their associated histone modifications from preexisting nucleosomes. However, biochemical analyses and recent experiments in mammalian cells have raised the idea that preexisting H3–H4 tetramers might split into dimers, resulting in mixed nucleosomes composed of “old” and “new” histones. It is unknown to what extent different genomic loci might utilize such a mechanism and under which circumstances. Here, we address whether tetramer splitting occurs in a locus-specific manner by using sequential chromatin immunoprecipitation of mononucleosomes from yeast cells containing two differentially tagged versions of H3 that are expressed “old” and “new” histones. At many genomic loci, we observe little or no nucleosomal cooccupancy of old and new H3, indicating that tetramer splitting is generally infrequent. However, cooccupancy is detected at highly active genes, which have a high rate of histone exchange. Thus, DNA replication largely results in nucleosomes bearing exclusively old or new H3–H4, thereby precluding the acquisition of new histone modifications based on preexisting modifications within the same nucleosome. In contrast, tetramer splitting, dimer exchange, and nucleosomes with mixed H3–H4 tetramers occur at highly active genes, presumably linked to rapid histone exchange associated with robust transcription.
The packaging of eukaryotic DNA into chromatin influences many DNA-associated processes, including transcriptional regulation. Within the chromatin fiber, each basic nucleosome unit bears important chemical and structural information. The mechanisms by which nucleosomes are assembled on DNA during replication, transcription, DNA repair, etc., can thus impact not only the integrity of chromatin structure but also patterns of gene expression and epigenetic inheritance. The H3–H4 tetramer core of each nucleosome is the more stable component and contains most of the relatively persistent and functionally important histone methylation marks. Much attention has therefore been given to the questions of how the H3–H4 tetramers are formed and maintained on chromatin.
Early studies attempted to distinguish between a conservative assembly model, by which old and new histones form separate tetramers on replicating DNA, and a semiconservative assembly mechanism, by which existing tetramers are split into H3–H4 dimers, followed by association of new H3–H4 dimers to complete each nucleosome core (1–3). In the semiconservative model, the resulting tetramers would bear a mixture of old and new histones, allowing transmission of epigenetic information within the basic nucleosome unit. Though mechanistically attractive, the mixed tetramer model received little support from a variety of studies, which failed to detect old and new H3–H4 dimers within the same nucleosome (2, 4–8). A notable exception was an analysis of active chromatin from chicken cells suggesting substantial levels of mixed tetramers (9).
More recently, biochemical analyses demonstrated that H3 and H4 associate with predeposition histone chaperones as dimers, indicating that dimers, rather than tetramers, are the basic assembly units of the nucleosome core (10). As an important implication, these findings highlighted the possibility that deposition of new H3–H4 dimers, following splitting of existing old tetramers, might generate mixed old–new tetramers in the genome, according to the semiconservative assembly model. This scenario attracted significant attention, raising the questions of whether and to what extent tetramer splitting occurs, at which genomic regions, and during which replication-dependent or -independent processes (1–3, 11). This issue is also affected by the fact that many organisms have distinct H3 variants that are deposited during DNA replication (H3.1) or independently of DNA replication (H3.3) by different deposition machineries (12).
A recent analysis, published during the last stages of the present work, showed that all of the canonical H3.1 and most of the variant H3.3 in a human cell line are incorporated by the conservative assembly model, consistent with earlier observations (13). However, a minor fraction of H3.3 is present in mixed old–new tetramers, indicating that tetramer splitting is specific to the H3 variant that incorporates outside of replication. Tetramer splitting of H3.3 nucleosomes appeared to be inhibited, but not eliminated, by inhibitors of DNA replication, although this conclusion is based on differences between relatively low levels of tetramer splitting. Furthermore, it is unclear why DNA replication should affect tetramer splitting of a histone variant that is not known to be deposited by the DNA replication machinery. Lastly, because this analysis was performed on bulk chromatin, the extent to which tetramer splitting occurs at various regions of the genome and its mechanistic connection to other cellular processes remain unclear.
In the yeast Saccharomyces cerevisiae, a single H3 isoform homologous to H3.3 is incorporated both during and outside of replication. Importantly, experiments involving inducible expression of a tagged H3 indicate that the rate of H3 incorporation is not uniform in the genome as expected simply as a consequence of DNA replication. Instead, the rate of H3 incorporation differs dramatically among different loci in the genome, with promoters and highly transcribed coding regions showing much higher levels of H3 exchange than other genomic regions (14–16). These highly dynamic regions of chromatin presumably reflect the activity of nucleosome-remodeling complexes and histone chaperones that are recruited by activator proteins (17–21) and elongating RNA polymerase II (21–24). As the same H3 isoform incorporates at a variety of loci with different nucleosome exchange rates and transcriptional levels, yeast chromatin provides a good model for examining possible region- and transcription-dependent properties of nucleosome core assembly mechanisms.
Here, we assay the extent of tetramer splitting in a locus-specific manner by developing a dual-regulation experimental design, in which two differentially tagged versions of H3 are expressed in the same yeast cell as “old” and “new” histones. Sequential chromatin immunoprecipitation (ChIP) on mononucleosomes shows that old and new H3 usually do not cooccupy the same nucleosome, supporting the conservative assembly model as a major histone deposition mechanism. Interestingly, at loci showing rapid exchange of H3, cooccupancy of new and old H3 is observed, indicating that transcription-associated processes permit tetramer splitting and generate chimeric old-new tetramers at regions of dynamic chromatin.
Results
Experimental Design for Detecting Tetramer Splitting at Distinct Genomic Loci.
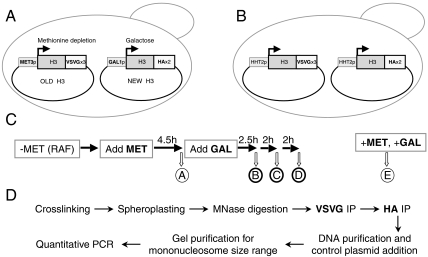
Our experimental setup utilizes two differentially tagged versions of H3, placed under inducible promoters and regulated to constitute old and new histones (Fig. 1A). The old H3 contains three copies of the VSV glycoprotein (H3–VSVG) and is expressed from the MET3 promoter, while H3 with two HA tags (H3–HA), expressed from the GAL1 promoter, functions as the new histone. Plasmids containing each version of inducible C-terminally-tagged H3 were introduced together into an S. cerevisiae strain lacking the endogenous genes for H3. Each tagged version of H3 is able to support viability when expressed as the only copy of H3, confirming its functionality. An additional positive control strain was generated, in which both tagged versions of H3 are under the control of the native promoter and thus coexpressed (Fig. 1B).
Fig. 1.
Experimental design. (A) The experimental strain YKY69 lacks the endogenous H3 genes and contains two centromeric plasmids expressing H3–VSVG from the methionine-repressible MET3 promoter and H3–HA from the galactose-inducible GAL1 promoter. (B) The control strain YKY64 is similar to YKY69, except that the two tagged versions of H3 are under the control of the native HHT2 promoter, resulting in their coexpression. (C) YKY69 cells are initially grown in medium lacking methionine and containing raffinose, leading to expression of only H3-VSVG (“old” histone). Methionine is added for 4.5 h to repress the MET3 promoter and terminate H3-VSVG expression (sample A). Cells are then treated with galactose to induce H3–HA expression (“new” histone) for 2.5 h, 4.5 h, and 6.5 h (samples B, C, and D, respectively). Sample E consists of YKY69 cells grown for 20 h in medium containing both methionine and galactose, resulting in expression of H3–HA. (D) Procedure for performing sequential ChIP of mononucleosomes.
The experimental strain was initially grown in the absence of methionine, to induce the MET3 promoter, resulting in expression of old H3–VSVG (Fig. 1C). Because raffinose was used as a carbon source, the GAL1 promoter regulating H3–HA remained uninduced at this time. To repress the MET3 promoter and terminate H3–VSVG expression, methionine was added, and cells were allowed to grow for 4.5 h, which was almost a generation. Following this period of MET3 promoter repression, galactose was added to induce the GAL1 promoter and thus initiate new H3–HA synthesis. Samples for analysis were taken just before (negative control) and at 2.5 h, 4.5 h, and 6.5 h after galactose addition. As another negative control, cells grown overnight in the presence of methionine and galactose represented the situation where only new H3–HA is induced.
The critical experiment was to perform sequential ChIP (25) on mononucleosomes isolated from the various samples. Specifically, formaldehyde-crosslinked chromatin was treated with micrococcal nuclease to produce mainly mononucleosome-sized chromatin fragments, which were subjected to two steps of immunoprecipitation to obtain DNA fragments associated with both VSVG- and HA-tagged H3 (Fig. 1D). Following reversal of the crosslinks, DNAs from single and sequential immunoprecipitations and inputs were separated on a gel, and mononucleosome-sized DNA fragments were isolated and analyzed by quantitative PCR. To control for differences in sample recovery, the DNAs were mixed with a constant low amount of digested nonyeast plasmid DNA prior to gel purification; PCR signals from a plasmid-generated 148-bp fragment were used for normalization.
Control Experiments.
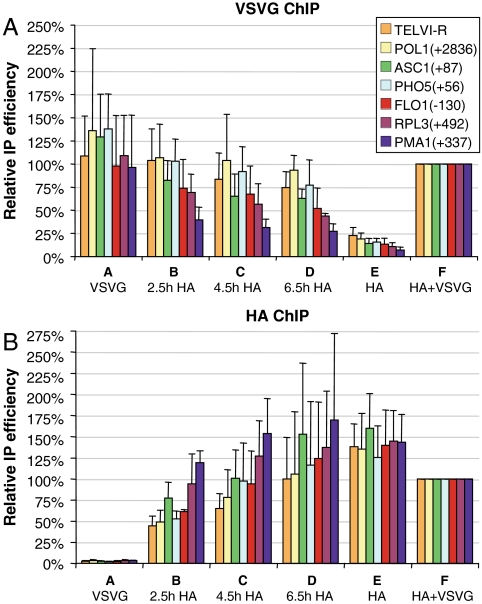
In accord with the experimental design, we observe an initially high level of old H3–VSVG prior to galactose induction (Fig. 2A, sample A), followed by a gradual loss (samples B–E). In contrast, levels of the new H3–HA are initially very low (Fig. 2B, sample A) and increase throughout the time course (samples B–E). The faster loss of the old H3–VSVG at some loci is complemented by an accelerated incorporation of the new H3–HA, and this is expected from the higher rates of replication-independent histone exchange at active gene ORFs and promoters (14–16). These dynamic regions with high nucleosome exchange rates include PMA1(+337), followed by RPL3(+492). Modest exchange is observed at FLO1(−130), PHO5(+56), and ASC1(+87). Despite these differences in H3–HA incorporation rate, the most static loci (TELVI-R and POL1) show substantial association of 30%, 50%, and 75% of the final levels at 2.5 h, 4.5 h, and 6.5 h, respectively. This induction time course should thus enable the detection of potential mixed tetramers at the various loci. In the positive control strain in which H3–VSVG and H3–HA are coexpressed from the native H3 promoter, both tagged H3 derivatives are incorporated at high levels (Fig. 2 A and B, sample F).
Fig. 2.
Association of old and new H3 at different loci. YKY69 cells were treated to induce “old” H3-VSVG expression, followed by its repression, and then by induction of “new” H3–HA expression, as described in Fig. 1C. In samples A and E, only a single H3 form was induced and substantially incorporated (VSVG- and HA-tagged, respectively), while the chromatin of samples B–D contained both H3 forms, incorporated at different times. Sample F represents the control strain YKY64, coexpressing the two tagged H3 forms from their native promoter. The graphs show the average results of ChIP analysis using VSVG (A) or HA (B) antibodies. H3 occupancy was analyzed at a telomeric locus (TELVI-R) and six different genes, with the positions relative to the translation start site indicated.
Generally Little Mixing of Old and New H3 in the Same Nucleosome.
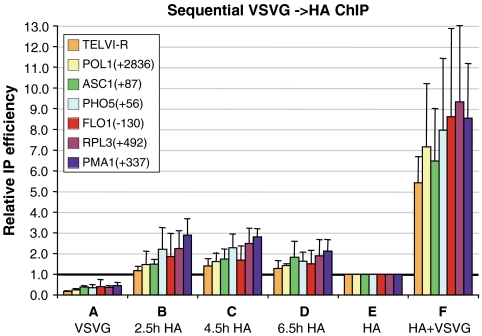
Sequential ChIP on mononucleosomes was used to determine the nucleosomal cooccupancy of H3–VSVG and H3–HA at different genomic loci during the galactose induction time course (Fig. 3). When coexpressed from the native H3 promoter (i.e., the positive control strain; sample F), the two H3 derivatives show high cooccupancy values that far exceed those seen when only one H3 version was induced (control samples A and E). This confirms our ability to detect the presence of the two H3 derivatives within the same nucleosome (i.e., a mixed H3–H4 tetramer) over a substantial dynamic range. Sample E, in which only the GAL1 promoter is induced, shows higher cooccupancy levels than sample A, due to residual expression of H3–VSVG from the repressed MET3 promoter. The cooccupancy values of sample E were thus taken as the experimental background, and only significantly higher values were scored as positive signals resulting from cooccupancy of the induced H3–VSVG and H3–HA.
Fig. 3.
Nucleosome cooccupancy of old and new H3. Samples from the experiments described in Figs. 1C and 2 were subjected to two rounds of immunoprecipitation using VSVG and HA antibodies. The graphs show the average results of sequential ChIP analysis, relative to the values of the single-induction sample E, regarded as experimental background. H3 cooccupancy was analyzed at a telomeric locus (TELVI-R) and six different genes, with the positions relative to the translation start site indicated.
In contrast to the high cooccupancy of H3–VSVG and H3–HA when coexpressed, the same proteins expressed sequentially as old and new H3 forms show generally low cooccupancy levels (samples B–D). Thus, old and new H3, assayed at seven different genomic loci, are not frequently present within the same nucleosome. These data suggest that most H3 incorporation events in the yeast genome do not involve tetramer splitting.
Mixing of H3–H4 Dimers Within Regions of Dynamic Histone Exchange.
While cooccupancy levels of old and new H3 are mostly not detected above the experimental background, some loci show values that are clearly higher. Of those, the highest values are obtained at PMA1(+337) at 2.5 h, 4.5 h, and 6 h (P-values < 0.01, < 0.005, and < 0.025, respectively, by Welch’s unpaired t-test), followed by RPL3(+492) at 2.5 h and 4.5 h (< 0.05 and < 0.025), and then PHO5(+56) at 4.5 h (< 0.025). Interestingly, the cooccupancy levels of the different loci generally correlate with their nucleosome exchange rate. The highest cooccupancy levels are observed at the most rapidly exchanging PMA1(+337) locus, and reach approximately 25% of the level seen upon coexpression of the different tagged H3 forms (after subtracting the experimental background). Thus, substantial levels of tetramer splitting occur at highly dynamic nucleosomes.
In principle, cooccupancy measurements simply reflect the levels of mixed H3–H4 tetramers at each genomic region. As such, cooccupancy will depend on the relative levels of the two H3 derivatives at a given locus at a given time, but it should not depend on the rate of histone exchange per se. For a given locus, maximal cooccupancy should occur when both H3 derivatives are present at 50% of the maximal level, and cooccupancies will gradually decrease as the association levels of the individual H3 derivatives are more discordant. Thus, the best comparison of cooccupancy values among different loci involves measurements where the relative association of the two H3 derivatives is similar, rather than measurements at a single time point. For example, clear cooccupancy at the dynamic PMA1 locus is observed at 2.5 h, when the level of the new H3–HA reaches 83% of full association. However, other loci that show similar H3–HA incorporation at later times (e.g., POL1, ASC1, and FLO1) do not have the same cooccupancy levels. More generally, conditions for examining maximal cooccupancy are achieved at all loci, yet cooccupancy is only observed at active genes (Fig. 3), which show high rates of H3 exchange (Fig. 2). Thus, tetramer splitting occurs preferentially at transcriptionally active genes and correlates well with dynamic histone exchange.
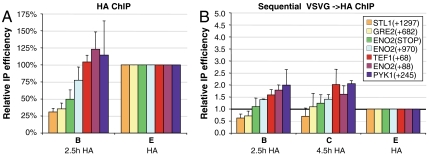
To further examine the relationship between histone exchange rate and mixing of old and new H3, we analyzed seven additional loci at genes expected to have low (STL1 and GRE2) or high (TEF1, ENO2, and PYK1) expression levels under our experimental conditions. These loci differ in their H3 exchange rate, as indicated by new H3–HA incorporation at 2.5 h, with STL1(+1297) and GRE2(+682) being less dynamic, as predicted (Fig. 4A). At the three ENO2 positions analyzed, H3–HA incorporation levels correlate with proximity to the gene’s start site. As shown in Fig. 4B, cooccupancy of H3–VSVG and H3–HA at 2.5 h and 4.5 h of galactose induction (samples B and C) relative to the experimental background (single-induction sample E) increases in accord with the increase in the rate of H3–HA incorporation levels (Fig. 4A). Thus, near-background levels of cooccupancy are observed at the relatively static STL1(+1297), GRE2(+682), and ENO2(STOP) loci, followed by higher levels at the more dynamic ENO2(+970), and even higher levels at the most dynamic loci TEF1(+68), ENO2(+88), and PYK1(+245). These results further indicate that, while tetramer splitting is generally an infrequent event, it is more prevalent at regions of high nucleosome exchange, producing substantial levels of mixed tetramers composed of old and new histones.
Fig. 4.
Nucleosomal occupancy and cooccupancy of inducible H3 at active and inactive loci. Single and sequential ChIP samples from the experiments of Figs. 1, 2, and 3 were analyzed for association at additional loci. The graphs show the average results of (A) ChIP analysis using HA antibodies, as described in Fig. 2B, or (B) sequential ChIP using VSVG and HA antibodies, as described in Fig. 3. H3 occupancy was analyzed at different genes, with the positions relative to the translation start site or around the stop codon (STOP) indicated.
Discussion
The mechanisms by which nucleosome cores are formed, propagated, and exchanged on genomic DNA are fundamental and have been a subject of interest for many years. While H3–H4 tetramers had been initially considered stable units, more recent studies challenged this notion and reignited interest in the fate of DNA-associated tetramers during replication, transcription, and other chromatin-disrupting processes. This study provides the first locus-specific analysis of nucleosome core composition with respect to existing and newly assembled histones. Our results confirm that most H3–H4 tetramers remain intact on DNA but unexpectedly show that significant levels of mixed tetramers occur at rapidly exchanging regions. The observation of mixed H3–H4 tetramers suggests that replication-independent histone deposition processes permit tetramer splitting and dimer exchange at genomic regions showing high rates of histone exchange.
Conservative Assembly of Nucleosome Cores During DNA Replication.
Based on a variety of experiments involving bulk chromatin, it has been long believed that in S phase, nucleosomes deposited on newly synthesized DNA are generated from newly synthesized histones and do not involve histone subunits from preexisting nucleosomes (2, 4–8). A recent paper focusing on the histone variant H3.1, which is deposited specifically during DNA replication, revealed that newly synthesized H3.1 did not form mixed tetramers with preexisting H3 molecules, confirming that nucleosomes are conservatively inherited during DNA replication (13). Here, using a different approach that permits the analysis of individual genomic regions, we fail to detect mixed H3–H4 tetramers on many genomic regions. Because all genomic regions are replicated exactly once per cell cycle, replication-coupled nucleosome deposition should occur equally at all genomic regions. Hence, the absence of mixed H3–H4 tetramers at many genomic regions confirms conservative assembly during DNA replication. While mixed H3–H4 tetramers are observed on genomic regions showing high rates of histone exchange (see below), the mechanisms involved in such dynamic histone exchange are independent of DNA replication.
Taken together, all these results indicate that tetramer splitting and mixed H3–H4 tetramers are not generated by the process of DNA replication. Thus, preexisting and newly synthesized nucleosomes are distinct structural entities, with newly deposited nucleosomes not bearing the histone modification present on the preexisting nucleosomes. As a consequence, propagation of histone modification patterns through cell division cycles does not occur mechanistically within individual nucleosomes but rather between adjacent or nearby nucleosomes via chromatin-binding proteins (e.g., Sir proteins, HP1, or polycomb complexes) that function across more extended genomic regions.
Tetramer Splitting at Regions of Dynamic Chromatin.
Our locus-specific analysis reveals mixed H3–H4 tetramers at genomic regions that have high transcriptional activity and rapid rates of histone exchange. When comparing overall old and new H3 cooccupancy signals between groups of five dynamic [RPL3, PMA1, ENO2(+88), TEF1, and PYK1] and five static [TELVI-R, POL1, STL1, GRE2, and ENO2(STOP)] loci, the dynamic group scores significantly higher (P-value of 10-10). At the PMA1 locus, the level of mixed tetramers is approximately 25% of that obtained upon coexpression of the two tagged H3 derivatives, suggesting that tetramer splitting events occur at a substantial frequency at highly dynamic loci. We do not understand why only a minority of H3–H4 tetramers appear to be mixed, but it is possible that multiple rounds of dynamic exchange might convert mixed H3–H4 tetramers into tetramers composed solely of new H3 molecules. Importantly, the locus-specificity of mixed H3–H4 tetramers on regions of high histone exchange strongly suggests that tetramer splitting in yeast is independent of DNA replication.
A recent study, published during the late stages of this work, detected low levels of mixed H3–H4 tetramers in the bulk population of the H3.3 nucleosomes but not in H3.1 nucleosomes in a human cell line (13). In one respect, our results are in agreement with this independent study in that they both link tetramer splitting to dynamic chromatin that exchanges independently of DNA replication. However, the other study attributes most tetramer splitting events to replication-dependent H3.3 deposition, even though H3.3 deposition typically occurs in a replication-independent manner. This conclusion is based on the reduction of tetramer splitting by inhibitors of DNA replication, although the differences in tetramer splitting are quantitatively modest. DNA replication inhibitors may also have an indirect effect on H3.3 deposition, especially because the experiments in the other study involve a relatively long time frame, unlike the relatively short time course used in our experiments. It is also possible that yeast and mammalian cells may differ with respect to the mechanism of tetramer splitting. Notably, our observation of tetramer splitting at active genes is consistent with an early observation of mixed tetramers in a chromatin fraction enriched in transcribed DNA in chicken cells (9).
Nucleosome Assembly and Disassembly Through Dimer Displacement.
There are two known mechanisms of dynamic histone exchange in yeast cells, both of which involve recruitment of nucleosome-remodeling complexes and histone chaperones to specific genomic regions. Recruitment is mediated either by activator proteins bound at promoter regions (17–21) or by elongating Pol II (and associated factors) at highly transcribed genes (21–24). We presume that the nucleosome-remodeling complexes and/or histone chaperones responsible for rapid histone exchange differ from those used during DNA replication. The restriction of tetramer splitting to regions of high nucleosome exchange may be explained by differences in the biochemical properties of these factors or by other differences between DNA replication and transcription.
The histone chaperones Asf1, HIR, FACT, and Spt6 are important for transcription-coupled exchange of histones H3 and H4 (16, 26–29). The crystal structure of Asf1 bound to an H3–H4 dimer prompted a “strand capture” model, in which Asf1 splits H3–H4 tetramers into dimers during nucleosome disassembly by interacting with the H4 tail (30), and this mechanism is supported by biochemical analyses (3, 31). Our detection of mixed tetramers at sites of dynamic chromatin provides in vivo support for such a strand capture mechanism, although the presumptive chaperone(s) mediating the observed tetramer splitting is unknown. Mixed tetramer formation at such dynamic regions might reflect the near simultaneous presence of multiple histone chaperone molecules at these loci.
Upon passage of Pol II, chaperone-mediated removal of an H3–H4 dimer could lead to several outcomes: (i) destabilization of the other H3–H4 dimer resulting in complete nucleosome eviction; (ii) reassociation of the original chaperone-bound H3–H4 dimer, leading to the initial (old) nucleosome; (iii) deposition of a new H3–H4 dimer, by the same or different chaperone, leading to a mixed H3–H4 tetramer. In any event, the enhanced presence of mixed tetramers at sites of active transcription suggest that nucleosome perturbation by Pol II involves transfer of parental H3–H4 as dimers. As suggested previously (1), such a mechanism of partial nucleosome unraveling and dimer displacement would cause less disruption of chromatin structure than full tetramer removal.
Implications of Tetramer Splitting.
By promoting histone exchange, the process of transcription can generate unique nucleosome cores containing old and new histones at active genomic regions. Such mixed tetramers possess the potential for transferring chemical or structural information within a nucleosome, although the functional role for these mixed tetramers awaits further investigation. More generally, the finding of tetramer splitting at specific genomic loci may have broader significance, because multiple assembly factors and histone chaperones mediate H3–H4 exchange in concert with other cellular processes such as DNA repair and DNA amplification. In the case of Pol II transcription, tetramer splitting may also depend on environmental conditions, and in this regard, stress-activated kinases (e.g., Hog1, Fus3, PKA) are recruited specifically to stress-activated coding regions (32, 33). It is also conceivable that, in certain cell types or developmental stages, specific H3–H4 exchange processes might generate elevated levels of mixed tetramers at distinct parts of the genome. Thus, while tetramer splitting occurs very infrequently (if at all) during replication-associated histone deposition, it clearly occurs at transcription-associated sites, and it may have important roles in other biological contexts.
Materials and Methods
DNAs and Yeast Strains.
To generate centromeric TRP1 plasmids expressing C-terminally tagged HHT2 and untagged HHF2 from their native promoters, two restriction sites were first introduced at the HHT2 C terminus in pJH18 (pRS314/HHT2–HHF2)(34). PCR fragments containing three VSVG epitopes from ZM475 (35) or two HA epitopes from pMPY–3xHA (36) were then inserted into these sites, generating pHHF2–HHT2VSVx3 and pH4H3HAx2, respectively. When introduced into a histone shuffle strain as the only source of H3, the latter two plasmids were able to support viability well, confirming the functionality of the resulting tagged H3 proteins. The HHT2–HHF2 locus of pH4H3HAx2 was then transferred into pRS316 (CEN URA3) to generate p316H4H3–HAx2. p414MET3–H3VSVGx3+H4 is based on pRS414 (CEN TRP1) and contains the MET3 promoter from pJR1811 (37) driving expression of HHT2 with three C-terminal VSVG tags, as well as HHF2 expressed from its native promoter. YCp33GAL–H3HAx2 is based on YCplac33 (CEN URA3) and contains the GAL1 promoter (-461 to -1) driving expression of HHT2 with two C-terminal HA tags.
The yeast strains YKY64 and YKY69 lack the endogenous genes for H3 and H4 and express tagged versions of H3 and untagged H4 from plasmids. YKY64 was generated by first transforming pHHF2–HHT2VSVx3 into the histone shuffle strain WZY42 (38) and selecting on 5–FOA-containing plates for cells that lost the original URA3 plasmid encoding H3 and H4. A second plasmid p316H4H3–HAx2 was then transformed to produce the final strain. YKY69 was similarly generated using p414MET3–H3VSVGx3+H4 and YCp33GAL–H3HAx2, except that the cells were grown on methionine-free plates before performing the 5–FOA selection and thereafter. This strain was normally grown without methionine to induce H3–VSVGx3 expression from the MET3 promoter.
Cell Growth.
The experimental YKY69 culture was grown overnight in synthetic complete medium lacking methionine (for MET3 induction), uracil, and tryptophan and containing 2% raffinose as a carbon source. Methionine was then added to 2 mM to repress H3–VSVG expression from the MET3 promoter, and the cells were allowed to grow for 4.5 h (a generation being approximately 5 h). To induce H3–HA expression from the GAL1 promoter, 2% galactose was added, followed by continued cell growth. Samples for analysis were taken before and at different times after galactose addition. An additional YKY69 culture was grown for 20 h in the above medium containing both methionine and galactose. The control strain YKY64 was grown overnight in the above medium.
Preparation of Nucleosomal Extracts.
Yeast cultures of 400 ml (OD600 0.6–1.0) were crosslinked with 1% formaldehyde for 5 min at room temperature, quenched by adding 60 ml of 2.5 M glycine, and washed twice. Subsequent spheroplasting and micrococcal nuclease (MNase) treatment were done essentially as described (39). Briefly, cells were digested in 40 ml Buffer Z by adding 600 μl Zymolyase 20 T (25 mg/ml; US Biological) and incubating at 30 °C for 45–60 min with shaking. Spheroplast pellets were spun, washed, and resuspended in NP-S buffer. Each sample was split into three 800 μl aliquots, which were treated with 25–100 U/ml of MNase (USB) for 35 min at 37 °C. The digestion was terminated by placing the reactions on ice and adding EDTA to 10 mM, followed by 800 μl of Adjust-FA buffer (100 mM HEPES-KOH pH 7.5, 250 mM NaCl, 2 mM EDTA, 2% Triton X-100, 0.2% sodium deoxycholate, 0.2% SDS, and 2 mM phenylmethylsulfonyl fluoride). Samples were centrifuged at 4 °C for 30 min at 14,000 rpm, and the supernatants were removed and frozen at -80 °C. To assess the extent of MNase digestion, a 150 μl aliquot of each reaction was taken, and the purified DNA was run on a 1.5% Agarose gel. Reactions where mononocleosomal DNA (∼150 bp) was the predominant form were used for subsequent ChIP experiments.
Sequential Chromatin Immunoprecipitation (ChIP) on Mononucleosomes.
Sequential ChIP was performed essentially as described previously (25). Briefly, 400 μl nucleosomal extracts were incubated with Agarose-conjugated anti-VSVG (Sigma A1970; 30 μl of 1∶1 suspension) and 550 μl FA lysis buffer for 90 min at room temperature with rotation. Following stringent washes, the immunocomplexes were eluted from the beads by heating for 10 min at 68 °C in buffer containing 1% SDS. A second round of immunoprecipitation was performed for 90 min using 25 μl anti-HA (F-7, Santa Cruz) and 25 μl bed volume protein A-Sepharose, followed by washes and elution as above. For single ChIP experiments, 40 μl nucleosomal extracts were used.
Immunoprecipitated and input samples were decrosslinked by heating. The resulting DNA was purified through Qiagen columns, eluted in 30 μl TE, and mixed with 40 pg of an irrelevant plasmid digested to produce a 148-bp fragment, which served as a recovery control. The DNA mixtures were run on a 1.5% Agarose gel, and mononucleosomal fragments (90–190 bp) were excised, purified using the Qiagen gel extraction kit, and analyzed by real-time quantitative PCR (40). To control for differences in DNA recovery during gel purification, the genomic signals of each DNA sample were normalized to that of the 148-bp fragment. Normalized signals from immunoprecipitations were then divided by those of the inputs to produce the relative IP efficiency. For each genomic locus, the values were expressed relative to those of a given control sample. They represent the average and standard deviation of four independent experiments (three for Fig. 4).
In sequential ChIP experiments, sample E was regarded as the experimental background and set as 1. The highest cooccupancy levels in Fig. 3, seen at PMA1 (samples B and C), were compared to those of the positive control, where both H3 forms are coexpressed for a long time (sample F), by first subtracting the background signal (sample E) from each value, and then dividing the experimental cooccupancy (samples B and C) by that of the positive control (sample F). The resulting approximately 25% value provides an estimate for the capacity to form mixed old–new tetramers at highly dynamic regions. Unpaired Welch’s or Student’s t-tests were used for calculating one-tailed P-values. In the combined P-value for the difference between grouped dynamic and static loci, each group consisted of 34 observations, from samples B and C (as in Figs. 3 and 4) at five genomic loci.
Acknowledgments.
We thank David Allis, Arthur Hsu, Zarmik Moqtaderi, Masayasu Nomura, Jasper Rine, Sharon Roth, and Alain Verreault for strains and plasmids. We also thank Joe Geisberg, Zarmik Moqtaderi, and Chris (Koon-Ho) Wong for protocols and helpful advice. This work was supported by a grant to K.S. from the National Institutes of Health (GM30186).
Footnotes
The authors declare no conflict of interest.
References
- 1.Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Annunziato AT. Split decision: What happens to nucleosomes during DNA replication? J Biol Chem. 2005;280:12065–12068. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 3.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 4.Leffak IM, Grainger R, Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977;12:837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- 5.Prior CP, Cantor CR, Johnson EM, Allfrey VG. Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: a system for studying changes in nucleosomes assembled in vivo. Cell. 1980;20:597–608. doi: 10.1016/0092-8674(80)90306-2. [DOI] [PubMed] [Google Scholar]
- 6.Jackson V. Deposition of newly synthesized histones: Hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry. 1988;27:2109–2120. doi: 10.1021/bi00406a044. [DOI] [PubMed] [Google Scholar]
- 7.Jackson V. In vivo studies on the dynamics of histone-DNA interaction: Evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990;29:719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- 8.Yamasu K, Senshu T. Conservative segregation of tetrameric units of H3 and H4 histones during nucleosome replication. J Biochem. 1990;107:15–20. doi: 10.1093/oxfordjournals.jbchem.a122999. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Leffak M. Assembly of active chromatin. Biochemistry. 1986;25:2055–2060. doi: 10.1021/bi00356a033. [DOI] [PubMed] [Google Scholar]
- 10.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 11.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 13.Xu M, et al. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 14.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 15.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Deckert J, Struhl K. Histone acetylation at promoters is differentially affected by activators and repressors. Mol Cell Biol. 2001;21:2726–2735. doi: 10.1128/MCB.21.8.2726-2735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 19.Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 20.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Workman JL. Nucleosome displacement in transcription. Gene Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 22.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 24.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geisberg JV, Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 2004:32–e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 27.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Kim HJ, Seol JH, Han JW, Youn HD, Cho EJ. Histone chaperones regulate histone exchange during transcription. EMBO J. 2007;26:4467–4474. doi: 10.1038/sj.emboj.7601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamai A, Puglisi A, Strubin M. Histone chaperone spt16 promotes redeposition of the original H3–H4 histones evicted by elongating RNA polymerase. Mol Cell. 2009;35:377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 30.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 32.Proft M, et al. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell. 2006;23:241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 33.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 34.Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 35.Moqtaderi Z, Struhl K. Expanding the repertoire of plasmids for PCR-mediated epitope tagging in yeast. Yeast. 2008;25:287–292. doi: 10.1002/yea.1581. [DOI] [PubMed] [Google Scholar]
- 36.Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 37.Fox CA, Ehrenhofer-Murray AE, Loo S, Rine J. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science. 1997;276:1547–1451. doi: 10.1126/science.276.5318.1547. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu CL, et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan X, Lamarre-Vincent N, Wang Q, Struhl K. Extensive chromatin fragmentation improves enrichment of protein binding sites in chromatin immunoprecipitation experiments. Nucleic Acids Res. 2008;36:e125. doi: 10.1093/nar/gkn535. [DOI] [PMC free article] [PubMed] [Google Scholar]