Abstract
Toll-like receptors (TLRs) play a crucial role in several innate immune responses by regulating autophagy, but little is known about how TLR signaling controls autophagy. Here we demonstrate that p62/SQSTM1 is required for TLR4-mediated autophagy, which we show as selective autophagy of aggresome-like induced structures (ALIS). Treatment with LPS or Escherichia coli induced LC3+ dot-like structures, and their assembly, but not lysosomal degradation, occurred independently of classic autophagic machinery. Microscopic and ultrastructural analyses showed that p62 is a component of the induced LC3+ dots and these TLR4-induced p62+ structures resemble ALIS. The levels of p62 mRNA and protein were increased in TLR4-activated cells and knockdown of p62 suppressed the ALIS formation and LC3-II conversion. The accumulation of p62 and ALIS required activation of Nrf2 by reactive oxygen species-p38 axis-dependent TLR4/MyD88 signaling, suggesting a link between innate immune and oxidative-stress responses. These findings indicate that TLR4-driven induction of p62 plays an essential role in the formation and the autophagic degradation of ALIS, which might be critical for regulating host defense.
Toll-like receptors (TLRs) are pattern recognition receptors that have evolved to detect infection by recognizing the conserved pathogen-associated molecular patterns and trigger innate immune responses to defend against invading microorganisms (1, 2). Engagement of TLR4 with Gram-negative bacteria, such as Escherichia coli, via bacterial membrane component LPS, triggers the assembly of Toll/IL-1R (TIR) domain-containing adaptors, such as MyD88 (myeloid differentiation primary response gene 88) and TRIF (TIR domain-containing adaptor inducing IFN-β). MyD88-dependent TLR4 signaling ignites activation of cellular signaling molecules, IRAK4 (IL-1R–associated kinase 4) and TRAF6 (TNFR-associated factor 6), the downstream kinases, IKK, p38, and JNK, leading to the induction of inflammatory cytokines, including IL-1β and IL-6. In contrast, TRIF-mediated pathways mainly regulate antiviral type I IFN responses through TRAF3-TBK1-IKKi and the activation of a transcription factor IFN-regulating factor 3. Nuclear factor erythroid 2-related factor 2 (Nrf2), a transcriptional factor that is responsible for cellular defense against oxidative stress also contributes to TLR4-mediated innate immune responses (3).
Recent research has implicated evolutionarily conserved macroautophagy (hereafter referred to as autophagy) in innate immune responses (4, 5). Autophagy is a bulk degradation system by which cytoplasmic materials are engulfed into double-membrane vesicles, known as autophagosomes, and delivered to lysosomes for degradation (6, 7). In the past decade, the basic steps involved in autophagy have been elucidated. Autophagy-related gene products, such as ATG5 and ATG7, mediate the conjugation of phosphatidylethanolamine (PE) to microtubule-associated protein 1 light-chain 3 (LC3), an essential step in autophagy, by mechanisms similar to ubiquitin-conjugation (6, 7). The lipidation of Gly120 of LC3 with PE converts the soluble form (LC3-I) to another (LC3-II) that specifically associates with autophagosomes (8).
In addition to regulating autophagy (9–11), stimulation with LPS induces formation of ubiquitin-positive aggresome-like structures in cytosol that were referred to as ALIS (aggresome-like induced structures) in macrophages and DALIS in dendritic cells (12–15). However, the underlying mechanisms by which TLR4 signaling controls autophagy and ALIS are largely unknown. Signaling adapter p62 (also known as SQSTM1) can bind to LC3 and ubiquitinated proteins, and plays a key role in selective autophagy of ubiquitinated protein aggregates and organelles (16–20). Here we identify TLR4-mediated autophagy as a selective autophagy of ALIS and demonstrate that p62 plays an essential role in this process. Our findings reveal that, as a physiological innate immune response to TLR4-stimulation, transcriptional induction of p62, its accumulation in ALIS, and elimination of ALIS by autophagy occurs in macrophages. Moreover, we present evidence that p38- and reactive oxygen species (ROS)-mediated Nrf2 activation contributes to this process.
Results
Formation of TLR4-Mediated LC3+ Dot-Like Structures Occurs Independently of Classic Autophagic Machinery.
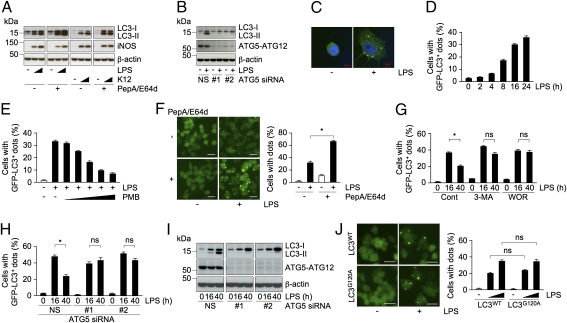
Engagement of TLRs with microbial components is reported to promote autophagy in macrophages (9–11); however, very little is known about the signaling pathways that connect TLRs to autophagy. To understand this process, we first evaluated the status of LC3-II and the accumulation of LC3+ structures in TLR4-activated macrophages. Treatment of RAW264.7 cells with TLR4 ligand, LPS, or E. coli, resulted in the accumulation of LC3-II in a dose-dependent manner, with a modest increase in the level of LC3-I (Fig. 1A). Cotreatment with lysosomal protease inhibitors pepstatin A and E64d (PepA/E64d) further increased the levels of LC3-II, but not that of inducible NOS that accumulated in response to TLR4 activation (Fig. 1A), suggesting elimination of LC3-II via lysosomal degradation pathways. To evaluate role of ATG5 and ATG7 in the accumulation of LC3-II, we estimated LC3-II levels in cells in which expression of ATG proteins was suppressed. Two siRNAs against ATG5 and ATG7 were effective in suppressing expression at the protein level (Fig. 1B and Fig. S1A). Suppression of either ATG5 or ATG7 reduced LPS-induced LC3-II accumulation (Fig. 1B and Fig. S1A), indicating participation of classic autophagic machinery in the LC3-II conversion in stimulated macrophages. Because LC3-II selectively associates with autophagosomes, we immunostained RAW cells using anti-LC3 antibody and found association of endogenous LC3 with dot-like structures upon LPS stimulation (Fig. 1C).
Fig. 1.
Formation of TLR4-mediated LC3+ dots occurs independently of classic autophagic machinery. (A and B) Immunoblots of extracts from RAW cells treated with LPS (0.1 or 1 μg/mL) or K12 (0.5 or 5 × 107) in the presence or absence of PepA/E64d for 24 h (A), or cells transfected with ATG5 siRNAs (NS, #1, or #2) and treated with LPS (B) probed for the indicated proteins. (C) Confocal images of cellular localization of endogenous LC3 in cells treated with LPS. (Scale bars, 5 μm.) (D–J) Percentage of cells with GFP-LC3+ dots. RAW cells expressing GFP-LC3WT (D–J) or -LC3G120A (J) were treated with LPS at the indicated times (D), along with a series of threefold dilutions of PMB (E), in the presence or absence of PepA/E64d (F), in the absence (Cont) or presence of the indicated compounds for 16 h (G), at the indicated times after cells transfected with ATG5 siRNAs (H), and for 24 h (J). The fluorescent images (F, Left and J, Left) and immunoblots of extracts from cells transfected with ATG5 siRNAs (I) are also included. (Scale bars, 20 μm.) The results shown are means ± SD; *P < 0.005; ns, not significant.
To investigate TLR4-mediated autophagosome formation, we established RAW cells stably expressing GFP-tagged LC3 (GFP-LC3). Unlike transient transfection of GFP-LC3, which has been reported to induce nonspecific aggregates, stable expression showed uniform GFP staining without any puncta, allowing reliable quantification of dot formation in autophagy-activated cells (21). Treatment of these cells with LPS resulted in the accumulation of GFP-LC3+ dots in a time-dependent manner (Fig. 1D and Fig. S1B). Incubation with polymyxin B (PMB), an antibiotic that interferes with LPS-TLR4 binding, blocked the formation of these dots (Fig. 1E), indicating specificity to TLR4-initiated signaling. Treatment of LPS- or E. coli-stimulated RAW cells expressing GFP-LC3 with PepA/E64d resulted in an increase in the number of cells with GFP-LC3+ dots and in an enhancement in the size of the dots (Fig. 1F and Fig. S1C), suggesting that the induced structures are cleared via lysosomal degradation pathways. Collectively, these results indicate that TLR4 signaling controls autophagy in macrophages.
Next, to verify how LPS stimulation triggers the formation of the LC3+ dot-like structures, we examined the effect of 3-methyladenine (3-MA) or wortmannin (WOR), two compounds known to prevent the generation of autophagosomes by inhibiting PI3K activity (8). Surprisingly, coincubation with LPS and 3-MA, or WOR, did not reduce the number of cells with GFP-LC3+ puncta in 16 h after stimulation, but blocked the decrease in the number of cells with dots at 40 h (Fig. 1G), raising a possibility that classic autophagic machinery is involved in the degradation, but not in the generation, of the LC3+ dots. To investigate such a possibility, we examined formation of LPS-induced structures in GFP-LC3 expressing RAW cells in which the expression of either ATG5 or ATG7 is suppressed using siRNA. Consistent with the results observed in cells treated with 3-MA or WOR, the loss of either ATG5 or ATG7 caused no significant change in the number of cells with GFP-LC3+ dots at 16 h after stimulation, whereas it prevented the decrease observed at 40 h in nonspecific (NS) siRNA-treated cells (Fig. 1H and Fig. S1D). Suppression of either of the ATG proteins blocked the accumulation of LC3-II at both 16 or 40 h of LPS stimulation (Fig. 1I and Fig. S1E), suggesting that LPS-induced LC3+ dot formation does not require conversion of LC3-I to -II. Consistent with such a notion, stimulation with LPS of RAW cells expressing GFP-LC3G120A, a single point mutation of Gly120 to Ala of LC3 that prevents conjugation with PE of LC3, resulted in puncta formation similar to that observed in RAW cells expressing GFP-LC3WT (Fig. 1J). These results collectively support the notion that TLR4-mediated LC3+ dot formation, but not their lysosomal clearance, occurs independently of classic autophagic machinery.
TLR4 Signaling Induces p62 mRNA, Protein, and Formation of ALIS That Are Substrates of Autophagy.
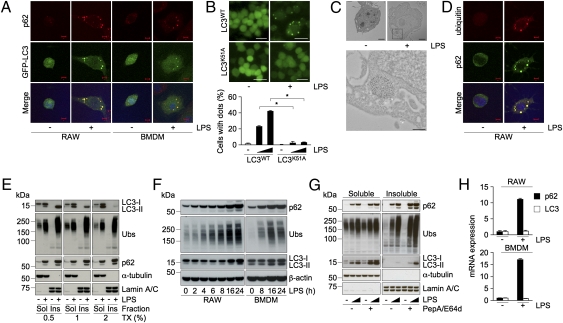
Because LPS-induced LC3+ dot formation does not require the classic autophagic machinery, we focused on molecules that could potentially affect their assembly and hypothesized that one such molecule is the LC3-interacting protein p62 (18–20). To determine involvement of p62, we first stained LPS-stimulated GFP-LC3 expressing RAW cells with anti-p62 antibody. Whereas p62 staining showed diffused pattern in unstimulated cells, it showed strong punctate staining with dots in LPS-stimulated cells (Fig. 2A). Interestingly, most of the p62-containing dots colocalized with inducible GFP-LC3+ puncta (Fig. 2A). We also explored the dot formation in mouse primary bone marrow-derived macrophages (BMDMs) expressing GFP-LC3. Activation of BMDMs with LPS also resulted in the accumulation of puncta that were positive for both p62 and GFP-LC3 (Fig. 2A), suggesting that both p62 and LC3 are present in the same compartment in activated macrophages. Because LC3 could interact directly with p62, and Lys51 of LC3 plays a critical role in the association with p62 (20), we hypothesized that interaction with p62 is essential for the formation of LC3+ structures. To explore this idea, we evaluated the LC3+ dot formation in RAW cells expressing a Lys51 to Ala mutant LC3, GFP-LC3K51A. Although expression of neither GFP-LC3WT nor -LC3 mutant affected the accumulation of p62+ dot-like structures, GFP-LC3K51A, but not -LC3WT or G120A, failed to accumulate into dot-like structures in LPS-stimulated cells (Fig. 2B and Fig. S2A), indicating that LC3-recruitment into dots is mediated by p62.
Fig. 2.
LPS-mediated transcriptional up-regulation of p62 controls ALIS formation and autophagy. (A) Confocal images of cellular localization of p62 in GFP-LC3 expressing RAW cells (Left) and BMDMs (Right) stimulated with LPS (1 μg/mL) for 16 h. (Scale bars, 5 μm.) (B) Fluorescent images (Upper) and percentage of cells with GFP-LC3+ dots (Lower) of RAW cells expressing GFP-LC3WT or -LC3K51A treated with LPS. (Scale bars, 20 μm.) (C) Immunoelectron micrographs of LPS-stimulated RAW cells. Cells treated with LPS were immunolabeled for p62 (Upper). (Scale bars, 2 μm.) (Lower) Magnified image of selected region. (Scale bar, 0.5 μm.) (D) Confocal images of localization of ubiquitin and p62 in RAW cells stimulated with LPS. (Scale bars, 5 μm.) (E–G) Immunoblots of detergent-soluble (Sol) and -insoluble (Ins) fractions of 0.5, 1, or 2% Triton X-100 (TX)-containing buffers from RAW cells treated with LPS (E), of the detergent (2%)-soluble fractions from RAW cells (Left) and BMDMs (Right) treated with LPS for the indicated times (F), and of the detergent-soluble and -insoluble fractions from RAW cells treated with LPS in the presence or absence of PepA/E64d (G), probed for the indicated proteins. Ubs, ubiquitinated proteins. (H) The levels of p62 mRNA in RAW cells (Upper) and BMDMs (Lower) stimulated with LPS for 16 and 2 h, respectively. The results shown are means ± SD; *P < 0.005.
To characterize the nature of LPS-induced dot-like structures ultrastructurally, we performed immunoelectron microscopic analysis using anti-p62 antibody. In LPS-stimulated RAW cells, we observed association of p62 mainly with aggregate-like structures in the cytoplasm without clear membrane (Fig. 2C), structures that appeared morphologically similar to “p62-body” in nonphagocytic cells (17–19). In phagocytes, LPS was reported to induce ubiquitin-positive aggregate-like structures (ALIS/DALIS) that were detergent-insoluble (13, 14), and to our knowledge no association of p62 or LC3 with LPS-induced ALIS has ever been reported. Thus, we evaluated whether the LPS-induced p62+/LC3+ dot-like structures contained ubiquitinated proteins. LPS-stimulated RAW cells were immunostained with anti-p62 antibody together with FK2, which recognizes ubiquitin on proteins. Untreated cells showed faint staining for ubiquitin, whereas cells activated with LPS exhibited a strong signal as clear punctate staining (Fig. 2D). Notably, nearly all of the LPS-induced ubiquitin-positive dots colocalized with p62+ puncta (Fig. 2D). Furthermore, we also observed colocalization of ubiquitin-positive structures with LPS-induced GFP-LC3+ dots in both RAW cells and BMDMs (Fig. S2B). Because ubiquitinated proteins in ALIS were reported to be largely detergent-insoluble (13, 14), we evaluated the detergent-solubility of p62 along with ubiquitinated proteins from RAW cells by fractionation and immunoblot analysis. Consistent with earlier reports, the detergent-insoluble fraction from LPS-stimulated cells included ubiquitinated proteins, and in addition these fractions also contained substantial amount of p62 that was not solubilized with even 2% Triton X-100 (Fig. 2E). In contrast, we could detect LC3-II mostly in the detergent-soluble fraction with 2%, but not 0.5%, Triton X-100 (Fig. 2E), suggesting detergent labile association between LC3 and p62. These results collectively indicate that the structures formed in macrophages in response to TLR4 stimulation consist of p62, LC3, and ubiquitinated proteins and are likely ALIS. Accordingly, we hereafter refer TLR4 signal-induced LC3+ dot-like structures as ALIS.
If the accumulation of p62 in ALIS were the result of translocation of the basal p62 from the detergent-soluble to -insoluble compartment, one would expect decrease in p62 level in the detergent-soluble fraction. Surprisingly, we observed a robust increase in the levels of p62 in both soluble and insoluble fractions of LPS-stimulated cells (Fig. 2E), suggesting physiological up-regulation of p62. Consistent with such a possibility, we found that LPS stimulation could increase p62 levels in the detergent-soluble fraction in a time-dependent manner that was attenuated by cotreatment with PMB (Fig. 2F and Fig. S2C). Importantly, the increases in p62 protein could be detected in BMDMs as well (Fig. 2F), indicating that stimulation of TLR4 results in the accumulation of p62 protein in macrophages. Moreover, in these cells we also found substantial increase in the levels of ubiquitinated proteins and LC3, other components of ALIS (Fig. 2F).
Because p62 is reported to be a substrate for autophagy (17–20), we next investigated whether p62, soluble or detergent-insoluble, is targeted for selective autophagic degradation. Whereas little difference in the levels of the detergent-soluble p62 and ubiquitinated proteins with or without lysosomal inhibition was observed in LPS-stimulated cells, increase in the levels of detergent-insoluble p62 and ubiquitinated proteins was clearly noted in the presence of PepA/E64d (Fig. 2G), suggesting that ALIS are selective targets for lysosomal degradation. Intriguingly, in the presence of PepA/E64d we could detect only LC3-II in the detergent-insoluble fraction (Fig. 2G), suggesting that ALIS-associated LC3-I is converted to LC3-II that integrates into autophagosomal membranes. Confirmatively, an increase in the levels of LC3-II in the detergent-soluble fraction upon lysosomal inhibition was detected (Fig. 2G). These results collectively suggest that ALIS assembled in stimulated macrophages are substrates for autophagic degradation.
To evaluate if accumulation of p62 protein was the result of increased p62 transcription, we quantified p62 mRNA levels using real-time RT-PCR. Although little change in LC3 mRNA was detected, p62 mRNA increased more than 10-fold after LPS stimulation in both RAW cells and BMDMs (Fig. 2H). Collectively, these results suggest that TLR4 signaling transcriptionally up-regulates p62 expression, resulting in the formation of ALIS, which are substrates for autophagy in macrophages.
p62 Is Required for TLR4-Mediated ALIS Formation in Macrophages.
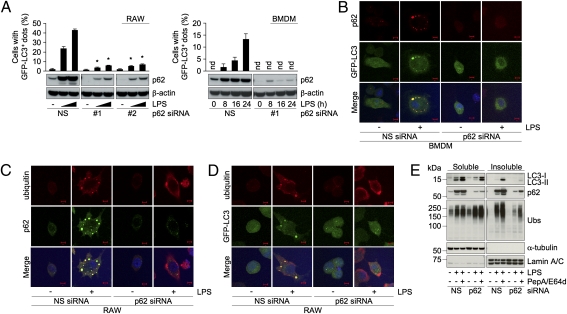
To investigate the role of p62 in TLR4-mediated ALIS formation, we suppressed the expression of p62 using siRNAs and examined the effect on LPS-induced GFP-LC3+ dot formation as a marker of ALIS. Two siRNAs were effective in suppressing induced p62 expression in LPS-stimulated macrophages (Fig. 3A). Almost no cells with LPS-induced GFP-LC3+ puncta were observed in p62 siRNA-transfected RAW cells and BMDMs compared with NS siRNA-transfected cells (Fig. 3A). Absence of LPS-induced LC3+ dots, which was confirmed by confocal analysis (Fig. 3B and Fig. S3A), indicates that ALIS formation and recruitment of LC3 into ALIS is dependent on p62 in macrophages. Next, p62 knockdown cells were explored for the presence of the ubiquitin-positive structures by immunostaining. Suppression of p62 also blocked accumulation of ubiquitin-positive foci in both RAW and RAW cells expressing GFP-LC3 (Fig. 3 C and D). As in the case of LPS stimulation, treatment with E. coli also induced ubiquitin-positive and LC3+ dots in p62-dependent manner (Fig. S3B). We also estimated the accumulation of ubiquitinated proteins by immunoblot analysis. Whereas knockdown of p62 did not affect the levels of the ubiquitinated proteins in unstimulated cells, in LPS-treated cells suppression of p62 greatly diminished their accumulation in the detergent-insoluble fraction (Fig. 3E), further confirming requirement of p62 for ALIS assembly. Treatment with PepA/E64d could not restore the ubiquitinated proteins to the detergent-insoluble fraction in these cells (Fig. 3E). Next, to follow autophagic flux in p62 knockdown cells, we examined the levels of the LC3-II in stimulated RAW cells treated with or without PepA/E64d. Correlating with attenuation of ALIS formation (Fig. 3A), we found reduced levels of LC3-II in the detergent-soluble fraction in p62 knockdown RAW cells (Fig. 3E). Furthermore, in p62 siRNA-transfected cells lysosomal inhibition could not restore LC3-II to the levels found in NS siRNA-transfected cells both in detergent-soluble and detergent-insoluble fractions (Fig. 3E). These results collectively suggest that up-regulation of p62 plays an essential role in the generation of ALIS and its degradation by autophagy in TLR4-activated macrophages.
Fig. 3.
Requirement of p62 for LPS-induced selective autophagy of ALIS. (A–E) Cells transfected with p62 siRNAs (NS, #1, or #2) were treated with LPS (1 μg/mL) for 16 h. (A) Percentage of cells with GFP-LC3+ dots (Upper) and immunoblots of the extracts for the indicated proteins (Lower) in RAW cells (Left) and BMDMs (Right) expressing GFP-LC3. The results shown are means ± SD; * Indicates statistical significance compared with NS (P < 0.005). nd, Not detectable. (B–D) Confocal images of cellular localization of the indicated molecules in BMDMs (B) and RAW cells (C and D). (Scale bars, 5 μm.) (E) Immunoblots of the detergent-soluble and -insoluble fractions from RAW cells treated with or without PepA/E64d probed for the indicated proteins. Ubs, ubiquitinated proteins.
Activation of p38-Nrf2 Axis by MyD88-Dependent TLR4 Signaling Is Required for p62 Up-Regulation.
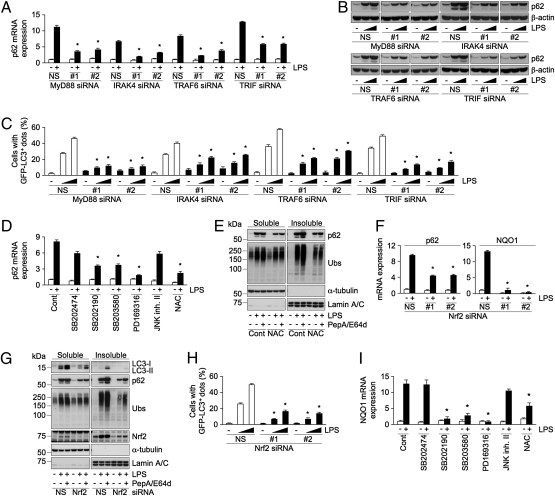
Because p62 is transcriptionally induced by stimulation with LPS (Fig. 2H), to determine which of the several signaling molecules that regulate TLR4 signaling controls p62 induction we used either RNAi to suppress the expression or specific inhibitors that block enzymatic activity. Knockdown of MyD88 with two siRNAs reduced LPS-induced accumulation of p62 both at mRNA and protein levels (Fig. 4 A and B), indicating the participation of MyD88 in this process. Similar to the effect observed in LPS-activated MyD88 knockdown cells, suppression of IRAK4 or TRAF6, downstream molecules of MyD88, attenuated the LPS-induced accumulation of p62 (Fig. 4 A and B). Because TRIF has been shown to be involved in MyD88-dependent TLR4 signaling (22), we examined whether TRIF is required for MyD88-dependent p62 up-regulation. Knockdown of TRIF reduced the induction of p62 (Fig. 4 A and B). Effectiveness of the suppression of the signaling molecules was confirmed by reduction in the induction of IL-6 mRNA and at the protein levels (Fig. S4 A and B). Because LPS-induced ALIS formation is dependent on p62 level, we checked the effect of knockdown of either of these molecules on LPS-induced GFP-LC3+ dot formation. In agreement with the reduction in p62 up-regulation, suppression of any of MyD88, IRAK4, TRAF6, and TRIF reduced the formation of GFP-LC3+ dots (Fig. 4C), suggesting that MyD88-dependent TLR4 signaling is required for induction of p62 and resulting ALIS formation.
Fig. 4.
MyD88-dependent activation of p38 and Nrf2 is required for p62 induction. (A–C) Cells transfected with siRNAs (NS, #1, or #2) for the indicated genes were stimulated with LPS (1 μg/mL, 16 h). The levels of mRNA (A) and protein (B) of p62 in RAW cells, and percentage of cells with GFP-LC3+ dots (C) in GFP-LC3 expressing RAW are shown. (D and E) RAW cells incubated without (Cont) or with the indicated inhibitors were stimulated with LPS. The levels of p62 mRNA (D) and immunoblots of the detergent-soluble and -insoluble fractions probed for the indicated proteins (E) are shown. Ubs, ubiquitinated proteins. (F–H) Cells transfected with Nrf2 siRNAs (NS, #1, or #2) were stimulated with LPS. The levels of mRNAs of p62 and NQO1 (F), immunoblots of the cellular extracts probed for the indicated proteins (G), and percentage of cells with GFP-LC3+ dots (H) are shown. (I) RAW cells incubated without (Cont) or with the indicated inhibitors were stimulated with LPS. The levels of NQO1 mRNA are shown. The results shown are means ± SD; *statistical significance compared with NS or Cont (P < 0.005).
Next, to identify kinases responsible for p62 induction, we focused on p38 and JNK, which are downstream kinases activated by TRAF6. Treatment with p38 inhibitors, such as SB202190, SB203580, and PD169316, substantially reduced LPS-induced p62 mRNA expression, as well as IL-6 mRNA (Fig. 4D and Fig. S4C). In contrast, treatment with negative control inhibitor, SB202474, or JNK inhibitor II (JNK inh. II) had very little effect (Fig. 4D and Fig. S4C). Treatment with inhibitors of p38, but not JNK, reduced LPS-induced GFP-LC3+ dot formation, and accumulation of ubiquitinated proteins in the detergent-insoluble fraction (Fig. S4 D and E). These results suggest a role for p38, but not JNK, in TLR4-mediated induction of p62 and ALIS formation.
Given that LPS-induced ROS selectively activates p38, but not JNK (23), we speculated that the ROS-p38 axis might be involved in the induction of p62 and ALIS formation. To examine the possibility, we evaluated the effect of an antioxidant on LPS-induced p62 mRNA and ALIS formation in RAW cells. Treatment with N-acetyl-l-cysteine (NAC) reduced LPS-induced p62 mRNA and protein (Fig. 4 D and E) and prevented accumulation of ubiquitinated proteins in the detergent-insoluble fractions, and GFP-LC3+ dot formation in a dose-responsive manner (Fig. 4E and Fig. S4F). Consistent with the notion of a reduction in the formation of ALIS, treatment with lysosomal inhibitors could not restore the levels of ubiquitinated proteins in detergent-insoluble fractions in NAC-treated cells (Fig. 4E). These results indicate that ROS-p38 axis contributes to LPS-induced ALIS formation.
ROS is also a critical regulator in the activation of Nrf2, which regulates the expression of several antioxidant proteins, but also p62 (24, 25). Thus, we hypothesized that Nrf2 might function as a trancriptional regulator of TLR4-mediated p62 up-regulation. To test this theory, we suppressed the expression of Nrf2 using two siRNAs and estimated the levels of p62 mRNA. Treatment of control cells with LPS substantially increased the mRNA of p62 along with that of NAD(P)H:quinone oxidoreductase (NQO1) and peroxiredoxin 1 (Prdx1), known Nrf2 target genes (Fig. 4F and Fig. S4G) (25, 26), demonstrating activation of Nrf2 in LPS-stimulated cells. Suppression of Nrf2 effectively diminished LPS-induced mRNA expression of not only NQO1 and Prdx1, but also that of p62 (Fig. 4F and Fig. S4G), indicating that p62 is transcriptionally up-regulated by Nrf2 upon LPS activation. As reported (3), loss of Nrf2 had a little enhancing effect on IL-6 mRNA (Fig. S4G). Furthermore, we examined the effect of suppression of Nrf2 expression on the accumulation of p62, ubiquitinated proteins, and LC3-II in LPS-stimulated RAW cells treated with or without lysosomal inhibitors. Knockdown of Nrf2 diminished LPS-induced accumulation of p62 both in the detergent-soluble and -insoluble fractions (Fig. 4G). Similar to the case of LPS, treatment with E. coli also accumulated p62 both in the detergent-soluble and -insoluble fraction in Nrf2-dependent manner (Fig. S4H). We could observe a decrease in the accumulation of ubiquitinated proteins and LC3-II in the detergent-insoluble and -soluble fractions, respectively, in Nrf2 knockdown cells (Fig. 4G). In these cells, consistent with reduced p62 level, a substantial decrease in the number of cells with LPS-induced GFP-LC3+ dots were noted (Fig. 4H). Moreover, LC3-II level in PepA/E64d-treated Nrf2 knockdown cells was much less than that was observed in control cells (Fig. 4G), demonstrating contribution of Nrf2-p62 axis in TLR4-mediated selective autophagy of ALIS. Finally, to investigate involvement of ROS-p38 axis in LPS-induced Nrf2 activation, we evaluated the effect of NAC and p38 inhibitors on LPS-induced expression of NQO1 and Prdx1 mRNAs. Unlike negative control inhibitor or JNK inh. II, treatment with either NAC or p38 inhibitors resulted in a substantial reduction in Nrf2-target gene expression (Fig. 4I and Fig. S4I), demonstrating the contribution of ROS-p38 axis to TLR4-mediated Nrf2 activation.
Discussion
In this study, we demonstrate that LPS or E. coli-induced LC3+ dot formation occurs independently of classic autophagic machinery and these LC3+ dots are indistinguishable by immunofluorescence, and ultrastructural and biochemical analyses from ALIS/DALIS structures reported earlier (12–15). Furthermore, our study shows that LC3-I associates with ALIS and p62-dependent ALIS formation is required for the conversion of LC3-I to -II, raising the possibility that the ALIS-associated LC3-I is selectively converted to LC3-II in the process of engulfment of ALIS and its autophagic degradation. Thus, we believe that TLR4-mediated autophagy reported in macrophages (9–11) is selective autophagy of ALIS.
Our study proposes that macrophages in response to pathogen recognition facilitate ROS-p38-Nrf2 activation leading to the accumulation of p62 and the formation of ALIS. Such a notion is supported by the reported accumulation of p62 or ALIS in cells exposed to chemical-induced oxidative-stress conditions (12, 25). Because Keap1 (Kelch-like ECH-associated protein 1) tightly regulates ROS-mediated Nrf2 activation, it is possible that Keap1 plays a role in TLR4-mediated ALIS formation and their degradation. Under normal conditions, Keap1 promotes proteasomal degradation of Nrf2, whereas under oxidative-stress conditions ROS-mediated Keap1 inhibition results in the activation of Nrf2 (27, 28). Recently, noncanonical activation of Nrf2 in autophagy-deficient cells, in which aberrant accumulation of p62 results in entrapment of Keap1 in the p62+ detergent-insoluble fraction, has been reported (29, 30). Thus, it is plausible that in infected cells the physiological accumulation of p62/ALIS and its elimination by autophagy ensure appropriate innate immune responses via Nrf2 activation because loss of function of Nrf2 leads to deleterious innate immune responses in mice (3). Therefore, our results provide a link between evolutionarily conserved innate immune and oxidative-stress responses.
The presence of ubiquitinated proteins in ALIS (17–19) suggests that p62 recruit ubiquitinated proteins to ALIS and signal-dependent ubiquitination events occur in ALIS. Moreover, because ubiquitinated proteins of DALIS are considered as one of the determining sources of antigen peptides presented by MHC class I in dendritic cells (13), p62/ALIS-dependent autophagy might control antigen presentation in phagocytes. Identification of specific ubiquitinated proteins and the E3 ligases responsible for the ubiquitination in the TLR4 signal-induced ALIS may help in understanding other antimicrobial functions of ALIS.
Recently, several groups have proposed that p62 in epithelial cells targets intracellular Salmonella typhimurium and the ActA2 deletion mutant of Listeria monocytogenes for autophagy (31, 32). Given the diverse functions attributed to p62, a contribution of ALIS to TLR4 responses other than that linked to bacterial elimination cannot be ruled out. Our findings strengthen the importance of ALIS in host immune defense, and reveal a broader scope for p62 in innate immune responses against microbial infection.
Experimental Procedures
Autophagic process was evaluated by immunoblot and microscopic analyses. Extracts from control and stimulated cells prepared in the 2% Triton X-100 buffer and were subjected to immunoblot analysis. Cells with GFP-LC3 puncta were quantified using the ImageJ program, and 100 to 500 cells were counted and the percentage of cells with GFP-LC3+ dots was calculated. Detailed experimental procedures are described in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Drs. T. Kitamura for providing retrovirus vector, B. Taylor for cell sorting, S. Garfield for confocal analysis, K. Hartman for sequencing, K. Nagashima for electron microscopic analysis, and J. Ashwell and R. Bosselut for critical review of the manuscript. This work was supported by the intramural research program of the National Institutes of Health, Center for Cancer Research, National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014156108/-/DCSupplemental.
References
- 1.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 3.Thimmulappa RK, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010;189:925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szeto J, et al. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006;2:189–199. doi: 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- 13.Lelouard H, et al. Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature. 2002;417:177–182. doi: 10.1038/417177a. [DOI] [PubMed] [Google Scholar]
- 14.Lelouard H, et al. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J Cell Biol. 2004;164:667–675. doi: 10.1083/jcb.200312073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canadien V, et al. Cutting edge: Microbial products elicit formation of dendritic cell aggresome-like induced structures in macrophages. J Immunol. 2005;174:2471–2475. doi: 10.4049/jimmunol.174.5.2471. [DOI] [PubMed] [Google Scholar]
- 16.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 20.Ichimura Y, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 21.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: Caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 23.Hsu HY, Wen MH. Lipopolysaccharide-mediated reactive oxygen species and signal transduction in the regulation of interleukin-1 gene expression. J Biol Chem. 2002;277:22131–22139. doi: 10.1074/jbc.M111883200. [DOI] [PubMed] [Google Scholar]
- 24.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Ishii T, et al. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 26.Itoh K, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 27.Wakabayashi N, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 28.Itoh K, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau A, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 31.Zheng YT, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 32.Yoshikawa Y, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.