Abstract
Intraerythrocytic malaria parasites can obtain nearly their entire amino acid requirement by degrading host cell hemoglobin. The sole exception is isoleucine, which is not present in adult human hemoglobin and must be obtained exogenously. We evaluated two compounds for their potential to interfere with isoleucine utilization. Mupirocin, a clinically used antibacterial, kills Plasmodium falciparum parasites at nanomolar concentrations. Thiaisoleucine, an isoleucine analog, also has antimalarial activity. To identify targets of the two compounds, we selected parasites resistant to either mupirocin or thiaisoleucine. Mutants were analyzed by genome-wide high-density tiling microarrays, DNA sequencing, and copy number variation analysis. The genomes of three independent mupirocin-resistant parasite clones had all acquired either amplifications encompassing or SNPs within the chromosomally encoded organellar (apicoplast) isoleucyl-tRNA synthetase. Thiaisoleucine-resistant parasites had a mutation in the cytoplasmic isoleucyl-tRNA synthetase. The role of this mutation in thiaisoleucine resistance was confirmed by allelic replacement. This approach is generally useful for elucidation of new targets in P. falciparum. Our study shows that isoleucine utilization is an essential pathway that can be targeted for antimalarial drug development.
Human malaria afflicts ∼250 million people annually and results in nearly 1 million deaths (1). The most deadly of the four human malaria-causing parasites, Plasmodium falciparum is increasingly acquiring resistance to all available chemotherapeutics agents (2). To expand the repertoire of antimalarial targets, we focused on proteins within the isoleucine acquisition pathway. A complete amino acid supply is crucial for rapidly developing blood-stage parasites. Most amino acid biosynthetic pathways are absent in parasites (3). However, multiple pathways exist to ensure amino acid acquisition: they may be taken up directly from the medium or may be obtained from hemoglobin degradation. Isoleucine is unique in that it is the only amino acid that is not present in human adult hemoglobin. It is the sole amino acid that must be supplied exogenously to growing parasites (4).
Aminoacyl-tRNAs (AA-tRNAs) are ribosome substrates that pair codons with the cognate amino acids during the elongation phase of protein synthesis. AA-tRNAs are synthesized by the 3′-esterification of tRNAs with the appropriate amino acid by aminoacyl-tRNA synthetases (AARSs) (5). The accuracy of AARSs is critical for the overall fidelity of protein translation. The cytosol, the apicoplast, and the mitochondria are translationally active compartments of apicomplexan parasites and require charged tRNAs; however, only the cytosol and the apicoplast contain AARSs, while mitochondria are thought to import charged tRNAs (6).
The P. falciparum genome of malaria parasites encodes two potential isoleucyl-tRNA synthetases (IRSs) with accession numbers PF13_0179 and PFL1210w. The predicted enzymes are distantly related with sequence identity of only 20%. The predicted sequence of PF13_0179 is more similar to human IRS, with a sequence identity of 47.5%, whereas that of PFL1210w is more similar to bacterial IRS; the enzyme shares 29.3% identity with the IRS from Streptobacillus. The PF13_0179 product is predicted to be cytoplasmic, whereas the PFL1210w product is predicted to be targeted to the apicoplast (6). We refer to PF13_0179 as cyto-IRS and PFL1210w as api-IRS.
We report that in P. falciparum the isoleucine analog thiaisoleucine specifically interacts with the cyto-IRS, whereas mupirocin inhibits the activity of the api-IRS. Both compounds kill parasites and the midnanomolar IC50 values for mupirocin validate the drug as a lead compound. Point mutations in compound-resistant parasites were identified by genomic analysis including high-density tiling microarrays. Confirmation of the role of a mutation in thiaisoleucine resistance by allelic replacement illustrates the utility of this approach to validate drug targets.
Results
Thiaisoleucine and Mupirocin Interact with Isoleucine Utilization Targets.
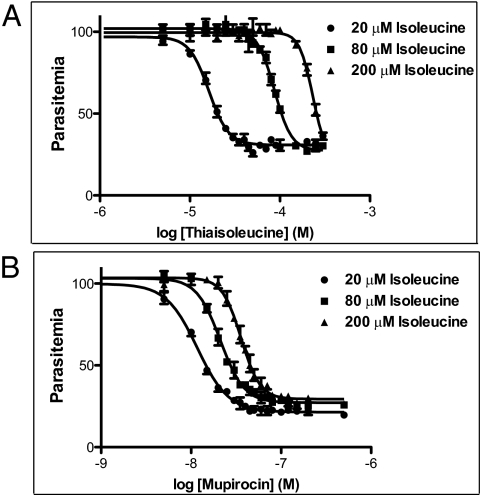
Thiaisoleucine is an analog of isoleucine that has substitution of the γ-methylene group by a sulfur atom (SI Appendix, Fig. S1 A and B). The compound kills blood stage parasites at micromolar concentrations (4). The potency of thiaisoleucine was attenuated by increasing the isoleucine concentration in the media in which parasites were maintained (Fig. 1A). This suggests that thiaisoleucine directly competes with isoleucine for a target. Mupirocin (SI Appendix, Fig. S1C) is a compound in clinical use for treatment of methicillin-resistant Staphylococcus aureus infections. This compound killed parasites at midnanomolar concentrations (Fig. 1B). Like thiaisoleucine, IC50 values for mupirocin increased with increasing extracellular isoleucine concentrations, although the magnitude of the IC50 change was smaller.
Fig. 1.
Potency of thiaisoleucine and mupirocin depends on exogenous isoleucine concentration. Parasite growth in the presence of 20 μM, 80 μM, or 200 μM isoleucine and either thiaisoleucine (A) or mupirocin (B). Growth of triplicate cultures at increasing concentration of drug was measured by flow cytometry. Data points are shown as mean ± range. Curves show least-square fits of data points. IC50 values and SEs are provided in SI Appendix, Table S1.
Kinetics of Parasite Death.
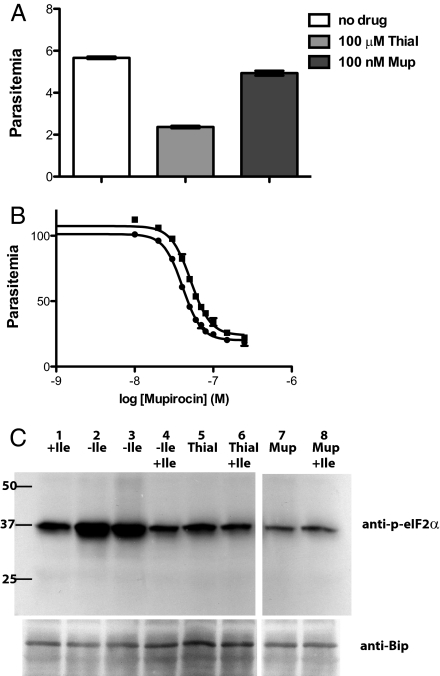
The kinetics of death was different for thiaisoleucine and mupirocin. Ring-stage parasites treated with thiaisoleucine were inhibited in development and did not reinvade successfully even after removal of drug (Fig. 2A). Mupirocin-treated cultures appeared normal for more than one complete asexual cycle but failed to develop invasion-competent parasites in the second cycle. This “delayed death” phenotype is similar to what is observed for other compounds that act against apicoplast targets such as clindamycin and azithromycin (7). Ultimate growth inhibition by mupirocin was not affected by removal of the drug in the first cycle (Fig. 2B). The failure of parasites to recover from drug treatment after removal of either thiaisoleucine or mupirocin indicates that the drugs act irreversibly. The distinct kinetics of parasite killing by the two compounds suggests that they inhibit different isoleucine utilization targets.
Fig. 2.
Thiaisoleucine and mupirocin kill parasites by different mechanisms, and neither agent elicits a starvation response. (A) Thiaisoleucine-treated parasites do not reinvade efficiently, whereas mupirocin-treated parasites initially appear unaffected. Synchronous ring-stage parasites grown in 20 μM isoleucine were incubated with either 100 μM thiaisoleucine (ThiaI) or 100 nM mupirocin (Mup) or no compound (triplicate cultures for each condition). Compounds were removed after 16 h by washout into high (200 μM) isoleucine media. Parasite growth was assessed by flow cytometry after an additional 32 h. Data are shown as mean ± SD. (B) Removal of mupirocin at the schizont stage does not alter the drug potency. A concentration range of mupirocin was added to six ring-stage cultures. At the late schizont stage cultures were washed 3× and placed back into either medium without drug (three replicates; ●, data mean) or medium containing mupirocin (three replicates; ■, data mean). Parasitemia was measured after control parasites had reinvaded twice. Data points are shown as mean ± range. Errors are very small. IC50 values and SEs are provided in SI Appendix, Table S2. (C) Neither thiaisoleucine nor mupirocin elicits an amino acid starvation response. Immunoblot detecting either phosphorylated-eIF2α (anti–p-eIF2α) or the endoplasmic reticulum protein BiP, which served as a loading control. Parasites were isoleucine starved for 75 min (2–4) followed by reincubation with 80 μM isoleucine for an additional 35 min (4). Parasites in lane 1 were maintained at 80 μM isoleucine throughout. Those in lanes 5 and 6 were incubated with 100 μM thiaisoleucine and those in lanes 7 and 8 were incubated with 80 nM mupirocin for 75 min. Those in lanes 6 and 8 were reincubated with an additional 80 μM isoleucine for 35 min. The initial isoleucine concentration for all samples was 80 μM. This blot is representative of two independent experiments.
When parasites are starved of isoleucine, they rapidly sense the amino acid deficit and respond by phosphorylation of eIF2α to shut off general translation (8). Increased eIF2α phosphorylation is a sensitive indicator of starvation. It occurs at isoleucine concentrations that fail to support optimal growth (<20 μM) (SI Appendix, Fig. S2) (4). Neither compound elicited this starvation response in treated parasites (Fig. 2C).
Selection of Thiaisoleucine-Resistant Parasites.
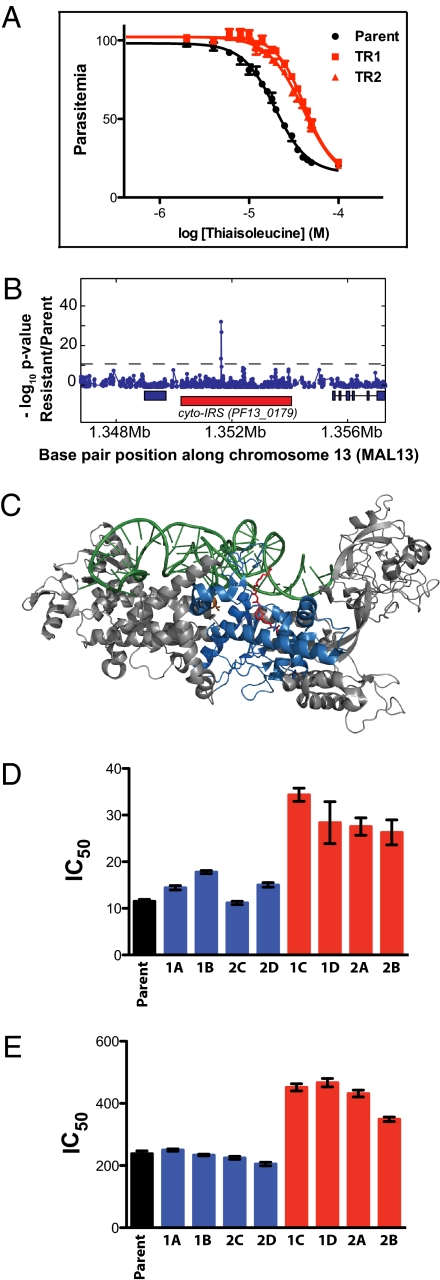
To determine the target of thiaisoleucine, we selected resistant parasites. We used 4 × 107 parasites in a single-step selection, as described elsewhere (9). Resistant parasites growing in 20 μM isoleucine were selected with 20 μM thiaisoleucine (IC80). Several different parasite strains were used, but only 3D7 yielded resistant parasites. Drug treatment cleared the culture of detectable parasites, and surviving parasites became visible at day 19 after drug addition, approximately the amount of time that it would take for a single parasite to overgrow the culture. Resistant parasites were cloned by limiting dilution and assessed for ability to grow in thiaisoleucine-containing media. All clones analyzed (25 total) were approximately twofold resistant (Fig. 3A). Similarly to WT parasites (Fig. 1A), the IC50 values of resistant parasites increased with higher isoleucine concentrations (SI Appendix, Table S3). This suggests that thiaisoleucine interacts with a target that requires isoleucine for activity.
Fig. 3.
Thiaisoleucine-resistant parasites contain a mutation in the cyto-IRS gene, and the mutation is responsible for the resistance. (A) Sensitivity to thiaisoleucine of the 3D7 parent and two resistant clones (TR1 and TR2). Resistant parasites were selected in 20 μM thiaisoleucine/20 μM isoleucine. Clones were then grown in 20 μM isoleucine with varying concentrations of thiaisoleucine and parasitemia was normalized to a no inhibitor control. Data points of triplicates are shown as mean ± range. IC50 values and SEs are provided in SI Appendix, Table S3. (B) z test analysis of microarray. Genomic DNA of clone TR1 was digested with DNaseI and hybridized to a 25-mer oligonucleotide tiling microarray. Plot shows −log P value for z test (blue line) performed with TR1 versus 3D7 reference. The red rectangle below the z test indicates the position of PF13_0179; blue rectangles indicate neighboring genes. A single SNP in PF13_0179 (the gene encoding cyto-IRS) is detected. The A2430T mutation results in a L810F change. (C) Model of IRS from S. aureus [pdb: 1FFY (24)]. The active site is marked by bound mupirocin in red, tRNA in green, and conserved residues of the Rossmann fold in blue. The residue mutated in thiaisoleucine-resistant parasites (L810F) corresponds to a valine residue in the bacterial protein (V622; shown in orange). This model was generated with MacPyMOL (DeLano Scientific LLC). Note that cyto-IRS is only distantly related to the structure shown and that its closer homologs do not bind mupirocin. (D) and (E) Sensitivity of WT and 8 allelic replacement clones toward thiaisoleucine in 20 μM (D) or 180 μM isoleucine (E). Clones expressing WT cyto-IRS are shown in blue (1A, 1B, 2C, and 2D); clones expressing L810F cyto-IRS are shown in red (1C, 1D, 2A, and 2B). Error bars indicate SEs. Numeric values are given in SI Appendix, Table S4.
Thiaisoleucine-Resistant Parasites Carry a Mutation in the Gene for the Cytoplasmic Isoleucyl-tRNA Synthetase.
DNA isolated from two thiaisoleucine-resistant parasite clones was analyzed using high-density tiling microarray. This array contains ∼6 million 25-mer probes that are complementary to the 26 million bp P. falciparum genome and spaced on average every two to three bases throughout nonredundant coding regions. This high coverage means that the majority of newly emerged SNPs in a drug-resistant clone can be readily detected as a loss of signal at multiple consecutive probes covering the SNP. The probability of a genomic change is calculated from the number of consecutive probes showing a hybridization difference relative to the parental reference genome (10). The hybridizations of the two clones were compared with two 3D7 reference hybridizations using a P value cutoff of 1 × 10−10. The putative polymorphisms were filtered to include only those in which more than one probe displayed loss of hybridization.
The microarray analysis revealed subtelomeric deletions as well as seven putative polymorphisms in the resistant clone (Dataset S1). We analyzed the putative polymorphisms by sequencing. Two potential mutations in chromosomes 1 and 7 were subtelomeric, did not reside in predicted genes, and could not be sequenced. Of the remaining five putative polymorphisms, four contained WT sequences. The other genomic change was on chromosome 13 in PF13_0179, a gene encoding a predicted cytoplasmic IRS (Fig. 3B). Sequencing showed that the hybridization difference was the result of a A2430T mutation, resulting in the change of amino acid 810 from L to F. The amino acid mutated in the resistant lines is located in the Rossmann fold, a highly conserved and well-structured domain of IRSs (Fig. 3C) (11). This domain binds isoleucine and ATP and is responsible for activating and transferring the amino acid to the tRNAIle. Mutations in this domain have been observed in bacterial IRSs that carry antibiotic resistance (12, 13). We hypothesized that the mutation in the predicted IRS (PF13_0179) is likely responsible for thiaisoleucine resistance.
Confirmation of the Mutation Role in Resistance.
To confirm that the mutation of L810F in PF13_0179 is responsible for resistance to thiaisoleucine, we performed allelic replacement with a single crossover homologous recombination construct encoding the amino acid change (SI Appendix, Fig. S3). In addition, a C-terminal GFP tag was introduced. This approach produces a single copy of a fluorescently tagged protein that is expressed at the native locus under control of the native promoter (14). In addition to the L810F change, the construct was engineered to carry a synonymous KasI restriction site just upstream of the allelic replacement codon that allows for easy screening of transfectants. Depending on the site of homologous recombination, parasites will express either the WT or the mutant enzyme (SI Appendix, Fig. S3A). The allelic replacement vectors contained a human dihydrofolate reductase (hDHFR) cassette that allows selection of plasmid-containing parasites with the antifolate WR99210. Two independent transfections of WT 3D7 parasites were performed. Transfected parasites were never exposed to thiaisoleucine; WR99210 was used for selection. Using PCR, we analyzed 40 clones, 15 of which were WT at the KasI locus, whereas 25 were mutant. A subset (2 WT and 2 mutant from each transfection for a total of eight clones) was sequenced and the expected integration was confirmed by Southern blot (SI Appendix, Fig. S3B). Sensitivity to thiaisoleucine was determined at two isoleucine concentrations. All parasite clones containing the introduced L810F mutation tolerated approximately twice the concentration of drug compared with untransfected parasites or compared with transfected parasites expressing WT enzyme (Fig. 3 D and E). Other than resistance to thiaisoleucine, we could not detect a phenotypic difference between the parental strain and thiaisoleucine-resistant selected parasites or parasites carrying the allelic replacement. The mutation was not deleterious to asexual blood-stage parasites, as it was preserved over 4 mo of continuous culturing.
Mupirocin Targets the Apicoplast Isoleucyl-tRNA Synthetase.
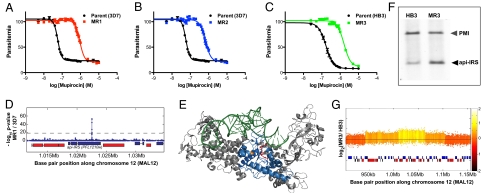
To identify the target of mupirocin, drug-resistant parasites were selected. Three independent selections yielded mupirocin-resistant parasites. Selections 1 and 2 were carried out using the 3D7 strain at 500 nM (1) or 150 nM mupirocin (2). Selection 3 was performed using the HB3 strain at 300 nM mupirocin. HB3 is naturally about threefold more resistant to mupirocin than all other strains investigated. Resistant parasites became noticeable ∼20 d after drug addition in each case and were cloned and analyzed for ability to replicate at increasing mupirocin concentrations. Clones MR1, MR2, and MR3 from selections 1–3 above, respectively, were found to be 18-, 13-, and 10-fold resistant to mupirocin compared with the respective parental clone (Fig. 4 A– C). DNA from cloned, resistant MR1 was hybridized to the tiling microarray. Comparison with two reference hybridizations of WT 3D7 using a P value cutoff of 1 × 10−10 and the filter step described above revealed six potential polymorphisms in addition to several subtelomeric deletions (Dataset S2). We analyzed the putative polymorphisms by sequencing and found that only two of the six potential mutations were real. One mutation was in the gene PFD1005c at bp 2048. The change of T to C at this position results in an amino acid mutation of L 683 to P. PFD1005c encodes an erythrocyte membrane protein (PfEMP1) that is an adhesion molecule. We considered it unlikely for PfEMP1 to contribute to mupirocin resistance. The other mutation was a nonsynonymous change of C to T at bp 4034 in the gene PFL1210w (Fig. 4D). This gene encodes api-IRS and the mutation changes proline 1233 to serine. Like the L810F mutation in cyto-IRS, api-IRS P1233 is located in the highly conserved Rossmann-fold of AARSs (Fig. 4E). A change of proline to serine may destabilize the helix that P1233 is predicted to initiate and could rearrange side chains critical for mupirocin binding.
Fig. 4.
Mupirocin-resistant parasites contain mutated api-IRS or amplifications of the gene. (A) Sensitivity to mupirocin of the 3D7-parent (black) and clone MR1 (P1233S) (red). (B) Sensitivity to mupirocin of the 3D7-parent (black) and clone MR2 (ΔT86) (blue). (C) Sensitivity to mupirocin of the HB3-parent (black) and clone MR3 (green). Clone MR3 does not contain a mutation. (D) High-resolution microarray. Plot of −log P value for z test (blue line) performed with clone MR1 versus 3D7 reference shows a single SNP in api-IRS. The location of the api-IRS gene (PFL1210w) is indicated with the blue rectangle below the z test result. Neighboring genes are shown as red or blue rectangles. (E) Model of IRS from S. aureus [pdb: 1FFY (24)]. Active site is marked by bound mupirocin in red; tRNA is in green; and conserved residues of the Rossmann fold are in blue. Residue mutated in the mupirocin-resistant line MR1 (P1233S) corresponds to proline 606 (shown in orange). This model was generated with MacPyMOL. (F) Southern blot of NcoI, EcoRV, ScaI digested total DNA from mupirocin-sensitive HB3 and clone MR3. Api-IRS and plasmepsin I (PMI) were detected with gene-specific probes. Gray arrowhead marks the 6.5-kb plasmepsin I (PMI) fragment. Black arrowhead marks 3.8-kb api-IRS fragment. This blot is representative of three experiments. (G) Plot of the log2 ratio of the intensity of each unique probe in MR3 versus HB3. The probe log ratios were colored by the moving average over a 500-bp window as indicated in color bar. A large (∼150-kb) copy number variation surrounding PFL1210w (marked with green asterisks) is present. A smaller amplification (∼50-kb) encompassing the gene within the CNV is observed as well.
Comparing MR2 to 3D7 using genome-scanning microarray analysis showed subtelomeric deletions in MR2 that frequently arise in response to long-term culturing as well as seven potential SNPs in nonantigenic protein-coding regions (Dataset S3). We determined by sequencing that only three of the seven potential SNPs were real mutations. Two reside in predicted proteins of unknown function (MAL7P1.154a and PFL2410w) and one in PFL1210w, the api-IRS gene (SI Appendix, Fig. S4). Of the three proteins with mutations in the MR2 clone, only PFL1210w contains an apicoplast-targeting sequence. The mutation in PFL1210w is a deletion of bp256-258, corresponding to the deletion of threonine 86. We were unable to determine the mechanism by which ΔT86 decreases sensitivity to mupirocin. T86 is unlikely to localize to the enzyme active site. It resides in the C-terminal end of the predicted apicoplast targeting sequence, adjacent to intron 1. We considered the possibility that Δ256–258 may result in altered splicing. mRNA was prepared and analyzed but the splicing of the mutant was found to be unchanged. SI Appendix, Fig. S5A, illustrates the api-IRS gene structure and position of the Δ256–258 and C4034T mutations.
Genome-scanning of MR3 detected a gene amplification surrounding PF1210w as well as an amplification within the amplification (Fig. 4G), in addition to subtelomeric deletions and SNPs in genes encoding variant surface antigens (Dataset S4). Sequencing confirmed that there were no mutations in the api-IRS gene in clone MR3. However, Southern blotting did show that MR3 has an increased number of gene copies (Fig. 4F). Importantly, none of the subtelomeric deletions or SNPs in MR1, MR2, and MR3 were common among the three independent resistance selections, whereas all three strains contained lesions with PFL1210w. Other than resistance to mupirocin, we could not detect a phenotypic difference between selected and WT parasites. Neither the deletion of T86 nor the P1233S change was deleterious to asexual blood-stage parasites, as the mutations were preserved over 3 mo of continuous culturing.
Attempted Allelic Replacement of api-IRS.
We attempted to introduce the P1233S mutation of mupirocin-resistant parasites into the parental strain by allelic replacement. The C-terminal portion of the gene (bp2815–5503) was amplified by PCR and cloned into the allelic replacement construct described above for cyto-IRS. The C4034T mutation was introduced into one targeting construct. To facilitate screening of transfectants a HindIII site at bp4013 was mutated in the construct carrying the C4034T change (SI Appendix, Fig. S5B). Both WT and mutant constructs were transfected into both 3D7 and mupirocin-resistant clone MR2 parasites (ΔT86). WR99210-resistant parasites were obtained and analyzed after two rounds of WR99210 cycling. Three to six independent transfections were examined for each. PCR amplification indicated correct targeting of the construct to the api-IRS locus. However, none of the 68 recombinants contained the mutated HindIII site at bp4013 or the C4034T change (SI Appendix, Fig. S5B). Thus, homologous recombination always occurred C-terminal to the mutations.
We considered the possibility that either or both mutations could impart a growth disadvantage and be subject to negative selection. To test this hypothesis, we transfected the MR1 clone (which has the C4034T api-IRS mutation) with the WT allele. Targeting constructs contained intron 3 (SI Appendix, Fig. S5) or had intron 3 deleted (SI Appendix, Fig. S6). Again, the allelic replacement constructs integrated into the api-IRS genes (SI Appendix, Fig. S6B) but none of the 40 recombinants had replacement of C4034T (SI Appendix, Fig. S5C). We conclude that the region upstream of bp4034 is inaccessible to homologous recombination.
Two Plasmodium IRSs Localize to Distinct Compartments in the Parasite.
We used fluorescence microscopy to experimentally determine the location of the two predicted P. falciparum IRSs. To localize PF13_0179 in the parasite, the C-terminally GFP-tagged version of the enzyme described above was used. GFP fluorescence was observed in the parasite cytoplasm, confirming that the gene is expressed during the erythrocytic stages and that it is localized in the proper compartment for the bulk of protein synthesis (Fig. 5A). Fluorescence intensity and localization were identical for the WT and the thiaisoleucine-resistant version of the enzyme. Expression was maximal in trophozoite and early schizont-stage parasites.
Fig. 5.
Localization of two IRSs: PF13_0179 is cytoplasmic, PFL1210w is in the apicoplast. (A) Live fluorescence microscopy of a doubly infected RBC expressing C-terminally tagged PF13_0179. The enzyme is expressed from the native promoter. (B) Live fluorescence microscopy of a schizont-stage parasite episomally expressing the N-terminal 92 amino acid of PFL1210w fused to GFP. (C) Immunofluorescence microscopy of the construct used in B using α-GFP and α-acyl carrier protein.
The predicted coding region for PFL1210w (1696 aa) is larger than that of PF13_0179 (1272 aa). In addition to a number of insertions that are distributed throughout the sequence, PFL1210w also carries an N-terminal extension that encodes a predicted signal peptide and a transmembrane domain. The PATS prediction program gives a high likelihood of apicoplast-targeting with a score of 0.996 (15). PFL1210w appears to be expressed at low levels in erythrocytic stages: mass spectrometry detected only one peptide of the large protein (16), and we could not detect fluorescence upon expressing the gene with a C-terminal GFP tag using the native promoter. To investigate the localization of PFL1210w, we fused the N-terminal 92 amino acids in frame with GFP and expressed the construct under a strong (hsp86) promoter (17). In live parasites, the GFP signal was present in ribbon-like structures characteristic for the apicoplast and it became more abundant in later stages of parasite development (Fig. 5B). The GFP signal colocalized with the well-characterized apicoplast-resident protein acyl carrier protein by immunofluorescence (Fig. 5C). This supports a function for PFL1210w in the apicoplast.
The delayed death phenotype of mupirocin-treated parasites (Fig. 2B) also suggests that the drug target is localized in the apicoplast. Delayed death has been described in detail for parasites of the phylum Apicomplexa (7, 18). It is observed when apicoplast gene transcription or translation is inhibited. Hallmarks of delayed death include failure to develop invasion-competent merozoites in the second cycle and no effect in the first cycle of parasite growth. Furthermore, washout of drug in the first cycle does not alter drug potency. Mupirocin has been well characterized as a tight-binding competitive inhibitor of bacterial IRSs (19), which show sequence similarity to P. falciparum api-IRS.
Discussion
We have evaluated two compounds potentially active against Plasmodium isoleucine utilization to gain insight into the utility of this pathway as a drug target. We succeeded in raising drug-resistant parasites for both compounds and have shown that the two compounds interact with different enzymes. Thiaisoleucine-resistant parasites maintain mupirocin sensitivity, and mupirocin-resistant parasites are WT in their response to thiaisoleucine (SI Appendix, Table S6). Mupirocin is potent against malaria parasites and is the most effective delayed-death drug described thus far. It is a clinically approved drug, but is restricted to topical use because it is rapidly hydrolyzed in body fluids at its ester bond (20). Still, the potential for new, improved mupirocin analogs with good activity against malarial api-IRS is high. Thiaisoleucine is not a drug lead; the amino acid analog has limited potency against parasites and likely has a low therapeutic index. Nevertheless it is a valuable tool to study isoleucine acquisition and indicates that simple isoleucine analogs can be incorporated by the cyto-IRS, resulting in parasite toxicity.
Identification of drug targets by selection of compound-resistant parasites raises an important issue. If resistance develops, does this mean that a particular enzyme is not a good target? We believe that resistance developed in the laboratory should not be considered a deterrent to drug development. Malaria parasites develop rapid resistance to most agents. Drugs such as chloroquine, to which resistance cannot be achieved with single-point mutants, are rare. Consequently, monotherapy is never acceptable, and multiple new targets are urgently needed, as all present drugs are failing.
Drug resistance might arise from unexpected and undetected mutations elsewhere in the genome and validation must be carried out. We succeeded in validating the interaction of cyto-IRS with thiaisoleucine by recapitulating resistance in WT parasites after introduction of a single amino acid change into the cyto-IRS gene. Unfortunately, we could not directly validate that a single amino acid change in api-IRS gives rise to mupirocin resistance. It is unclear why homologous recombination in api-IRS always occurred in the C-terminal portion of the gene. Based on the position of the mutation in the targeting construct, we expected ∼30% of clones to carry the change. Mechanisms other than length of homologous sequence must at least partially regulate recombination sites. The api-IRS gene contains a number of low complexity regions. These regions may be important for frequency of recombination. Most likely, low complexity sequences in api-IRS also prevented us from obtaining full-length clones for overexpression in the parasites. Our assessment of the mupirocin target therefore relies on the repeated appearance of mutations and amplifications in the api-IRS gene in independent drug selections. Tiling microarray analysis on three independently obtained mupirocin-resistant parasite clones showed that changes in only one gene, PFL1210w, the gene encoding api-IRS, are in common among the resistant parasites, a finding that is highly unlikely to occur without cause. In bacteria, mupirocin resistance results from mutations in the IRS gene, uptake of a plasmid encoding a homologous IRS, or increased copy numbers of the gene (13). Similar mechanisms appear to permit growth of malaria parasites in mupirocin: we characterized two clones with point mutations in the api-IRS gene and one clone with a gene amplification.
Although cyto-IRS interacts with thiaisoleucine, inhibition of the synthetase is not likely the cause of parasite death. Inhibition of IRS activity would be expected to result in accumulation of uncharged tRNAIle similar to amino acid starvation. However, thiaisoleucine-treated parasites do not initiate an amino acid starvation response (Fig. 2C). Parasites may instead be poisoned by thiaisoleucine incorporation into newly translated proteins. Consistent with this idea is the limited recovery of parasites after thiaisoleucine removal (Fig. 2A). Previous studies in other organisms have described the following: (i) thiaisoleucine competes with isoleucine for binding to the synthetase; (ii) tRNAIle can be charged with thiaisoleucine; (iii) the analog can be incorporated into polypeptides; and (iv) neither the ribosome run-off nor the protein chain elongation is affected by substitution (21). Given the high conservation of Plasmodium and mammalian IRSs, it is probable that thiaisoleucine acts similarly in the parasite.
New drugs against malaria are urgently needed. Two recent publications describing thousands of compounds active against blood-stage malaria give us hope (22, 23). However, elucidation of the compound targets is important for drug development. This paper describes an approach to identify and validate chemo-targets in P. falciparum. The natural mutational frequency of malaria parasites is high enough to isolate drug-resistant parasites. Sites of nonsynonymous changes or gene amplification in resistant parasites can then be identified with high-density microarray. By genetically replacing WT with mutant versions of the target gene, validation of the mechanism of drug resistance can be achieved.
Materials and Methods
Reagents.
dl-thiaisoleucine and mupirocin were obtained from Sigma. Other reagents are described in SI Appendix.
Vector Construction, Culturing of Parasites, Transfections. Southern and Western Blotting, and Fluorescence Microscopy.
Details of vector construction, parasite culturing, transfections. Southern and Western blotting. and fluorescence microscopy are given in SI Appendix.
Selection of Resistant Parasites.
For thiaisoleucine-resistance selection, the compound was added at 20 μM to asynchronous parasite cultures (parasitemia of 1%) that had been maintained in media containing 20 μM isoleucine for 4 wk before the experiment. Several laboratory strains of P. falciparum were used in the selection (HB3, Dd2, W2, and 3D7), and for each selection ∼4 × 107 parasites were used. After addition of thiaisoleucine, parasite growth medium was changed twice daily for 6 d,and once daily thereafter. Every 6 d, parasites were diluted 1:1. Several selections were attempted, but only one with 3D7 yielded resistant parasites. Mupirocin-resistance selection was performed in normal RPMI medium (382 μM isoleucine). All successful selections started with 1 × 106 asynchronous parasites (parasitemia of 0.5%). Selections 1 and 2 were carried out using the strain 3D7 and 500 nM or 150 nM mupirocin. Selection 3 was carried out with the strain HB3 and 300 nM mupirocin. Surviving parasites became visible 21, 23, or 21 d after drug addition for selection 1, 2 or 3, respectively. No resistant parasites were obtained with strains Dd2 and W2. From each selection, parasites were cloned, and at least two clones were characterized. Clones from the same selection did not show differences, and we report data from one clone from each selection (MR1 from selection 1, MR2 from selection 2, and MR3 from selection 3). Media changes and parasite dilutions were performed as for the thiaisoleucine selection.
Tiling Microarray Analysis on Resistant Parasites.
Tiling microarray analysis was performed as previously described (10). For TR1, TR2, MR1, and MR3, the z test in the polymorphism detection used a P value cutoff of 1 × 10−10 and reference 3D7 or HB3 hybridizations. For MR2 a P value cutoff of 1 × 10−5 and three different reference 3D7 hybridizations were used to identify putative SNPs. Predicted polymorphisms that had decreased relative hybridization intensity in more than one probe were considered. Additional methods are described in SI Appendix.
IC50 Determinations.
IC50 determinations were performed with synchronous ring-stage parasite cultures. For experiments requiring special isoleucine concentrations, parasites were maintained at 20 μM isoleucine for at least 5 d before the start of the experiment. For experiments with cyto-IRS allelic replacement clones, WR99210 was omitted for ∼2 d before the experiments. Data analysis was performed with the software package Prism. No constraints were used in any of the analyses. Additional details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Shalon Ledbetter for sharing reagents and for helpful discussions about isoleucine starvation, Barb Vaupel for assistance with molecular cloning, Rusiko Turabelidze for help with parasite culturing, and David Plouffe for microarray hybridizations. E.A.W., N.V.D., and S.E.B. were supported by grants from the W. M. Keck Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011560108/-/DCSupplemental.
References
- 1.World Health Organizaton . World Malaria Report 2008. Geneva: World Health Organization; 2008. pp. 1–78. [Google Scholar]
- 2.Dondorp AM, et al. Artemisinin resistance: Current status and scenarios for containment. Nat Rev Microbiol. 2010;8:272–280. doi: 10.1038/nrmicro2331. [DOI] [PubMed] [Google Scholar]
- 3.Payne SH, Loomis WF. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot Cell. 2006;5:272–276. doi: 10.1128/EC.5.2.272-276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Istvan ES, Gluzman IY, Gross J, Goldberg DE. Plasmodium falciparum ensures its amino acid supply with multiple acquisition pathways and redundant proteolytic enzyme systems. Proc Natl Acad Sci USA. 2006;103:8840–8845. doi: 10.1073/pnas.0601876103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 6.Pino P, et al. Mitochondrial translation in absence of local tRNA aminoacylation and methionyl tRNA Met formylation in Apicomplexa. Mol Microbiol. 2010;76:706–718. doi: 10.1111/j.1365-2958.2010.07128.x. [DOI] [PubMed] [Google Scholar]
- 7.Fichera ME, Bhopale MK, Roos DS. In vitro assays elucidate peculiar kinetics of clindamycin action against Toxoplasma gondii. Antimicrob Agents Chemother. 1995;39:1530–1537. doi: 10.1128/aac.39.7.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fennell C, et al. PfeIK1, a eukaryotic initiation factor 2alpha kinase of the human malaria parasite Plasmodium falciparum, regulates stress-response to amino-acid starvation. Malar J. 2009;8:99. doi: 10.1186/1475-2875-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rathod PK, McErlean T, Lee PC. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:9389–9393. doi: 10.1073/pnas.94.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dharia NV, et al. Use of high-density tiling microarrays to identify mutations globally and elucidate mechanisms of drug resistance in Plasmodium falciparum. Genome Biol. 2009;10:R21. doi: 10.1186/gb-2009-10-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nureki O, et al. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- 12.Antonio M, McFerran N, Pallen MJ. Mutations affecting the Rossman fold of isoleucyl-tRNA synthetase are correlated with low-level mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:438–442. doi: 10.1128/AAC.46.2.438-442.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulander W, Maisnier-Patin S, Andersson DI. Multiple mechanisms to ameliorate the fitness burden of mupirocin resistance in Salmonella typhimurium. Mol Microbiol. 2007;64:1038–1048. doi: 10.1111/j.1365-2958.2007.05713.x. [DOI] [PubMed] [Google Scholar]
- 14.Klemba M, Beatty W, Gluzman I, Goldberg DE. Trafficking of plasmepsin II to the food vacuole of the malaria parasite Plasmodium falciparum. J Cell Biol. 2004;164:47–56. doi: 10.1083/jcb200307147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuegge J, Ralph S, Schmuker M, McFadden GI, Schneider G. Deciphering apicoplast targeting signals—feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene. 2001;280:19–26. doi: 10.1016/s0378-1119(01)00776-4. [DOI] [PubMed] [Google Scholar]
- 16.Aurrecoechea C, et al. PlasmoDB: A functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37(Database issue):D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo I, Oksman A, Goldberg DE. Fatty acid acylation regulates trafficking of the unusual Plasmodium falciparum calpain to the nucleolus. Mol Microbiol. 2009;72:229–245. doi: 10.1111/j.1365-2958.2009.06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim L, McFadden GI. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci. 2010;365:749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakama T, Nureki O, Yokoyama S. Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J Biol Chem. 2001;276:47387–47393. doi: 10.1074/jbc.M109089200. [DOI] [PubMed] [Google Scholar]
- 20.Thomas CM, Hothersall J, Willis CL, Simpson TJ. Resistance to and synthesis of the antibiotic mupirocin. Nat Rev Microbiol. 2010;8:281–289. doi: 10.1038/nrmicro2278. [DOI] [PubMed] [Google Scholar]
- 21.Busiello V, Di Girolamo M, De Marco C. Thiaisoleucine and protein synthesis. Biochim Biophys Acta. 1979;561:206–214. doi: 10.1016/0005-2787(79)90503-3. [DOI] [PubMed] [Google Scholar]
- 22.Gamo FJ, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 23.Guiguemde WA, et al. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silvian LF, Wang J, Steitz TA. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.