Abstract
Bacterial metabolism is characterized by a remarkable capacity to rapidly adapt to environmental changes. We restructured the central metabolic network in Escherichia coli to force a higher production of NADPH, and then grew this strain in conditions favoring adaptive evolution. A six-fold increase in growth capacity was attained that could be attributed in multiple clones, after whole genome mutation mapping, to a specific single mutation. Each clone had an evolved NuoF*(E183A) enzyme in the respiratory complex I that can now oxidize both NADH and NADPH. When a further strain was constructed with an even higher degree of NADPH stress such that growth was impossible on glucose mineral medium, a solid-state screening for mutations restoring growth, led to two different types of NuoF mutations in strains having recovered growth capacity. In addition to the previously seen E183A mutation other clones showed a E183G mutation, both having NADH and NADPH oxidizing ability. These results demonstrate the unique solution used by E. coli to overcome the NADPH stress problem. This solution creates a new function for NADPH that is no longer restricted to anabolic synthesis reactions but can now be also used to directly produce catabolic energy.
Keywords: adaptive evolution, NADPH metabolism, complex I
Evolutionary microbiology under environmental or metabolic constraints is an emergent field of research. Although it is known that growing microorganisms are not static over either short or long periods, the evolutionary process called adaptive evolution developed to adapt to constraints is poorly understood at the genetic, biochemical, and metabolic levels.
Experiments of adaptive evolution have been conducted in the laboratory to understand the behavior of Escherichia coli in response to different environmental or metabolic constraints: (i) wide serial subculturing for over 20 years (more than 40,000 generations) on glucose-limited mineral medium (1), (ii) growth for more than 700 generations on glycerol, a maladapted substrate although a complete metabolic pathway for this substrate was already present (2), and (iii) culture over 600–800 generations of single gene deletion mutants affected in metabolic key branch points [phosphoglucose isomerase (Δpgi), phosphoenolpyruvate carboxylase (Δppc), triose phosphoisomerase (Δtpi), phosphotransacetylase (Δpta)] to evaluate the effect of targeted metabolic perturbations on the structure of the metabolic network (3). The main conclusions of this work highlighted the strong robustness of the genetic and metabolic networks of E. coli and indicated that evolution never invented a new solution such as a reassignment of enzyme function to adapt to the environmental or metabolic constraints. Instead, the main feature of evolution was an increased in the capacity of already active pathways or the activation of latent pathways.
The objective of the present work was to study the ability of E. coli to adapt to and overcome a strong unbalance and an imposed rigidity of the redox state of cellular metabolism. Cellular metabolism results from an intricate biochemical network that is delicately balanced and regulated to enable growth under fluctuating environmental conditions. The release of energy from growth substrates, and the capture of this energy for the biosynthesis of cell components, involves redox reactions in which electrons and/or hydrogen atoms are transferred between donor and acceptor molecules. Thus, the pyridine nucleotides, NADH and NADPH occupy central points in redox metabolism. Although NADH and NADPH are chemically similar, NADPH has a lower redox potential than NADH and they are involved in distinct biochemical functions. NADPH primarily drives anabolic reactions, whereas NADH has a catabolic role (4). To fulfill these distinct roles, these two redox couples are generally not in thermodynamic equilibrium and the NADPH to NADP+ ratio is in a more reduced state than the NADH to NAD+ ratio. NADPH is mainly produced in the oxidative branch of the pentose phosphate pathway (PPP) or by the isocitrate dehydrogenase in the tricarboxylic acid (TCA) cycle. Moreover, E. coli possesses two transhydrogenases that play important roles in NADPH metabolism (4): (i) the membrane-bound proton translocating transhydrogenase PntAB that is used to produce NADPH from NADH and (ii) the soluble energy independent transhydrogenase UdhA that is mainly involved in NADH production from NADPH. With both isoforms, E. coli can counterbalance situations of catabolic under deprivation or overproduction of NADPH, whereas organisms lacking transhydrogenases can not tolerate imbalances between catabolic NADPH production and anabolic NADPH consumption (5, 6). In this study we designed an E. coli strain in which we both (i) restructured its central metabolic network to force a higher production of NADPH, and (ii) eliminated the possibility to produce NADH from NADPH, and then use excess NADPH to produce energy. We will report the unique solution used by E. coli to overcome the imposed redox constraints and the manner in which existing metabolic dogmas can be challenged in a synthetic biology type of metabolic design. This solution creates an unexpected function for NADPH that is no longer restricted to anabolic synthesis reactions but can now be also used to produce directly catabolic energy.
Results
Design of an E. coli Strain Exhibiting a High Intracellular NADPH/NADP+ Ratio.
To create an E. coli strain with a high intracellular NADPH/NADP+ ratio, we used rational metabolic engineering to limit the oxidation of NADPH for the production of NADH or reduced quinone, and to favor the reduction of NADP+ in the PPP. Key modifications were introduced in E. coli central metabolism to force cells to increase NADPH. Glucose flux was redirected through the PPP by deleting the pgi and edd genes, which encode phosphoglucose isomerase, the first enzyme of glycolysis (EMP), and 6-phosphogluconate dehydrogenase, the first enzyme of the Entner–Doudoroff pathway (EDP). NADPH oxidation to produce NADH or reduced quinone was eliminated by deleting the udhA and qor genes encoding soluble transhydrogenase and quinone oxidoreductase (Fig. S1) (7). The engineered E. coli strain Δpgi∷FRT, Δedd∷FRT, Δqor∷FRT, ΔudhA∷FRT was successfully constructed and named NA23 (Table S1). During the oxidation of glucose to acetyl-CoA, this strain produced 2 mol of NADPH and 2 mol of NADH per mole of glucose, while a wild-type strain using only the EMP produces 4 mol of NADH per mole of glucose. Furthermore this strain was unable to use excess NADPH to produce energy by the combined use of the soluble transhydrogenase and the proton-pumping NADH:ubiquinone oxidoreductase (Nuo).
Phenotypic Analysis of the NA23 Mutant Strain.
During aerobic growth on glucose mineral medium, the NA23 mutant displayed both a low specific growth rate (μ = 0.04 ± 0.01 h-1) and a low specific glucose consumption rate (qs = 0.9 ± 0.1 mmol·g-1·h-1) compared to the wild-type strain (μ = 0.6 ± 0.03 h-1 and qs = 10.4 ± 0.7 mmol·g-1·h-1). No acetate was produced, suggesting that the metabolic perturbations did not lead to carbon overflow, and indicating a kinetic limitation for glucose catabolism (Table 1). As expected, the NADPH/NADP+ ratio was 2-fold higher for the NA23 mutant (2.05 ± 0.13) than for the WT strain (1.05 ± 0.08) validating the metabolic engineering strategy. A 3-fold lower NADH/NAD+ ratio was observed for the NA23 strain compared to the WT strain (0.11 ± 0.01 versus 0.31 ± 0.04) (Table 1).
Table 1.
Aerobic growth parameters of unevolved, evolved, and wild-type mutants during exponential growth on glucose
| Strain | Specific genotype | Historic | Growth rate (μ) | Biomass yield (YX/S) | Glucose consumption rate (qS) | Acetate production rate (νA) | NADPH/NADP+ ratio | NADH/NAD+ ratio |
| h-1 | g·g-1 | mmol·g-1·h-1 | mmol·g-1·h-1 | |||||
| NA 23 | MG1655 ΔudhA Δpgi Δqor Δedd | unevolved mutant | 0.04 ± 0.01 * | 0.22 ± 0.01 | 0.9 ± 0.1 | 0.02 ± 0.01 | 2.05 ± 0.13 | 0.11 ± 0.01 |
| NA 23E04 | MG1655 ΔudhA Δpgi Δqor Δedd nuoF * | evolved mutant | 0.24 ± 0.02 | 0.27 ± 0.02 | 3.1 ± 0.2 | 0.01 ± 0.01 | 1.85 ± 0.02 | 0.14 ± 0.01 |
| Wild-type (WT) | MG1655 | 0.6 ± 0.03 | 0.34 ± 0.01 | 10.4 ± 0.7 | 2.6 ± 0.3 | 1.05 ± 0.08 | 0.31 ± 0.04 |
*Average ± S.D. for at least 3 independent experiments.
Adaptive Evolution of the E. coli (Δpgi∷FRT, Δedd∷FRT, Δqor∷FRT, ΔudhA∷FRT) Strain.
To evaluate the capability of the NA23 mutant to overcome the severe imbalance in its central metabolism, adaptive evolution was conducted by serially subculturing in glucose mineral medium, maintaining exponential phase. Two independent evolution experiments were conducted in parallel, and specific growth rates were measured for both populations. A rapid in vivo adaptive evolution of the NA23 mutant strain occurred in four serial subcultures (approximately 20 generations). Results for adaptive evolution in both cultures showed convergence of the improved phenotype. A maximum specific growth rate of 0.22 ± 0.01 h-1 was measured, which was six times higher than the original strain. Clones were isolated as colonies from both populations, and the specific growth rate on glucose mineral medium of 10 randomly selected clones was analyzed. Similar growth rates were measured for all of the clones (μ = 0.24 ± 0.02 h-1) and one (NA23E04) was arbitrarily selected for further characterization. Growth rates measured after adaptive evolution reached 40% of the wild-type MG1655 specific growth rate (Table 1).
Identification of Genetic Targets Responsible for E. coli Adaptive Evolution Using Genome-Wide Analysis.
To determine the genotype of the evolved strain NA23E04 responsible for its phenotype, genetic mutations were identified using comparative genome sequencing, (CGS, Roche Nimblegen). This method has been demonstrated to be fast and efficient in detecting multiple mutations present in one bacterium relative to another (8, 9). The comparison of the NA23E04 evolved clone and the MG1655 wild-type strain genomes revealed five signal differences, four corresponding to the four gene deletions. The other mutated region was PCR-amplified and sequenced. The single mutation was located in the nuoF gene (nuoF*), encoding the NuoF subunit of the catalytic domain of the proton-pumping NADH:ubiquinone oxidoreductase, and led to a change in one amino acid residue: glutamate at position 183 was replaced by alanine. CGS analysis strongly suggested that only one mutational event occurred during the adaptive evolution of the NA23 mutant on glucose mineral medium, so this mutation must be responsible for the increased fitness of the evolved strain.
Phenotypic Analysis of the Mutation at the Genome Level.
To confirm that nuoF* was the only mutation involved in the increased growth rate of the NA23 strain in glucose mineral medium, two recombinant strains were constructed using the gene replacement method described by Datsenko and Wanner (10). In one strain, the native nuoF gene was replaced with the evolved nuoF* gene from the NA23 mutant. In another, the wild-type nuoF gene was introduced into the evolved NA23E04 strain. Both clones were cultured in the same conditions in glucose mineral medium (Fig. S2). The NA23 unevolved mutant expressing the nuoF* evolved gene had the same maximum specific growth rate (μ = 0.23 h-1· ± 0.02) as the NA23E04-evolved clone (μ = 0.24 h-1 ± 0.02). Conversely, the NA23E04-evolved strain expressing the nuoF wild-type gene had a similar maximum specific growth rate (μ = 0.05 h-1 ± 0.01) as the NA23 mutant (μ = 0.04 h-1 ± 0.01). These results clearly confirmed that the single mutation in nuoF* was the unique mutation responsible for the adaptive evolution. To provide more insights into the physiological consequences of nuoF* mutation, the native nuoF gene was replaced with the evolved nuoF* gene in the wild-type MG1655 strain. The resulting reconstructed strain MG1655 nuoF*, named WT*, exhibited similar aerobic growth parameters on glucose mineral medium (μ = 0.58 ± 0.01 h-1, YX/S = 0.31 ± 0.01 g.g-1, qS = 10.4 ± 0.4 mmol·g-1·h-1, νA = 3.0 ± 0.3 mmol·g-1·h-1), assuming a neutral effect of the nuoF mutation in the WT genotype regarding growth phenotype even after evolution of this strain over 4 serial subcultures.
Analysis of nuoF Sequence in the Evolved Clones.
To determine if the nuoF mutation was responsible for the evolution in the 10 selected clones, the nuoF gene was sequenced in the nine remaining clones. The same nuoF* mutation leading to a E183A substitution was observed in all clones, indicating that this mutation occurred early in the culture of the NA23 strain. Assuming that at the end of the fourth subculture, most of the cells had the nuoF* mutation, we calculated when this mutation appeared in the culture, and found that it occurred during the first subculture on glucose mineral medium.
Physiological Consequences of NuoF* in Respiratory Chain.
NuoF is one of the three NuoE, F, and G subunits of the soluble NADH:ubiquinone oxidoreductase fragment of the respiratory chain complex I, which contains a total of 13 subunits, and couples the transfer of two electrons from NADH to ubiquinone with the membrane translocation of four protons (11). Complex I is composed of three domains: the soluble NADH:ubiquinone oxidoreductase fragment, the connecting fragment, and the membrane domain. The NuoBCD connecting fragment is required for proper assembly of the NADH soluble fragment, in particular for the incorporation of Fe-S clusters. The overexpression of NuoBCDEFG leads to a properly assembled soluble fragment (NuoEFG) that has an NADH:ubiquinone oxidoreductase activity that can transfer electrons from NADH to ferricyanide, an artificial electron acceptor, with the same efficiency as the entire complex (12). Bungert et al. developed a simple and fast method for overexpressing and purifying an active soluble NADH:ubiquinone oxidoreductase fragment using direct affinity chromatography, based on engineered Nuo subunits (13). Using the same strategy, NuoEFG-native and NuoEF*G-evolved soluble fragments were successfully produced and purified. Native and evolved purified soluble fragments were biochemically characterized by measuring their NADH/ferricyanide and NADPH/ferricyanide reductase activities (14). As is shown in Table 2, and as previously reported, enzyme kinetics revealed a high NADH/ferricyanide reductase activity for the native soluble form (kcat = 44 s-1), and a low activity when NADPH was used as electron donor (kcat = 13 s-1). The Km was 4 ± 1 μM for NADH, and 1150 ± 80 μM for NADPH, in the same range as previous reports (12, 15). In contrast, the evolved soluble fragment (NuoF*E183A) showed a high NADPH/ferricyanide reductase activity (kcat = 60 s-1) and a low Km for NADPH (16.7 ± 2.9 μM), which corresponded to a catalytic efficiency (3.6 ± 1 s-1·μM-1) for NADPH that has not previously been reported for NADH:ubiquinone oxidoreductase. However the catalytic efficiency using NADH was four times lower than the catalytic efficiency seen with the native fragment.
Table 2.
Enzymatic properties of wild-type and evolved purified soluble NADH:ubiquinone oxidoreductase fragments.
| Native fragment | NuoF*(E183A) fragment | NuoF*(E183G) fragment | ||
| Km (μM) | NADH | 4.3 ± 0.8 * | 7.9 ± 1.4 | 18.3 ± 2.9 |
| NADPH | 1150 ± 80 | 16.7 ± 2.9 | 8.4 ± 1.7 | |
| Kcat (s-1) | NADH | 43.8 ± 2.1 | 18.7 ± 1.3 | 61.3 ± 5.0 |
| NADPH | 13.2 ± 0.3 | 59.2 ± 6.0 | 29.2 ± 2.5 | |
| Kcat/Km (s-1 μM-1) | NADH | 10.1 ± 2.4 | 2.4 ± 0.6 | 3.4 ± 1.4 |
| NADPH | 0.011 ± 0.001 | 3.6 ± 1.0 | 3.5 ± 1.0 |
*Means ± S.D. for 2–3 independent experiments.
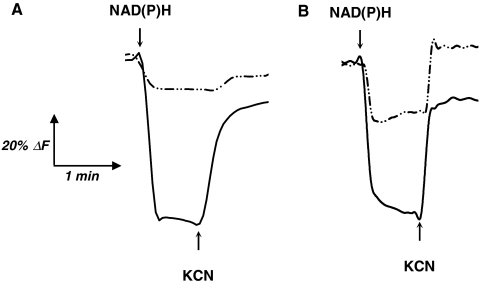
To complete the biochemical data and answer the question as to whether the electrons from NADPH access the respiratory chain, we used inside-out (ISO) vesicle preparations from parental (NA23) and evolved (NA23E04) strains and measured NAD(P)H oxidase activities under aerobic and anaerobic conditions (Table 3). As expected, NADPH oxidase activity in evolved ISO vesicles was significantly higher than in parental ISO vesicles. Experimental differences between NADH and NADPH oxidase activities could be explained by the NADH:dehydrogenase II (Ndh) contribution (16) that represents approximately 50% of total NADH oxidase activity (17). In absence of O2, all oxidases activities sharply decreased to zero demonstrating the O2 dependent oxidation of NADPH mediated by electron transfer through respiratory chain. To demonstrate that electron transfer from NADPH to the respiratory chain can pump protons, we evaluated the membrane potential (ΔΨ), component of the proton motive force ( ), generated in ISO vesicles during electron transfer from NAD(P)H through the respiratory chain. The ability to generate ΔΨ in ISO vesicles was studied using oxonol V as fluorescence probe. NADH and NADPH abilities to generate ΔΨ were compared between the parental NA23 and evolved NA23E04 strains (Fig. 1). Only membrane vesicles prepared from evolved strain were able to generate ΔΨ upon the addition of both NADH and NADPH (Fig. 1B), as indicated by oxonol V fluorescence. Weak ΔΨ generation in parental ISO vesicles upon NADPH addition (Fig. 1A) could be generated by the membrane bound pyridine nucleotide transhydrogenase (PntAB), functioning as a proton pump (18). Inhibition of terminal cytochrome oxidase by potassium cyanide caused ΔΨ collapse, near the initial level (Fig. 1B), except for parental ISO vesicles in the presence of NADPH (Fig. 1A), confirming the proton motive force generation by electron transfer from NADPH to O2 only occurs in evolved ISO vesicles.
), generated in ISO vesicles during electron transfer from NAD(P)H through the respiratory chain. The ability to generate ΔΨ in ISO vesicles was studied using oxonol V as fluorescence probe. NADH and NADPH abilities to generate ΔΨ were compared between the parental NA23 and evolved NA23E04 strains (Fig. 1). Only membrane vesicles prepared from evolved strain were able to generate ΔΨ upon the addition of both NADH and NADPH (Fig. 1B), as indicated by oxonol V fluorescence. Weak ΔΨ generation in parental ISO vesicles upon NADPH addition (Fig. 1A) could be generated by the membrane bound pyridine nucleotide transhydrogenase (PntAB), functioning as a proton pump (18). Inhibition of terminal cytochrome oxidase by potassium cyanide caused ΔΨ collapse, near the initial level (Fig. 1B), except for parental ISO vesicles in the presence of NADPH (Fig. 1A), confirming the proton motive force generation by electron transfer from NADPH to O2 only occurs in evolved ISO vesicles.
Table 3.
NAD(P)H oxidase activity in ISO vesicles
| NA23 | NA23E04 | |||
| Activity (μmol·min-1·mg-1)* | NADH | NADPH | NADH | NADPH |
| Aerobic oxidase | 0.47 ± 0.05* | 0.010 ± 0.003 | 0.71 ± 0.07 | 0.28 ± 0.03 |
| Anaerobic oxidase | 0.002 ± 0.001 | 0.006 ± 0.001 | 0.005 ± 0.001 | 0.004 ± 0.001 |
*Means ± S.D. for 3–4 independent measurements
Fig. 1.
(A) and (B) Generation of ΔΨ in at the respiratory chain ISO vesicles. The quenching of oxonol V fluorescence was used to monitor potential membrane generation in ISO vesicles prepared from NA23 (A) and NA23E04 (B) strains. Solid and dash-dot lines represent quenching upon NADH and NADPH, respectively.
Thus, the adaptive evolution of the NA23 mutant strain over the short period of 20 generations led to the selection of an evolved NADH:ubiquinone oxidoreductase that could use both NADH and NADPH as an electron donor to create a proton motive force. Moreover, the unique NADPH oxidation pathway of the NA23E04 evolved strain correlated with the decrease in the NADPH/NADP+ ratio from 2.05 to 1.85 (Table 1).
Evolution of E. coli Under Higher NADPH Stress.
In the initial strategy, we constructed a strain that produced 2 mol of NADPH and 2 mol of NADH per mole of glucose oxidized to acetyl-coA. To increase NADPH stress we constructed a strain that produced 3 mol of NADPH and 1 mol of NADH per mole of glucose oxidized to acetyl-coA, by replacing the pgi deletion in strain NA23 with a deletion of the pfkA and pfkB genes encoding phosphofructokinases (NA24 strain; Table S1). This strain was unable to grow in glucose mineral medium. As we previously found that an evolved Δpgi strain appeared during the first subculture, we applied a different strategy to select an evolved strain using the ΔpfkAB strain. We took 109 cells from LB medium and plated them on glucose mineral medium plates, obtaining eight colonies that all contained a mutation in the nuoF gene. Five had the same mutation as the NA23 evolved strain (NuoF*E183A) and three had another mutation, but at the same position (NuoF*E183G). The five clones with the E183A mutation had an average growth rate on glucose of μ = 0.08 ± 0.01 h-1 whereas the three others had an average growth rate of μ = 0.06 ± 0.03 h-1.
Biochemical characterization of the NuoEF*(E183G)G soluble fragment was performed as described above by measuring both NADH and NADPH ferricyanide reductase activities. As expected, the selected evolved soluble NADH:ubiquinone oxidoreductase fragment could use both NADH and NADPH as an initial electron donor, with a Km value for NADPH of 8.4 ± 1.7 μM (Table 2). A 320-fold increase in the catalytic efficiency of the evolved NuoEF*(E183G)G (3.5 ± 1 s-1·μM-1) soluble fragment compared to the native fragment was observed.
Enzyme Structure Model of NuoF, NuoF*E183A, and NuoF*E183G.
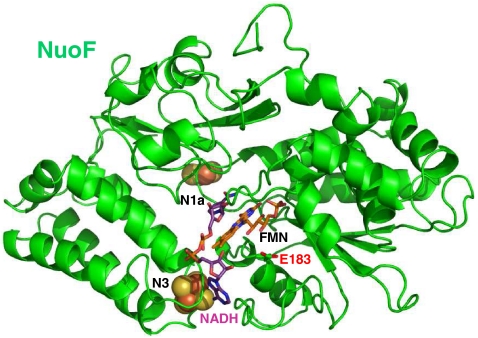
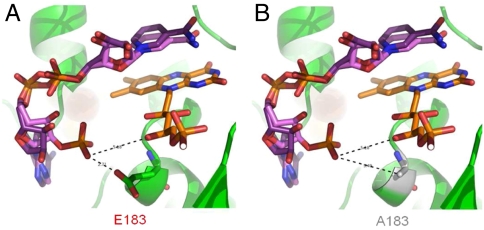
For additional biochemical characterization, and to investigate the relationship between enzyme structure and the catalytic mechanism of the evolved fragment, both E. coli NuoF and the two NuoF* subunits were subjected to modeling, with manual docking of NAD(P)H based on the recently solved crystal structure (19) of the hydrophilic domain of Thermus thermophilus complex I. The NuoF subunit has 49% identity to the T. thermophilus Nqo1 subunit, and both subunits contain an NADH-binding site, with flavin mononucleotide (FMN) as the primary electron acceptor, and an N3 tetranuclear cluster (20). All residues involved in binding FMN and NADH are strongly conserved. Fig. 2 shows the putative structure of the native E. coli NuoF subunit containing FMN, and indicating invariant residues including glutamate 183, which is involved in interaction with NADH. NADPH was docked manually in the same position as NADH (Fig. 3A) that was experimentally determined in the T. thermophilus Nqo1 subunit. It showed that the NADH cavity cannot accommodate an NADPH molecule because of both steric hindrance and repulsive effects with the charged glutamate side chain (Fig. 3A). On the other hand when glutamate 183 was replaced with alanine (Fig. 3B) or glycine, the negative phosphate positioning was favored by a longer distance of 5.79 Å from the NADPH phosphate oxygen and the absence of repulsive effect. These modeling results explained how NuoEF*G established an original NADPH reoxidation pathway in the cell.
Fig. 2.
Putative structure of the NuoF subunit from E. coli deduced from the T. Thermophilus Nqo1 structure. The two prosthetic [4Fe-4S] and [2Fe-2S] clusters from N3 and N1a, and the FMN cofactor are in black.
Fig. 3.
(A) and (B) Comparison of the FMN and NAD(P)H binding sites in the NuoF and NuoF* subunits. NADH and NADPH are superimposed and docked in front of FMN. Distances from the negatively charged NADPH phosphate to FMN and residue 183 are in angstroms. Sterically hindrance between the NADPH phosphate and glutamate 183 is seen. NADH is not obstructed and can be placed in front of FMN.
Analysis of the Conservation of the E183 Residue Among All the NuoF Sequences Available in the Genebank Protein Database.
We align all the NuoF protein sequences around the E183 amino acid residue (based on E. coli NuoF numbering) and analyzed the conservation of this residue. It was highly conserved among the NuoF proteins and when it was absent, like for example in the NuoF of Schizosaccharomyces pombe (accession number NM_001021761.1), the glutamate residue was replaced by an aspartate residue that should also exclude NADPH from entering the active site.
Discussion
In this work, the central metabolic network in E. coli was restructured to force a higher production of NADPH and it was coupled to the elimination of the natural metabolic pathway to produce NADH from NADPH. Two strains, NA23 and NA24 producing, respectively, 2 and 3 mol of NADPH from the oxidation of glucose to acetyl-CoA were rationally designed, constructed and independently grown on glucose mineral medium. The ability of these strains to adapt to, and overcome the NADPH stress was evaluated. Results illustrate that both strains were able to evolve rapidly and adapt to this metabolic constraint. The genetic basis of this fast adaptive evolution was identified and revealed that in each case, a single gene, nuoF, encoding a subunit of the water-soluble NADH:ubiquinone oxidoreductase fragment, was mutated at only one position in the entire genome sequence. Comparative biochemical and in vivo characterization of both native and evolved NADH:ubiquinone oxidoreductases were undertaken and showed that: (i) the evolved NADH:ubiquinone oxidoreductase soluble fragment had a higher NADPH ferricyanide reductase activity and a higher NADPH catalytic efficiency for NADPH than the native one (ii) the evolved NADH:ubiquinone oxidoreductase possessed a higher oxygen dependent NADPH oxidase activity than the native one, and (iii) the evolved NADH:ubiquinone oxidoreductase was able to transfer electrons from NADPH to respiratory chain and this was coupled to proton motive force generation that was not observed for the native complex I. Finally, modeling of the NuoF and NuoF* subunit structures supported the conclusion that both mutations in the nuoF gene exert their beneficial effect via the same mechanism. The presence of an amino acid residue carrying a neutral and short aliphatic side chain; i.e. alanine or glycine, at glutamate 183 in the binding pocket for NADH and FMN allows NADPH phosphate positioning. Glutamate 183, by excluding NADPH from the binding pocket, plays a key role in the strict NADH specificity of NADH:ubiquinone oxidoreductase. The fact that this selectivity resides in a single amino acid residue, highly conserved among the NuoF proteins, suggests that evolutionary forces in natural environments are such that microorganisms obtain a clear advantage from separate control of cofactor pools.
Thereby, the complete characterization of the improved phenotypes observed in both NA23 and NA24 evolved strains at genetic, biochemical and physiological levels clearly demonstrated that the mutation at only one position in NuoF is the unique fitness-enhancing mutation. The mutation led to the creation of an original metabolic pathway for energy generation, through the reassignment of an unexpected function to the NADH: ubiquinone oxidoreductase: a NADPH reoxidation function. Adaptive evolution under NADPH imbalance created a unique mechanism of NADPH oxidation that has never been described in nature. By evolving the strictly NADH-dependent NADH:ubiquinone oxidoreductase to an enzyme that used both NADH and NADPH, a direct catabolic role was created for NADPH. NADPH plays here an original role in a living cell: a dual anabolic and direct catabolic role. Indeed, NADPH is normally strictly used for anabolism and to protect cells from different oxidative stresses, whereas NADH has a catabolic role in providing energy to cells. As it is vital to all organisms to maintain a perfect balance between the catabolic NADPH formation and the anabolic demand, several natural mechanisms have been described to ensure such network-wide balancing (21). Transhydrogenases are mainly recognized to be involved in this phenomenon and, in organisms lacking transhydrogenases, the lack of cofactor specificity of the glucose-6-phosphate dehydrogenase appears to be a major mechanism to avoid catabolic NADPH overproduction. Any hypothesis suggesting a possible direct NADPH reoxidation pathway through the respiratory chain has never been described.
With regard to strain evolution, the observed phenomenon is an also unusual event: a reassignment of new function of an enzyme. Indeed, in most reported cases, adaptive evolution resulted from an increase in the capacity of already active pathways or the activation of latent pathways (3, 22). Moreover, a strong parallelism of the adaptive evolution process for independent endpoints was observed that is unusual. Adaptive parallelism was previously reported during bacterial evolution experiments (8, 23). However in these cases, bacterial evolution was parallel at the level of genes, but only rarely were substitutions identical at the base pair level. In the present study, the target of the evolution obtained was highly reproducible even in strains exhibiting different original genotypes, suggesting that modification of the NADH-binding pocket of NADH:ubiquinone oxidoreductase is the easiest way for cells to adapt and overcome a strong imbalance in NADPH redox state.
To conclude, this study demonstrates that surviving the imposed NADPH imbalance in absence of the soluble transhydrogenase UdhA clearly requires a new metabolic function for NADPH in E. coli: NADPH is now involved in direct energy generation through the respiratory chain. This work demonstrates again the strong robustness of both genetic and metabolic networks of E. coli and illustrates how existing metabolic dogmas can be challenged in a synthetic biology type of metabolic design.
Materials and Methods
Construction of mutant strains.
Chromosomal deletion, integration, and removal of antibiotic resistance genes were according to Datsenko and Wanner (10). Details are given in SI Text. Oligonucleotides used are listed in Table S1.
Adaptive Evolution.
Evolution of NA23 MG1655 Δpgi∷FRT Δedd∷FRT Δqor∷FRT ΔudhA∷FRT strains was in 50 mL of glucose minimal synthetic medium in 500 mL Erlenmeyer baffled flasks, at 37 °C, on a gyratory shaker at 200 rpm. Samples were frozen in 20% (v/v) glycerol solution from each serial evolution subculture. Details are given in SI Text.
Identification of Mutations at the Genome Level.
Ten cloned colonies were isolated from both populations at the fourth generation of evolution, with five from each population, and grown on glucose minimal medium to determine specific growth rates. NA23E04 was selected for mutation investigation by CGS (Roche Nimblegen) because of its higher growth rate. Genomic DNA from NA23E04 mutant and wild-type MG1655 strain was prepared using a QIAamp DNA Mini Kit (bacterial extraction protocol, Qiagen) and concentrated by vacuum centrifugation (Speedvac) if necessary before CGS analysis. Putative mutations from CGS were validated by PCR and sequencing.
Analytical Procedures and Physiological Parameters.
Cell growth was monitored as OD600. Glucose and acetate concentrations were determined by high-performance liquid chromatography. Separation was performed on a BioRad Aminex HPX-87H column (300 mm × 7.8 mm), and detection was done by refractometry. Operating conditions were: mobile phase 5 mM sulphuric acid, flow-rate 0.6 mL/ min, temperature 40 °C. Physiological parameters (maximum growth, specific glucose consumption and specific acetate production rates) were determined during exponential growth phase using a calculated correlation factor of 0.35 g cellular dry weight per OD600.
Determination of Intracellular Cofactor Concentrations.
NADPH, NADP+, NADH, and NAD+ extractions were by acid or alkaline lysis as described (24). The NAD(P)(H) cycling assay was adapted from Gibon and Larher (25) in microplate format. Details are given in SI Text.
Complex I Characterization
Construction of Expression Vectors.
Expression of NuoEFG NADH:ubiquinone oxidoreductase soluble fragments was performed using a pET11a/nuoB-G/NuoFc expression vector that added a Streptag II to the C-terminus of NuoF, kindly supplied by Professor Thorsten Friedrich (13). Details are given in SI Text.
Expression, Purification, and Biochemical Characterization of NuoEFG and NuoEF*G Soluble Fragments.
To express wild-type NuoEFG, evolved NuoEF*(E183A)G and NuoEF*(E183G)G soluble fragments, competent E. coli BL21(DE3) was transformed with pET11a/nuoB-G/NuoFc, pET11a/nuoB-G/NuoF*(E183A)c or pET11a/nuoB-G/NuoF*(E183G)c. Transformants were grown, harvested, and stored as described (12). Details are given in SI Text.
Preparation of Inside-Out Vesicles.
Inside-out (ISO) vesicles were prepared according to a modified procedure of Matsushita et al. (26). Details are given in SI Text.
Enzyme Assays on ISO Vesicles.
Each NAD(P)H oxidase activity of ISO vesicles was measured at 25 °C spectrometrically (26). Anaerobic assays were done in a JACOMEX anaerobic chamber with anaerobic buffer and ISO vesicles degassed by a gentle stream of nitrogen. Details are given in SI Text.
Determination of Membrane Potential (ΔΨ).
The generation of ΔΨ (inside-positive) in ISO vesicles was monitored at 25 °C by following fluorescence quenching of oxonol V (Fluka) (27). Details are given in SI Text.
NuoF Subunit Modeling.
A structure template from T. thermophilus (PBD-ID: 4FUG) with 49% sequence identity was used for E. coli NuoF subunit modeling. After careful examination for potential alignment errors, the automated comparative protein modeling program MODELLER 9v5 was employed. Figures were prepared with PyMOL (DeLano Scientific, http://pymol.sourceforge.net/).
Supplementary Material
Acknowledgments.
We thank Pr. Thorsten Friedrich (Institut für Organische Chemie und Biochemie, Freiburg, Germany) for the gift of the soluble NADH:ubiquinone oxidoreductase fragment expression plasmid. We also thank Dr. Nicholas Lindley (LISBP, Toulouse, France) for comments on the manuscript. This work was supported by the Association Nationale de la Recherche et de la Technologie/Délégation Régionale à la Recherche et à la Technologie and METabolic Explorer (Clermont-Ferrand, France).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010431108/-/DCSupplemental.
References
- 1.Philippe N, Crozat E, Lenski RE, Schneider D. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays. 2007;29:846–860. doi: 10.1002/bies.20629. [DOI] [PubMed] [Google Scholar]
- 2.Ibarra RU, Edwards JS, Palsson BO. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- 3.Fong SS, Nanchen A, Palsson BO, Sauer U. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J Biol Chem. 2006;281:8024–8033. doi: 10.1074/jbc.M510016200. [DOI] [PubMed] [Google Scholar]
- 4.Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem. 2004;279:6613–6619. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- 5.Nissen TL, Anderlund M, Nielsen J, Villadsen J, Kielland-Brandt MC. Expression of a cytoplasmic transhydrogenase in Saccharomyces cerevisiae results in formation of 2-oxoglutarate due to depletion of the NADPH pool. Yeast. 2001;18:19–32. doi: 10.1002/1097-0061(200101)18:1<19::AID-YEA650>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Canonaco F, et al. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett. 2001;204:247–252. doi: 10.1111/j.1574-6968.2001.tb10892.x. [DOI] [PubMed] [Google Scholar]
- 7.Thorn JM, Barton JD, Dixon NE, Ollis DL, Edwards KJ. Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH. J Mol Biol. 1995;249:785–799. doi: 10.1006/jmbi.1995.0337. [DOI] [PubMed] [Google Scholar]
- 8.Herring CD, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 9.Conrad TM, et al. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10:R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich T. The NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochim Biophys Acta. 1998;1364:134–146. doi: 10.1016/s0005-2728(98)00024-3. [DOI] [PubMed] [Google Scholar]
- 12.Braun M, Bungert S, Friedrich T. Characterization of the overproduced NADH dehydrogenase fragment of the NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochemistry. 1998;37:1861–1867. doi: 10.1021/bi971176p. [DOI] [PubMed] [Google Scholar]
- 13.Bungert S, Krafft B, Schlesinger R, Friedrich T. One-step purification of the NADH dehydrogenase fragment of the Escherichia coli complex I by means of Strep-tag affinity chromatography. FEBS Lett. 1999;460:207–211. doi: 10.1016/s0014-5793(99)01341-1. [DOI] [PubMed] [Google Scholar]
- 14.Friedrich T, et al. A small isoform of NADH:ubiquinone oxidoreductase (complex I) without mitochondrially encoded subunits is made in chloramphenicol-treated Neurospora crassa. Eur J Biochem. 1989;180:173–180. doi: 10.1111/j.1432-1033.1989.tb14629.x. [DOI] [PubMed] [Google Scholar]
- 15.Pohl T, et al. Nucleotide-induced conformational changes in the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) Biochem Soc Trans. 2008;36:971–975. doi: 10.1042/BST0360971. [DOI] [PubMed] [Google Scholar]
- 16.Jaworowski A, Mayo G, Shaw DC, Campbell HD, Young IG. Characterization of the respiratory NADH dehydrogenase of Escherichia coli and reconstitution of NADH oxidase in ndh mutant membrane vesicles. Biochemistry. 1981;20:3621–3628. doi: 10.1021/bi00515a049. [DOI] [PubMed] [Google Scholar]
- 17.Noguchi Y, et al. The energetic conversion competence of Escherichia coli during aerobic respiration studied by 31 P NMR using a circulating fermentation system. J Biochem. 2004;136:509–515. doi: 10.1093/jb/mvh147. [DOI] [PubMed] [Google Scholar]
- 18.Chang DY, Hou C, Bragg PD. Anomalous effect of uncouplers on respiratory chain-linked transhydrogenation in Escherichia coli membranes: Evidence for a localized proton pathway? Arch Biochem Biophys. 1992;293:246–253. doi: 10.1016/0003-9861(92)90392-a. [DOI] [PubMed] [Google Scholar]
- 19.Sazanov LA, Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 20.Morgan DJ, Sazanov LA. Three-dimensional structure of respiratory complex I from Escherichia coli in ice in the presence of nucleotides. Biochim Biophys Acta. 2008;1777:711–718. doi: 10.1016/j.bbabio.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 21.Fuhrer T, Sauer U. Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J Bacteriol. 2009;191:2112–2121. doi: 10.1128/JB.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua Q, Joyce AR, Palsson BØ, Fong SS. Metabolic characterization of Escherichia coli strains adapted to growth on lactate. Appl Environ Microbiol. 2007;73:4639–4647. doi: 10.1128/AEM.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci USA. 2006;103:9107–9112. doi: 10.1073/pnas.0602917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lilius EM, Multanen VM, Toivonen V. Quantitative extraction and estimation of intracellular nicotinamide nucleotides of Escherichia coli. Anal Biochem. 1979;99:22–27. doi: 10.1016/0003-2697(79)90039-3. [DOI] [PubMed] [Google Scholar]
- 25.Gibon Y, Larher F. Cycling assay for nicotinamide adenine dinucleotides: NaCl precipitation and ethanol solubilization of the reduced tetrazolium. Anal Biochem. 1997;251:153–157. doi: 10.1006/abio.1997.2283. [DOI] [PubMed] [Google Scholar]
- 26.Matsushita K, Ohnishi T, Kaback HR. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry. 1987;26:7732–7737. doi: 10.1021/bi00398a029. [DOI] [PubMed] [Google Scholar]
- 27.Yamada H, Tokuda H, Mizushima S. Proton motive force-dependent and -independent protein translocation revealed by an efficient in vitro assay system of Escherichia coli. J Biol Chem. 1989;264:1723–1728. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.