Abstract
Stromal interaction molecules (STIM)s function as endoplasmic reticulum calcium (Ca2+) sensors that differentially regulate plasma membrane Ca2+ release activated Ca2+ channels in various cells. To probe the structural basis for the functional differences between STIM1 and STIM2 we engineered a series of EF-hand and sterile α motif (SAM) domain (EF-SAM) chimeras, demonstrating that the STIM1 Ca2+-binding EF-hand and the STIM2 SAM domain are major contributors to the autoinhibition of oligomerization in each respective isoform. Our nuclear magnetic resonance (NMR) derived STIM2 EF-SAM structure provides a rationale for an augmented stability, which involves a 54° pivot in the EF-hand:SAM domain orientation permissible by an expanded nonpolar cleft, ionic interactions, and an enhanced hydrophobic SAM core, unique to STIM2. Live cells expressing “super-unstable” or “super-stable” STIM1/STIM2 EF-SAM chimeras in the full-length context show a remarkable correlation with the in vitro data. Together, our data suggest that divergent Ca2+- and SAM-dependent stabilization of the EF-SAM fold contributes to the disparate regulation of store-operated Ca2+ entry by STIM1 and STIM2.
Keywords: NMR structure, protein stability, STIM2, store-operated calcium entry
Together, stromal interaction molecules (STIM) and Orai proteins are the major components of store-operated Ca2+ entry (SOCE) where endoplasmic reticulum (ER) Ca2+ store depletion leads to an open plasma membrane (PM) Ca2+ release activated Ca2+ (CRAC) channel configuration, vital to myriad Ca2+-signaled cellular functions (1). STIMs are type I, predominantly ER-localized transmembrane proteins that function as Ca2+ sensors through luminal EF-hand and SAM domains (2–5) and activators of Orai-composed CRAC channels (6–11) through putative cytosolic coiled-coil domains (12–17) (Fig. 1A). SOCE initiation occurs upon Ca2+-depletion dependent STIM oligomerization (18, 19). Cytosolic CRAC influx ensues after translocation of these multimers to ER-PM junctions inducing recruitment of Orai to the same sites (20–22). Vertebrates translate two STIM isoforms that despite high amino acid conservation (Fig. S1) are distinct in Ca2+ sensitivity and roles in SOCE. STIM1 is vital in stimulus-induced CRAC entry (2–4), while STIM2 is imperative in intracellular Ca2+ homeostasis (23, 24). Both STIM1 and STIM2 are requisite in CRAC-induced immune cell activation, notwithstanding a lesser effect of STIM2 knockout (-/-) on measureable SOCE compared to STIM1 (-/-) in T-cells and fibroblasts (24, 25). STIM2 is partially active at resting ER Ca2+ concentrations, resulting in both store-dependent and—independent modes of CRAC channel activation (26). STIM2 plays an important role in neuronal Ca2+ signaling, although both isoforms have been identified in a variety of vertebrate cell types (27). The EF-hand together with the SAM domains (i.e., EF-SAM)s in the luminal region of all STIMs are responsible for Ca2+ sensing and initiating the molecular reorganization at ER-PM junctions responsible for SOCE (18, 19).
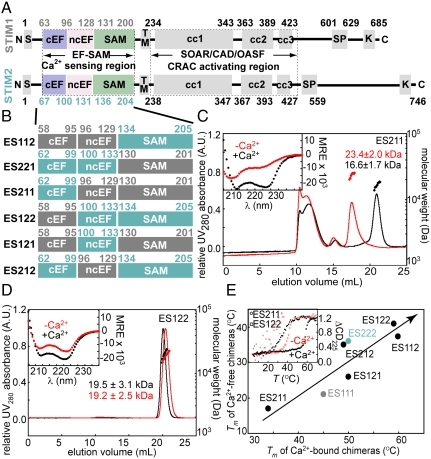
Fig. 1.
(A) Domain comparison of human STIM1 and STIM2. N, amino terminus; S, ER signal sequence; cEF, canonical EF-hand; ncEF, noncanonical EF-hand; SAM, sterile α motif; TM, transmembrane; cc, coiled coil; SP, Pro/Ser-rich region; K, Lys-rich region; and C, carboxy terminus. (B) STIM1 and STIM2 EF-SAM chimeric design. STIM1 and STIM2 components are indicated in gray and teal, respectively. (C) Far-UV CD spectra (inset) and gel filtration (left axis, solid lines) with in-line MALS determined molecular weights (right axis, filled circles) of Ca2+-depleted and -loaded ES211. (D) Far-UV CD spectra (inset) and gel filtration (left axis, solid lines) with in-line MALS determined molecular weights (right axis, filled circles) of Ca2+-depleted and -loaded ES122. (E) Correlation between the thermal stability of Ca2+ loaded and -depleted chimeras. The inset displays the thermal melts of ES211 and ES122. Red traces are Ca2+-depleted samples (i.e., 0.5 mM EDTA), while black traces are Ca2+-loaded (i.e., 10 mM CaCl2 added to the Ca2+-depleted samples) in C, D, and E.
In vitro, STIM2 EF-SAM exhibits distinct Ca2+-binding, folding, and stability characteristics compared to STIM1 (28, 29). These differences are vital to the Ca2+-sensing function of STIMs as Ca2+ dissociation, which occurs at different ER Ca2+ levels for STIM1 vs. STIM2 (23, 24), causes disruption of the EF-hand:SAM domain interaction and oligomerization of these luminal domains (19). However, these in vitro data are insufficient to explain the precise mechanistic nature of STIM functional distinctions. Here, we engineered STIM1/STIM2 EF-SAM chimeric fusions to delineate the structural basis for the differences observed between the isoforms in vitro and in live cells. We created both “super-stable” and “super-unstable” chimeras which exhibited discrete Ca2+ sensitivities and oligomerization properties in vitro and within the full-length STIM1 context. Using NMR spectroscopy, we solved the solution structure of human STIM2 EF-SAM to compare to our previously determined STIM1 structure and understand how this Ca2+-sensitive oligomerization switch region inimitably functions in vertebrates despite a very high sequence similarity (Fig. S1).
Results
STIM1/STIM2 EF-SAM Chimeras Have Distinct Biophysical Characteristics In Vitro.
The importance of the EF-SAM region in STIM1-mediated CRAC activation has been previously established by our (19) and other laboratories (18, 30, 31). In vitro, STIM1 EF-SAM is markedly destabilized upon Ca2+-depletion, subsequently undergoing partial unfolding-coupled oligomerization (5, 19). However, both STIM1 and STIM2 EF-SAM recombinant proteins have an inherent ability to oligomerize, albeit with STIM1 EF-SAM unfolding and oligomerizing considerably faster than STIM2 under similar solution conditions (28). STIM1 EF-SAM has a somewhat higher Ca2+ affinity than STIM2 (23) assessed by Ca2+-binding induced circular dichroic (CD) spectral changes (Fig. S2), underscoring a role for other structural factors in the stability differences between the isoforms and the need for more detailed biophysical and structural analyses.
First, we used a motif swapping approach to identify the key determinants of EF-SAM stability. We defined three major motifs as swapping candidates based on our STIM1 EF-SAM structure (19) and the high sequence homology between STIM1 and STIM2: (i) the canonical EF-hand, (ii) the noncanonical EF-hand, and (iii) the SAM domain. Subsequently, we engineered all combinations of STIM1/STIM2 EF-SAM chimeras into pET-28a vectors (Fig. 1B). Protein was attainable from every construct except ES221 (i.e., STIM2 canonical EF-hand motif and STIM2 noncanonical EF-hand motif fused to the STIM1 SAM domain) which showed no detectable expression using BL21(DE3) Escherichia coli cells, deficient in OmpT and lon proteases. ES221 did not appear to be sequestered in inclusions, because extraction using guanidine yielded no protein. Codon usage within each delineated motif and expression conditions were maintained from wild-type (i.e., induction with 0.5 mM IPTG at 25 °C), suggesting a greater instability and susceptibility to other E. coli proteases compared to the remaining chimeras.
Remarkably, each EF-SAM chimera that expressed in E. coli showed considerable α-helicity by far-UV CD in the presence of Ca2+; further, all these artificial EF-SAM domains had an innate structural sensitivity to the absence of Ca2+ characterized by reduced α-helicity (insets of Fig. 1 C and D and Fig. S3 A, C, and E). The Ca2+-loaded chimeras exhibited cooperative thermal unfolding transitions by CD, consistent with mutual unfolding of the EF-hand domains together with the SAM domain (Fig. 1E inset and Fig. S3 B, D, and F). Similar cooperative unfolding was observed in the Ca2+-depleted states, albeit at considerably lower temperatures (Fig. 1E inset and Fig. S3 B, D, and F). Overall, a strong correlation was observed between Ca2+-depleted and Ca2+-loaded EF-SAM stability where chimeras with the lowest Ca2+-loaded stability also maintained a decreased stability in the Ca2+-depleted state (Fig. 1E). The least stable chimera was ES211 with a Ca2+-depleted thermal melt exhibiting considerable scatter (Fig. 1E inset). This super-unstable EF-SAM chimera exhibited an apparent thermal unfolding midpoint (Tm) ∼ 4 and ∼19 °C lower than wild-type STIM1 and STIM2 EF-SAM in the Ca2+-depleted states, respectively. Our chimeric recombination also yielded super-stable EF-SAM with ES122 showing the greatest augmentation in Ca2+-depleted stability at ∼20 and 5 °C higher than wild-type STIM1 and STIM2, respectively (Table S1).
In order to evaluate the relationship between stability and oligomerization, we evaluated the quaternary structure of the EF-SAM chimeras in vitro using gel filtration with in-line multiangle light scattering (MALS) at 4 °C. The super-unstable ES211 chimera oligomerized in both the presence and absence of Ca2+, while the super-stable chimeras (i.e., ES122 and ES112) maintained a monomeric structure irrespective of Ca2+ levels (Fig. 1 C and D). The ES121 and ES212 chimeras which demonstrated stabilities most like wild-type STIM1 and STIM2 EF-SAM, respectively, also showed gel filtration-MALS profiles akin to the wild-type ancestors (Fig. S3 A, C, and E).
Overall, the Ca2+-depletion induced loss in structure and accompanying destabilization by the chimeras is consistent with the wild-type EF-SAMs; moreover, the oligomerization observed for the super-unstable ES211 and the STIM1-like chimeras along with the resistance to oligomerization by the super-stable and STIM2-like chimeras underscores the importance of stability in the oligomerization process.
Common Themes in STIM1 and STIM2 EF-SAM Structures.
We implemented conventional heteronuclear solution NMR spectroscopy to determine the three-dimensional structure of STIM2 EF-SAM. Purified recombinant STIM2 EF-SAM (Thr62-Gly205) exhibited a well-dispersed 1H-15N heteronuclear single quantum coherence (HSQC) spectrum in the presence of Ca2+ (Fig. S4A). The STIM2 EF-SAM structure was resolved using 2,708 nuclear Overhauser effect (NOE)-derived distance, 203 dihedral angle (i.e., 101 × ϕ and 102 × ψ), and 45 hydrogen bond restraints (Table S2). Ca2+-loaded STIM2 EF-SAM folds into a compact, highly α-helical structure with a backbone root mean square deviation (rmsd) of 0.50 ± 0.07 Å (Fig. S4B). The EF-hand domain contains two helix-loop-helix motifs (α1-loop1-α2, α3-loop2-α4), where loop1 coordinates the Ca2+ ion, evidenced by the downfield shifted 1H(N) Gly85 at ∼10.3 ppm (Fig. S4A) and loop2 stabilizes the canonical Ca2+ binding loop (i.e., loop1) via hydrogen-bonding [i.e., N(H) of Ile87: C(O) of Ile119]. The second, noncanonical EF-hand motif within STIM2 EF-SAM is not identifiable by sequence analyses and is structurally conserved in STIM proteins (19). A short helix (α5) links the EF-hand pair in sequence space to the SAM domain composed of five α-helices (α6 to α10) (Fig. 2A). The tertiary structure of STIM2 EF-SAM forms a surface electrostatic potential that is primarily acidic at neutral pH (Fig. 2B). The EF-hand pair contributes the majority of anionic charges to the potential map, while the SAM domain imparts considerable basic potential.
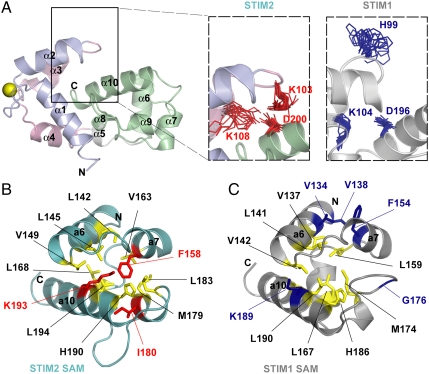
Fig. 2.
(A) Secondary structure motifs and globular fold of STIM2 EF-SAM. Motif color is consistent with Fig. 1A. The Ca2+ ion is shown as a yellow sphere. (B) Electrostatic surface potential of STIM2 EF-SAM at neutral pH. Basic potential is shown in blue, while acidic charge is in red. (C) Comparison of the EF-hand cleft hydrophobicity for STIM2 (teal) and STIM1 (gray). The hydrophobic clefts are shaded in yellow. The Lys103 and Trp128 which contribute to the cleft in STIM2 are shown in red, while the aligned residues in STIM1 are colored in blue. (D) Orientation of the EF-hand domains relative to the SAM domains in STIM1 and STIM2. The long axes of the helices (i.e., α2 and α10) used for the interhelical angle calculations are indicated. The STIM2 and STIM1 EF-SAM pdbIDs are 2L5Y and 2K60, respectively.
With greater than 85% sequence similarity, STIM2 EF-SAM is structurally homologous to STIM1 (i.e., backbone rmsd of 2.7 Å) (Fig. S5A). Both the canonical and noncanonical EF-hands adopt an “open” conformation characterized by interhelical angles of > 80°, facilitating the exposure of hydrophobic residues which form a concave nonpolar cleft. Each EF-hand motif and loop contributes hydrophobic residues to the formation of this cleft (i.e., canonical EF-hand: Leu72, Ile75, His76, Met79, and Phe85; noncanonical: Met100, Lys103, Lys108, Leu112, Ile119, Leu124, and Trp128) which serves as a dock (Fig. 2C) for a hydrophobic protrusion formed by the SAM domain of STIM2. The α10 helix of the STIM2 SAM domain minimally contributes Leu199 and Leu203 as hydrophobic anchors for interaction with the EF-hand domain cleft. The residue type and position of these anchors are conserved in STIM1; however, the α10 helix of the STIM2 SAM domain encodes an additional nonpolar Val201 at this end of the helix which is not conserved in STIM1 (Fig. S5B).
STIM1 and STIM2 Exhibit Variations in EF-SAM Structures.
Our present structure of STIM2 EF-SAM has illuminated some fascinating differences between the human isoforms. Most striking is the position of the STIM2 SAM domain which is rotated away from the canonical EF-hand motif such that α2 and α10 are in a primarily parallel conformation in STIM2 (i.e., ∼150° interhelical angle) compared to perpendicular in STIM1 (i.e., ∼94°) (Fig. 2 E and F). These interdomain conformational differences which augment the stability of STIM2 EF-SAM are promoted by at least two structural factors. First, the STIM2 EF-hand pair has a more extensive nonpolar cleft with the incorporation of Lys103 and Trp128 compared to the aligned His99 and Trp124 which are directed away from the STIM1 EF-hand cleft (Fig. 2 C and D). Secondly, Asp200 of the SAM α10 helix forms close ionic interactions with Lys103 located on the EF-hand loop between the α2 and α3 helices and Lys108 on α3 (Fig. 3A) which may pull the SAM domain away from the canonical EF-hand. In STIM1, the conserved α10 Asp (i.e., Asp196 in STIM1) and the α3 Lys (i.e., Lys104 in STIM1) also orient closely; however, the Lys103 aligns with a His99 in STIM1 that is directed away from α10 (Fig. 3A and Fig. S5B). Consistent with the more extensive charged interactions in STIM2, an Asp200Ala mutation decreases the EF-SAM Tm by ∼8 °C in STIM2 while the Asp196Ala variation in STIM1 decreases the Tm by ∼5 °C, resulting in similar mutant Ca2+-loaded stabilities (i.e., ΔTm ∼ 2 °C compared to ∼5 °C in the wild-type forms).
Fig. 3.
(A) Relative positions of STIM2 Lys103, Lys108, and Asp200 compared to aligned STIM1 His99, Lys104, and Asp196. The charged side chain positions of the 20 lowest energy structures for STIM2 are shown in red; the aligned residues of the 20 lowest energy structures for STIM1 are in blue. (B) Hydrophobic side chain packing in the STIM1 and STIM2 SAM domains. Residues in the hydrophobic cores are colored in yellow. The Lys193, Ile180, and Phe158 which enhance the STIM2 SAM domain (teal) hydrophobic core are shown in red. The aligned STIM1 SAM residues (i.e., Lys189, Gly176, and Phe154) as well as Val134 and Val138 which pack against Phe154 are in blue.
While a stronger EF-hand:SAM domain interaction due to more extensive hydrophobic and interdomain ionic interactions contribute to the increased stability of STIM2 EF-SAM compared to STIM1, it is of interest that chimeras which encode a STIM2 SAM domain are most stable (Fig. 1E) and most resistant to Ca2+-depletion induced oligomerization, regardless of the chimeric combination of the EF-hand pair. Our STIM2 EF-SAM structure also reveals differences in the composition of the SAM domain cores. The STIM2 SAM domain folds into a 5-helix bundle with 12 residues of nonpolar character at least 95% inaccessible to solvent (i.e., Leu142, Leu145, Val149, Phe158, Val163, Leu168, Met179, Ile180, Leu183, His190, Lys193, and Leu194); the STIM1 SAM domain only buries 9 hydrophobic residues (i.e., Val137, Leu141, Val145, Leu159, Leu167, Met174, His186, Leu190, and Ala194). The Ile180 which is buried in the STIM2 SAM domain is not conserved in STIM1 (i.e., Gly176) (Fig. S5B). This Ile180 is involved in a rearrangement of the core to include Phe158 and Lys193 in STIM2, whereas STIM1 excludes these conserved residues (i.e., Phe154 and Lys189) (Fig. 3 B and C). Introducing a Phe154Ala mutation into STIM1 EF-SAM reduces the Ca2+-loaded Tm by ∼11 °C because this Phe normally packs against Val134 and Val138 of α6 (Fig. 3C); however, the Phe158Ala variation in STIM2 dramatically destabilizes the Ca2+-loaded protein causing it to oligomerize and precipitate even at low temperatures (i.e., ∼4 °C). Hence, Phe158 is requisite for the global structural integrity of Ca2+-loaded STIM2 EF-SAM domain, while the STIM1 Phe154 plays a more local role in mediating hydrophobic contacts between the SAM α6 and α7 helices.
Stabilizing EF-SAM Domains within Full-Length STIM1 Inhibit CRAC Entry.
To further define the role of EF-SAM stability as well as the other STIM structural domains in the regulation of SOCE we engineered our most destabilizing (i.e., ES211) and stabilizing chimeras (i.e., ES122) along with wild-type STIM2 EF-SAM (i.e., ES222) into full-length STIM1 for functional analyses. Monomeric cherry fluorescent protein (mCherry) N-terminally fused to STIM1, STIM1-chimeras, and STIM2 were separately transfected into HeLa cells and the total internal reflective fluorescence (TIRF) localization was assessed before and after treatment with thapsigargin (TG). At resting ER luminal Ca2+, mCherry-STIM1 shows a diffuse distribution; however, TG-induced ER Ca2+ store depletion caused an extensive reorganization of STIM1 into puncta (Fig. S6). Cells expressing mCherry-STIM2 were partitionable into TG-sensitive and -insensitive groups. The TG-sensitive group demonstrated a similar response to store depletion as STIM1 expressing cells; however, the remaining cells exhibited a persistent and Ca2+ store-independent presence of puncta (Fig. S6). Cells overexpressing the STIM1-ES222 chimera were responsive to TG in a similar manner as wild-type STIM1, whereas cells expressing the super-unstable chimera (i.e., STIM1-ES211) displayed a constitutive presence of puncta; further, HeLa cells expressing the super-stable chimera displayed a diffuse distribution of mCherry at resting Ca2+ which markedly rearranged into puncta after TG treatment (Fig. S6). Remarkably, both the STIM1-ES222 and -ES122 full-length chimeras were sensitive to ER Ca2+ and formed morphologically similar puncta as wild-type STIM1.
We used electrophysiological measurements of CRAC current-density to examine the kinetics of channel activation in patch clamped HEK-293 cells cooverexpressing Orai1 and the mCherry-STIM proteins. After break-in with a pipette containing 20 mM EGTA (i.e., passive store depletion), STIM1 expressing cells showed an initial delay of ∼50 s followed by an inward developing current to a maximum of ∼8 pA/pF; further, the time to the maximal current after the initial delay was ∼100 s (Fig. 4A). Consistent with the TIRF observations, 7 of 11 STIM2-expressing cells demonstrated store-independent and maximal inward currents beginning at -8 pA/pF with no delay, while the remaining 4 of 11 cells were sensitive to Ca2+ store depletion as an inward developing current was observed with no delay and a time to maximal activation of ∼100 s (Fig. 4B). The STIM1–ES211 super-unstable expressing cells exhibited a maximal, constitutive inward rectifying current (i.e., ∼8 pA/pF) with no delay or further development implying the PM Ca2+ channels were in an open state, independent of ER Ca2+ levels (Fig. 4C). Cells expressing STIM1-ES222 displayed store-independent (i.e., 5 of 14 cells) or store-dependent (i.e., 9 of 14 cells) CRAC currents. The store-dependent currents exhibited a lag or delay from the time of break-in followed by the development of inward currents to a maximum inward density of ∼-7 pA/pF (Fig. 4D). The super-stable STIM1-ES122 chimera produced profiles which also exhibited a delay in activation; this delay was not different from wild-type STIM1; however, the time to maximal inward current-density after the delay was significantly longer than cells overexpressing wild-type STIM1 or STIM2 constructs (Fig. 4 E and F). The relatively small, basal outward currents did not change during CRAC measurements, and all inward rectifying currents were completely inhibited by the addition of La3+ to the external medium confirming a CRAC channel-dependent entry pathway. Overall, the electrophysiological data confirm that destabilization of EF-SAM activates CRAC entry, while stabilization of this domain prolongs the time to maximal activation.
Fig. 4.
Whole cell inward current plots of HEK-293 cells cooverexpressing Orai1 with wild-type mCherry-STIM1-EF-SAM (A), wild-type mCherry-STIM2-EFSAM (B), mCherry-STIM1-ES211 (C), mCherry-STIM1-ES222 (D), and mCherry-STIM1-ES122 (E). Data are means ± SEM, calculated from n, number of cells from at least three separate experiments. The definitions for delay and activation time are indicated in A. Gray bars indicate the addition of La3+ to the external medium. (F) Summary of delays in activation time and times to maximal inward current densities after normalization. P values were calculated using a two-tailed Student’s t-test, assuming equal variances and α = 0.5.
Discussion
In recent years, significant progress has been made in understanding protein folding coupled to target binding for a range of biological functions (32). We have previously suggested that STIM activation involves protein unfolding and homooligomerization, triggered by Ca2+ depletion of the EF-hand. Here, we have elucidated a basis for the distinct functional properties between STIM1 and STIM2 by defining the individual roles of the EF-hand and SAM domains in structural stability and oligomerization. While STIM1 and STIM2 possess distinct Ca2+ sensitivities (23) and cellular distribution (27), each motif within EF-SAM is remarkably interchangeable in chimeric combinations and some are even fully functional when engineered into full-length STIM1.
The Ca2+-binding induced stabilization of EF-hands is a well conserved feature of these common protein domains. For example, the isolated C- and N-terminal EF-hand domains of Troponin C (TnC), each undergo considerable stabilization upon Ca2+-binding (33). Despite the high sequence and structural homology to STIM1, the STIM2 EF-hand binds Ca2+ with lower apparent affinity than the STIM1 counterpart (Fig. S2) (23); however, STIM2 EF-SAM is considerably more stable than STIM1. Analogous to our STIM observations, the C-terminal EF-hand domain of TnC has a lower stability than the N-terminal domain, despite a higher Ca2+ affinity; further, the elevated affinity is attributable to the greater degree of folding (i.e., larger energetic barrier to Ca2+-induced unfolding) which occurs upon Ca2+ binding compared to the N-terminal domain (33). A similar phenomenon occurs in the STIM proteins, where the more stable apo EF-SAM and lesser Ca2+-induced folding of the STIM2 EF-hand contributes to a lower affinity. The combined lower Ca2+ affinity and higher stability has important regulatory consequences (see below).
The specific EF-hand domain translated in STIM proteins not only dictates the Ca2+ affinity, but also provides a mechanism for the autoinhibition of SAM oligomerization via an intramolecular association. Our present STIM2 EF-SAM structure reveals differences in this interaction for the human isoforms, revealing that the STIM2 EF-hand:SAM interaction is stabilized relative to STIM1 via increased cleft hydrophobicity, nonpolar residue packing within the SAM core, and interdomain ionic interactions; further, these structural factors contribute to the diminished oligomerization propensity for STIM2 EF-SAM compared to STIM1 (28). Although SAMs can exist as stable monomers, homooligomerization can occur through the N- and C-terminal helices or the so-called midloop (i.e., centrally located within SAM) and end-helix regions (i.e., C-terminally located within SAM) (34), structural features conserved in STIM proteins. However, the autoinhibition of SAM oligomerization via an intramolecular EF-hand association, as elucidated for STIMs, is unique among SAM domains.
Our in vitro chimera data suggest that the SAM domain is a key determinant of EF-SAM oligomerization propensity, where fusions expressing the STIM2 SAM show the highest stability and lowest oligomerization tendency, while those harboring the STIM1 SAM demonstrate the greatest oligomerization tendency. The least stable chimera, ES211 is constitutively oligomerized in vitro. Replacing wild-type STIM1 EF-SAM with this super-unstable chimera within the full-length context causes the STIM molecule to spontaneously oligomerize and activate CRAC channels independent of ER Ca2+ levels (Fig. 4C). On the other hand, the super-stable ES122 chimera is resistant to oligomerization, irrespective of Ca2+ at low temperature (i.e., ∼4 °C), and recombination within full-length STIM1 results in a significantly extended time to maximal activation of CRAC currents after ER Ca2+ store depletion (Fig. 4 E and F). Within our EF-SAM stability map (Fig. 1E), wild-type STIM1 and STIM2 position themselves in metastable states suggesting evolution selected for functionally optimized rather than stability maximized proteins.
The structural identity of the Ca2+-binding EF-hand also has a notable affect on the activation of SOCE, as engineering wild-type STIM2 EF-SAM (i.e., ES222) into the full-length STIM1 context and cooverexpression with Orai1 results in store-independent and store-dependent CRAC currents, similar to full-length STIM2. The partitionable groups are due to a greater sensitivity of the STIM2 canonical EF-hand to small fluctuations in basal ER Ca2+ levels (i.e., due to lower Ca2+ affinity). However, while the Ca2+ store-dependent fraction of wild-type STIM2 expressing cells exhibit no delay in the development of CRAC currents, the ES222 chimeric group demonstrates a visible delay (Fig. 4 B and D). The separable groups are only observed for ES222 when cooverexpressed with Orai1. These differences underscore the vital contributions of the local relative concentration levels and the structural identities of all CRAC channel signaling components, including other domains within STIM (13, 31) and Orai1, in mediating SOCE.
In conclusion, we propose that STIM proteins have evolved metastable EF-SAM domains as the molecular machinery that first responds to changes in ER Ca2+ levels. Ca2+-depletion induced destabilization of the EF-hand:SAM interaction induces partial unfolding-coupled oligomerization of this region (28), the initiation event in CRAC channel activation (18, 19, 21). Our present data show that EF-SAM within STIM1 and STIM2 differentially regulate CRAC channels through a divergent balance between EF-hand Ca2+ affinity and SAM domain stability (Fig. S7), in addition to the integrity of the EF-hand:SAM interaction. STIM1 is an effective agonist-induced CRAC channel activator via a high Ca2+ affinity and low SAM stability, making it less responsive to small variations in ER Ca2+, but highly reactive when Ca2+ depleted. STIM2 is more sensitive to small changes in ER Ca2+ for basal Ca2+ homeostasis (23), but also capable of responding to agonist-induced Ca2+ depletion (25). Weakly structured or disordered regions within proteins have important roles in cell signaling (32). The divergence in EF-SAM stability has implications in CRAC channel regulation: the instability of the STIM1 SAM domain and weakly structured apo EF-SAM, enhances the rate of association, perhaps via a larger radius of interaction (32, 35), while the more stable STIM2 SAM and more compact STIM2 EF-SAM attenuates homooligomerization, increasing the time to maximal CRAC channel activation.
Methods
Engineering of Chimeric Fusions.
Multiple rounds of polymerase chain reaction (PCR) were used to generate chimeric EF-SAM inserts, cloned into pET-28a using NheI and XhoI sites. Full-length mCherry-STIM proteins were in pCMV6-XL5. EF-SAM proteins were expressed in E. coli and purified as previously described (5, 29). Further details are in the SI Text.
Far-UV-CD and Gel Filtration-MALS.
CD data were acquired on a Jasco J-815 CD spectrometer (Jasco, Inc.) collected in 1 nm increments (20 nm/ min) using a 0.01 or 0.1 cm path length cuvette, 8 s averaging time, and 1 nm bandwidth at 20 °C. Spectra were corrected for buffer contributions. Gel filtration was performed with Superdex S200 10/300 GL columns at 4 °C. MALS measurements were performed in-line with the gel filtration using a miniDawn light and an Optilab rEX differential refractometer (Wyatt Technologies, Inc.). Molecular weight was calculated using the ASTRA software (Wyatt Technologies, Inc.) based on Zimm plot analysis and using a protein refractive index increment, dndc-1 = 0.185 L g-1.
Solution NMR.
The EF-SAM structure was determined as previously described (19). Data was collected on 500 and 600 MHz Inova (Varian, Inc.) or 800 MHz Avance (Bruker Biospin Ltd.) spectrometers equipped with triple resonance cryoprobes. Chemical shifts were assigned using XEASY after data processing with NMRPipe. CYANA was used for automated structure calculation based on > 87% complete chemical shift assignments and 15N-edited and 13C-edited NOESY peak lists. Water refinement of structures was performed using the RECOORD scripts in CNS. Further details are in the SI Text.
Mammalian Cell Culture and Transfection.
Cells were cultured on 35 mm (No 0) glass bottom plates (MarTek Corp.) in Dulbecco’s modified Eagle’s medium (HyClone Thermo Fisher Scientific, Inc.) supplemented with 5–10% vol/vol fetal bovine serum (Gibco, Inc.), 100 U mL-1 penicillin, and 100 μg mL-1 streptomycin (Sigma-Aldrich Co.) to 50–80% confluence. Transfection was performed with Lipofectamine LTX (Invitrogen, Inc.) or Transfectin (Biorad, Inc.).
Fluorescence Microscopy.
Fluorescence images were acquired on an Olympus IX70 (Olympus, Inc.) microscope with a 60× oil-immersion TIRF objective (N.A. 1.45) (Olympus, Inc.), illuminated by a 543.5 nm HeNe laser (05-LGP-193; 5 mW) (Melles Griot Laser Group) for mCherry. Excitation light was reflected by a 485-555-650TBDR dichroic and emitted light was passed through a 515-600-730TBEM filter (Omega Optical, Inc.). Images were digitized with a cooled Evolution QEi CCD camera (Media Cybernetics, Inc.).
CRAC Current Recordings.
Whole cell patch clamp experiments were performed at ambient temperature (i.e., ∼21–25 °C). Voltage ramps were applied every 5 s from a holding potential of 0 mV, covering a range of -90 to 90 mV over 1 s. The internal pipette solution contained (in mM): 145 Cs methane sulphonate, 8 NaCl, 3.5 MgCl2,10 Hepes, 20 EGTA, pH 7.2 and the extracellular solution contained (in mM): 145 NaCl, 5 CsCl, 1 MgCl2, 10 Hepes, 10 glucose, 10 CaCl2, pH 7.4. Applied potentials were corrected for a liquid junction of +12 mV resulting from a Cl- based bath solution and a sulphonate based pipette solution. All currents were leak-subtracted using 10 μM La3+.
Supplementary Material
Acknowledgments.
We thank Richard S. Lewis (Stanford) for providing mCherry-STIM1. We acknowledge a Canadian Institutes of Health Research operating grant (to M.I.), Canadian Foundation for Innovation grant (to M.I.) for an 800 MHz NMR spectrometer, and Natural Sciences and Engineering Research Council of Canada and Ontario Cancer Institute Knudson fellowships (to P.B.S.). The work was supported by Austrian Science Fund project P21118 (to C.R.). M.I. holds the Canada Research Chair in Cancer Structural Biology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank www.pdb.org (PDB ID code 2L5Y). The NMR chemical shifts and restraints have been deposited in the Biological Magnetic Resonance Bank (BMRB ID code 17289).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015125108/-/DCSupplemental.
References
- 1.Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- 2.Liou J, et al. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roos J, et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang SL, et al. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stathopulos PB, et al. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: an initiation mechanism for capacitive Ca2+ entry. J Biol Chem. 2006;281:35855–35862. doi: 10.1074/jbc.M608247200. [DOI] [PubMed] [Google Scholar]
- 6.Feske S, et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 7.Zhang SL, et al. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc Natl Acad Sci USA. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vig M, et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vig M, et al. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeromin AV, et al. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prakriya M, et al. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- 12.Huang GN, et al. STIM1 carboxyl-terminus activates native SOC, I(crac) and TRPC1 channels. Nat Cell Biol. 2006;8:1003–1010. doi: 10.1038/ncb1454. [DOI] [PubMed] [Google Scholar]
- 13.Muik M, et al. A cytosolic homomerization and a modulatory domain within STIM1 C terminus determine coupling to ORAI1 channels. J Biol Chem. 2009;284:8421–8426. doi: 10.1074/jbc.C800229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muik M, et al. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J Biol Chem. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- 15.Park CY, et al. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan JP, et al. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frischauf I, et al. Molecular determinants of the coupling between STIM1 and Orai channels: differential activation of Orai1-3 channels by a STIM1 coiled-coil mutant. J Biol Chem. 2009;284:21696–21706. doi: 10.1074/jbc.M109.018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luik RM, et al. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stathopulos PB, et al. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Luik RM, Wu MM, Buchanan J, Lewis RS. The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J Cell Biol. 2006;174:815–825. doi: 10.1083/jcb.200604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci USA. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ Levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bird GS, et al. STIM1 is a calcium sensor specialized for digital signaling. Curr Biol. 2009;20:1724–1729. doi: 10.1016/j.cub.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh-Hora M, et al. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat Immunol. 2008;9:432–443. doi: 10.1038/ni1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parvez S, et al. STIM2 protein mediates distinct store-dependent and store-independent modes of CRAC channel activation. Faseb J. 2007;22:752–761. doi: 10.1096/fj.07-9449com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berna-Erro A, et al. STIM2 regulates capacitive Ca2+ entry in neurons and plays a key role in hypoxic neuronal cell death. Science Signaling: The Signal Transduction Knowledge Environment. 2009;2:ra67. doi: 10.1126/scisignal.2000522. [DOI] [PubMed] [Google Scholar]
- 28.Stathopulos PB, Zheng L, Ikura M. Stromal Interaction Molecule (STIM) 1 and STIM2 calcium sensing regions exhibit distinct unfolding and oligomerization kinetics. J Biol Chem. 2009;284:728–732. doi: 10.1074/jbc.C800178200. [DOI] [PubMed] [Google Scholar]
- 29.Zheng L, Stathopulos PB, Li GY, Ikura M. Biophysical characterization of the EF-hand and SAM domain containing Ca2+ sensory region of STIM1 and STIM2. Biochem Biophys Res Commun. 2008;369:240–246. doi: 10.1016/j.bbrc.2007.12.129. [DOI] [PubMed] [Google Scholar]
- 30.Baba Y, et al. Coupling of STIM1 to store-operated Ca2+ entry through its constitutive and inducible movement in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2006;103:16704–16709. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Covington ED, Wu MM, Lewis RS. Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol Biol Cell. 2010;21:1897–1907. doi: 10.1091/mbc.E10-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suarez MC, et al. Free-energy linkage between folding and calcium binding in EF-hand proteins. Biophys J. 2008;95:4820–4828. doi: 10.1529/biophysj.108.135715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiao F, Bowie JU. The many faces of SAM. Science: The Signal Transduction Knowledge Environment. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 35.Pontius BW. Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem Sci. 1993;18:181–186. doi: 10.1016/0968-0004(93)90111-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.