Abstract
A number of heterologous enzymes have been investigated for cancer treatment and other therapeutic applications; however, immunogenicity issues have limited their clinical utility. Here, a new approach has been created for heterologous enzyme deimmunization whereby combinatorial saturation mutagenesis is coupled with a screening strategy that capitalizes on the evolutionary biology concept of neutral drift, and combined with iterative computational prediction of T-cell epitopes to achieve extensive reengineering of a protein sequence for reduced MHC-II binding propensity without affecting catalytic and pharmacological properties. Escherichia coli L-asparaginase II (EcAII), the only nonhuman enzyme approved for repeated administration, is critical in treatment of childhood acute lymphoblastic leukemia (ALL), but elicits adverse antibody responses in a significant fraction of patients. The neutral drift screening of combinatorial saturation mutagenesis libraries at a total of 12 positions was used to isolate an EcAII variant containing eight amino acid substitutions within computationally predicted T-cell epitopes—of which four were nonconservative—while still exhibiting kcat/KM = 106 M-1 s-1 for L-Asn hydrolysis. Further, immunization of HLA-transgenic mice expressing the ALL-associated DRB1*0401 allele with the engineered variant resulted in significantly reduced T-cell responses and a 10-fold reduction in anti-EcAII IgG titers relative to the existing therapeutic. This significant reduction in the immunogenicity of EcAII may be clinically relevant for ALL treatment and illustrates the potential of employing neutral drift screens to achieve large jumps in sequence space as may be required for the deimmunization of heterologous proteins.
Keywords: directed evolution, protein engineering
A variety of genetic and acquired human diseases can be treated by the systemic administration of enzymes catalyzing the depletion of metabolites that contribute to pathological states. Recombinant human enzymes are used extensively as replacement therapy for lysosomal storage disorders such as Gaucher, Fabry, and Pompe diseases (1). However, there are many diseases for which a human enzyme displaying the requisite catalytic and pharmacological properties for clinical use is unavailable. Therefore, heterologous enzymes, primarily of bacterial origin, have been evaluated for the treatment of a variety of disorders including phenylketonuria (2), gout (3), and a number of cancers that are sensitive to enzyme-mediated, systemic depletion of amino acids. Examples of the latter include a large fraction of hepatocellular carcinomas and metastatic melanomas that become apoptotic under conditions where the nonessential amino acid L-Arg in serum is depleted (4), central nervous system cancers that respond to L-Met deprivation (5), and acute lymphoblastic leukemia (ALL) for which enzyme-mediated L-Asn depletion is a critical step in the current clinical treatment approach (6–8).
A major impediment in the therapeutic application of heterologous enzymes is their immunogenicity, which results in the generation of antienzyme antibodies that in turn mediate a variety of adverse effects including hypersensitivity reactions, anaphylactic shock, and the inactivation and clearance of the enzyme itself (9). Masking of immunogenic epitopes via covalent modification with PEG can reduce protein immunogenicity (10), but eventually antigen-specific, and even PEG-specific, antibodies that contribute to therapeutic neutralization may be elicited toward PEGylated proteins (11, 12).
Protein immunogenicity can be ameliorated by mutating sequences likely to be recognized by the naive antibody repertoire (B-cell epitopes) or sequences that are bound by the major histocompatability complex (MHC)-II and thus can elicit T-cell-dependent immune responses. However, the identification and removal of B-cell epitopes is exceedingly difficult given their conformational nature, and is further complicated by our incomplete knowledge of naive antibody repertoires, and how they vary across different human populations. In contrast, there is extensive evidence from animal models, in vitro experiments, and early stage clinical studies, that the disruption of T-cell epitopes can reduce antibody responses in some therapeutic proteins (13, 14). T-cell receptors on CD4+ T cells recognize antigenic peptides (typically 13–25mers) presented in complex with MHC-II molecules on the surface of antigen-presenting cells (APCs). The MHC-II binding groove contains four well-defined pockets that accommodate the side chains of the P1, P4, P6, and P9 residues within a core 9mer region of the T-cell epitope, and these key residues largely determine binding affinity and specificity (15). Although the MHC-II locus is highly polymorphic, assays using APCs from volunteers representative of the major MHC-II haplotypes in human populations have been successfully deployed to identify T-cell epitopes that contribute to protein immunogenicity in a large fraction of patients. Alternatively, a plethora of in silico methods (16) have been developed over the past several years for the prediction of sequences that bind to various MHC-II alleles.
The removal of T-cell epitopes by mutagenesis has been used with some success in reducing the immunogenicity of humanized and chimeric antibodies (17–19). However, whereas these proteins contain, at most, a few relatively short potentially immunogenic sequences, heterologous enzymes that have not undergone immunological tolerance induction typically contain multiple T-cell epitopes, the removal of which thus necessitates extensive alteration of the polypeptide sequence in a manner that does not affect protein function. Further, enzyme catalysis is dictated not only by the active site residues, but also on a network of amino acids distributed throughout the protein (20). For this reason, the introduction of multiple amino acid substitutions that disrupt MHC-II binding but do not affect catalytic activity represents a significant challenge. This is particularly problematic when deimmunization requires the replacement of amino acids that are phylogenetically conserved and consequently, substitutions at these positions could impact protein stability or catalytic efficiency. Rational approaches for the incorporation of deimmunizing mutations into heterologous enzymes have previously proven to be effective (14, 21); however, the tolerable mutations necessary to reduce immunogenicity while concomitantly maintaining enzyme functionality may not always be readily determined by these means. Thus, an alternative strategy that utilizes high-throughput screening of combinatorial libraries could afford large jumps in sequence space, as may be required for the isolation of fully functional heterologous enzymes with reduced immunogenicity.
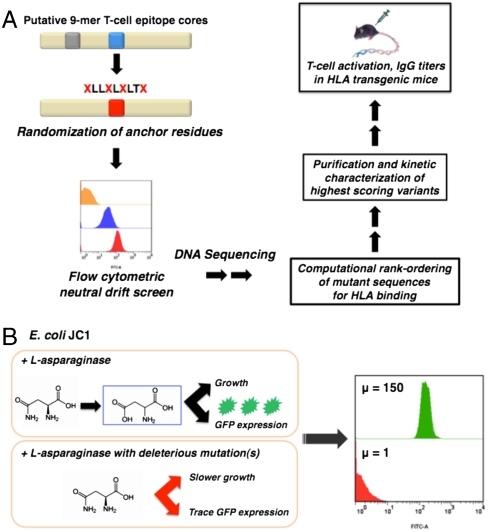
In this paper we take advantage of the evolutionary biology concept of neutral drift (22–24) for the combinatorial deimmunization of a therapeutic enzyme without loss of function. Neutral drift refers to the accumulation of mutations under selective conditions that do not ultimately impact protein function. For deimmunization, putative T-cell epitopes are first identified computationally (or experimentally), then key residues important for MHC-II binding are subjected to combinatorial randomization, and the resulting libraries are subjected to a neutral drift screen to isolate variants that retain WT function. The pools of neutral drift variants are evaluated for MHC-II binding and those that display scores indicative of compromised binding are purified and characterized biochemically. Finally, T-cell activation assays and antibody titers in transgenic mice homozygous for disease associated HLA alleles are used to evaluate T-cell epitope removal and immunogenicity, respectively (Fig. 1A). Although in this work we employed computational prediction of T-cell epitopes, the neutral drift screening methodology can be coupled to the experimental detection of sequences likely to bind MHC-II, using either haplotyped human peripheral blood mononuclear cell pools or relevant HLA-transgenic animals.
Fig. 1.
Deimmunization by combinatorial T-cell epitope removal using neutral drift. (A) Methodology for combinatorial T-cell epitope removal by neutral drift and subsequent evaluation of isolated variants using HLA-transgenic mice. (B) High-throughput neutral drift FACS screen for L-asparaginase. μ, geometric mean fluorescence.
The implementation of the strategy described above is critically dependent upon a high-throughput technology for the rapid isolation of mutations that have minimal or no effect on function. Unfortunately, neutral drift screens for most enzymes—let alone those of therapeutic significance—have not been developed, necessitating the use of surrogate screening methods that interrogate proteins for stability and expression rather than catalytic function (25). Manual assays, e.g., using a 96-well microtiter plate format, do not afford sufficient throughput for most purposes, whereas genetic selections based on complementation of auxotrophic strains to growth on selective media lack a necessary degree of quantitation. Thus, in many instances, the expression of clones displaying significant differences in catalytic activity does not result in noticeable differences in colony formation (26). Further, the extreme adaptability of biological systems can lead to growth via mechanisms that bypass the action of the expressed heterologous protein (27–29).
We have developed a simple and robust neutral drift screen readily applicable to a variety of important therapeutic enzymes that catalyze the depletion of amino acids or other metabolites important for disease states. Fig. 1B presents a schematic of the neutral drift screen as applied to the chemotherapeutic enzyme L-Asparaginase II (EcAII, EC 3.5.1.1). EcAII has been a cornerstone component of chemotherapeutic protocols for the treatment of ALL for over 40 y (30–33). In ALL, lymphoblasts lack or express low levels of L-asparagine synthetase (AS) (34) and therefore require the uptake of L-Asn from serum for cell proliferation (6). EcAII catalyzes the hydrolysis of L-Asn to L-Asp and ammonia with kcat/KM = 3.3 × 106 M-1 s-1 (as calculated in this study) resulting in the systemic depletion of serum L-Asn (7, 35, 36), which in turn induces apoptosis of ALL lymphoblasts (37, 38). However, antibody responses to EcAII have been reported in up to 60% of patients (39). The primary strategy for managing the disease in patients with adverse immune responses to EcAII is treatment with the Erwinia caratovora L-Asparaginase II, which although is non-cross-reactive with anti-EcAII antibodies (40), is also highly immunogenic and clinically inferior to EcAII with respect to both event-free survival and overall survival rates at 6 y (41).
Results
Development and Validation of Neutral Drift Screen.
To develop a neutral drift screen for EcAII, we first constructed Escherichia coli JC1 [MC1061 ΔaspCΔtyrBΔansAΔansBΔiaaA] in which the genes required for L-Asp biosynthesis (aspC, tyrB) and the three genes required for endogenous L-asparaginase enzymes were deleted. JC1 cells expressing a low level of recombinant EcAII formed normal size colonies when plated on minimal media plates with 19 amino acids (no L-Asp). In contrast, cells without plasmid or expressing the recombinant, inactive, EcAII-T12A point mutant formed pinpoint colonies, presumably because spontaneous hydrolysis of L-Asn provides a basal level of L-Asp for growth. The formation of pinpoint colonies by null mutants and the cross-feeding of L-Asp generated by low activity clones frustrated efforts to select neutral mutants on plates with selective media; in multiple attempts, similar size colonies formed by plating mutagenized enzyme were later found to encode enzymes with dramatically different L-Asn hydrolysis kinetics.
To enable an additional level of quantitation, cells were transformed with a plasmid expressing GFP under an IPTG inducible promoter, grown to late exponential phase in media containing all 20 amino acids, washed, and transferred for a short time period to media with 19 amino acids (no L-Asp) and IPTG to induce GFP synthesis. Following the addition of IPTG, GFP synthesis—and hence intracellular fluorescence—was dependent on the availability of L-Asp, which in turn was proportional to EcAII enzymatic activity. Utilizing this assay, we found that intracellular GFP fluorescence correlated well with the activity of a panel of recombinantly expressed EcAII variants displaying up to two orders of magnitude differences in catalytic efficiency. Thus, in contrast to the genetic selection approach described earlier, coexpression of a GFP reporter allowed for the discrimination of clones expressing asparaginases with varying degrees of enzymatic activity. Upon induction of GFP transcription from a strong promoter on a high copy number plasmid, the GFP mRNA accounts for a large fraction of the total cellular mRNA, therefore providing a time frame during which GFP synthesis correlates well with the rate of L-Asp generation, and thus with the relative asparaginase activity within the cell. For example, cells expressing EcAII-G57V exhibited a nearly 10-fold lower GFP fluorescence relative to cells expressing EcAII-G57A, which has approximately 15-fold higher catalytic efficiency (42) (Fig. S1A). Although the assay was able to easily eliminate low activity clones, cells expressing enzymes with high catalytic activity, such as EcAII-G57A [kcat/KM (L-aspartic acid β-hydroxomate, AHA, = 2.2 × 105 M-1 S-1 (42)], exhibited identical GFP fluorescence relative to WT EcAII [kcat/KM(AHA) = 8.2 × 105 M-1 S-1 (42)] indicating that the signal saturates for enzymes with kcat/KM within 3- to 4-fold of the WT enzyme catalytic efficiency. Nonetheless, enzymes with kcat/KM(L-Asn) > 106 M-1 s-1 should be more than sufficient for therapeutic purposes given that circulating L-Asn is depleted to negligible levels within minutes following the administration of a therapeutic dose of WT EcAII (43), and remain low for weeks afterward (32, 43). Therefore, even though enzymes with kcat/KM (L-Asn) up to 3- to 4-fold below that of the WT enzyme might result in marginally slower initial depletion of serum L-Asn, they should not affect the longer term maintenance of low serum L-Asn levels, which is the therapeutically relevant parameter. To validate the enrichment capabilities of the assay, three rounds of cell sorting produced a 6,000-fold enrichment of JC1 cells expressing WT EcAII from an initial mixture containing a 10,000-fold excess of JC1 cells expressing EcAII-T12A (Fig. S1B).
Computational Identification of Putative EcAII T-Cell Epitopes.
Putative EcAII T-cell epitopes were identified using the Immune Epitope Database (IEDB) consensus method (16). The protein sequence was parsed into overlapping 15mer peptide fragments (staggered by one residue) and within each fragment, 9-mer core regions were scored for predicted binding first to HLA-DRB1*0401, which shows strong association with childhood ALL in males (44) , and then to an additional seven HLA-DR alleles that collectively cover nearly 95% of the human population (45). Three 9-mer core regions that were scored with a consensus percentile rank (CPR) within the lowest 10% of the parsed peptide fragments as determined for binding to DRB1*0401 (CPR < 2) and that further received equivalently low scores for at least one other DRB1 allele were selected for T-cell epitope removal: M115RPSTSMSA, I216VYNYANAS, and V304LLQLALTQ (designated M115, I216, and V304 where these three residues correspond to the respective P1 positions).
Neutral Drift Screening of EcAII Libraries.
The P1, P4, P6, and P9 positions, which are most critical for the binding of peptides to the MHC-II binding groove (15), were subjected to saturation mutagenesis using the NNS (N = A, T, G, C; S = G, C) randomization scheme. Randomization and neutral drift screening were carried out sequentially, starting with M115 and continuing with I216 and finally V304 to (i) ensure complete library coverage for each individual epitope; (ii) evaluate the relative plasticity of different regions of the protein to amino acid substitutions, and (iii) simplify the structural interpretation of any observed changes in the activity of isolated mutants.
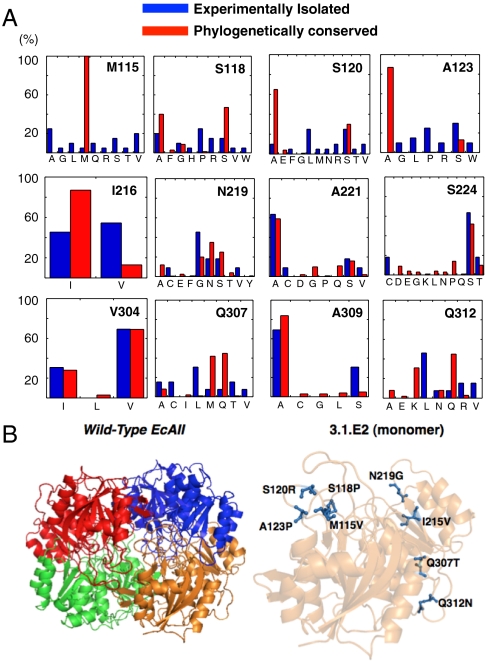
The 107 transformants of the M115 library (predicted theoretical diversity ≈10.56) were subjected to three rounds of FACS screening until the mean cell fluorescence of the sorted population was comparable to that of cells expressing the WT enzyme (Fig. S2A). In this instance, the high initial fluorescence of the library (μ = 90; Fig. S2A) suggested that amino acid substitutions at the targeted sites were generally tolerated. Following the final round of sorting, 120 individual clones selected at random were assayed in microtiter well plates using the colorimetric asparaginase substrate AHA. This secondary screen eliminated inactive clones that might have been inadvertently recovered during FACS enrichment. Active clones were found to display minor variations in AHA hydrolysis rates consistent with the notion that the FACS screen enriches variants with near WT activity. Fig. 2A shows the frequency of amino acid occupancy at M115, S118, S120, and A123. Interestingly, M115, which is absolutely conserved among the nearly 500 bacterial type II L-asparaginases in the database, could tolerate a variety of nonconservative substitutions. Analogous promiscuity was observed at both S120 and A123, which are also highly conserved phylogenetically. Evaluation of the isolated sequences using the IEDB consensus model revealed that the alteration of M115RPSTSMSA to V115RPPTRMSP results in over a 20-fold increase in CPR score for the DRB1*0401 allele, as well as increases in the CPR scores for five other HLA-DR alleles (Table 1). The resulting enzyme variant, EcAII M115V/S118P/S120R/A123P (designated as clone 1.1.C4), having four amino acid substitutions—three of which were nonconservative—displayed catalytic properties for the hydrolysis of L-Asn (kcat = 28 s-1, KM = 17 μM, kcat/KM = 1.6 × 106 M-1 s-1) that were nearly identical to those of the parental enzyme with only a twofold increase in KM.
Fig. 2.
Isolation of EcAII neutral drift mutants. (A) Residue plasticity of known bacterial type II asparaginases (IPR004550; n = 478) and of EcAII variants exhibiting WT activity isolated by neutral drift (n = 20 per library) at the 12 amino acids targeted for mutagenesis. Residue numbers correspond to positions in EcAII. (B) Location of the eight amino acid mutations differentiating wild-type EcAII and 3.1.E2. Images generated by PyMol (55). The location of these mutations in relation to the EcAII active site is shown in Fig. S5.
Table 1.
Computational prediction of T-cell epitopes in WT EcAII and the 3.1.E2 mutant by IEDB consensus method
| Minimum CPR |
|||
| HLA allele |
MRPSTSMSA (VRPPTRMSP) |
IVYNYANAS (VVYGYANAS) |
VLLQLALTQ (VLLTLALTN) |
| DRB1*0101 | 18.48 (n/a) | 5.6 (12.27) | 6.9 (n/a) |
| DRB1*0301 | 2.29 (2.24) | 1.07 (6.71) | 6.71 (15.89) |
| DRB1*0401 | 1.40 (30.75) | 0.27 (1.88) | 0.79 (2.42) |
| DRB1*0701 | 18.53 (n/a) | 0.56 (3.95) | n/a (18.91) |
| DRB1*0801 | 11 (9.9) | 5.17 (9.9) | 6.4 (9.9) |
| DRB1*1101 | 1.82 (6.94) | 1.12 (7.58) | 0.98 (0.94) |
| DRB1*1301 | 6.36 (6.41) | 0.38 (2.93) | 2.40 (13) |
| DRB1*1501 | 19.91 (27.58) | 3.14 (11.83) | 10.89 (9.32) |
| CPR ≤ 2 | 2 | 5 | 2 |
Minimum consensus percentile rank score (CPR) for the three targeted EcAII T-cell epitope core regions across eight common HLA-DR alleles (see SI Methods). The ALL-associated DRB1*0401 allele is shown in bold. Lower scores are indicative of higher predicted binding affinity. Sequence and scores for the 3.1.E2 mutant are shown in parentheses where mutations relative to WT EcAII are in bold. n/a, not predicted to bind.
The 1.1.C4 variant was then used as a template in the subsequent diversification of the P1, P4, P6, and P9 positions in I216VYNYANAS. The near background mean fluorescence of the initial library cell population (107 transformants) revealed that the overwhelming majority of amino acid substitutions at these residues are deleterious. Nonetheless, a population with near WT fluorescence was established after four rounds of FACS sorting (Fig. S2B). In contrast to the high degree of plasticity observed in the M115 core region, mutagenesis of the MHC-II anchor positions in the I216 core yielded mostly conservative amino acid substitutions (Fig. 2A). One variant of 1.1.C4 containing two mutations, a conservative change at I216V and a nonconservative one at N219G, displayed a 7-fold increase in CPR score of the modified I216 core region for DRB1*0401, and increases in the CPR scores for seven other common HLA-DR alleles (Table 1). Once again, these mutations did not affect the catalytic properties of the enzyme for the hydrolysis of L-Asn (kcat = 21 s-1, KM = 19 μM, kcat/KM = 1.1 × 106 M-1 s-1). This variant, designated 2.2.G10, was then used as a template for mutagenesis of the V304LLQLALTQ T-cell epitope core region (107 transformants). The final enzyme variant, designated 3.1.E2, further containing a nonconservative change at Q307T and a conservative Q312N substitution, showed a 3-fold increase in the CPR score for binding to DRB1*0401 and increased CPR scores for four other alleles (Table 1). The 3.1.E2 mutant contained a total of eight amino acid substitutions (Fig. 2B), but retained a kcat (L-Asn) identical to the parent enzyme with just a 3-fold increase in KM (L-Asn) (kcat = 24 s-1, KM = 23 μM, kcat/KM = 1.0 × 106 M-1 s-1). Further, 3.1.E2 displayed slightly reduced (33%) specific activity toward L-Gln hydrolysis that may be of therapeutic benefit (42), was stable in serum for over 10 d (Fig. S3); essentially identical to the WT EcAII (46), and could be expressed at a high yield ( > 30 mg/L shake flask culture) (Fig. S4). Finally, we note that because recombinant EcAII folds and expresses well, no mutants with low activity but compensatory expression levels were isolated in screening each library.
Evaluation of EcAII T-Cell Responses and Immunogenicity Using HLA-Transgenic Mice.
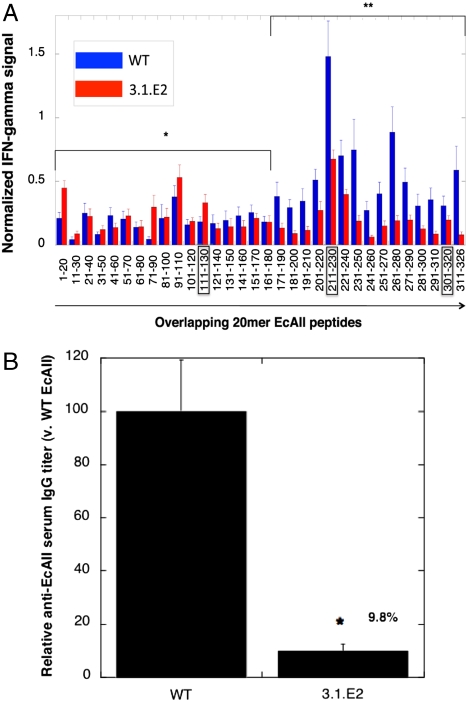
The immunogenicity of purified, low endotoxin preparations of WT EcAII and 3.1.E2 were evaluated in transgenic mice expressing human HLA-DRB1*0401 under the mouse MHC-II promoter and deficient in the endogenous murine MHC-II locus. As a stringent test of the potential for immunogenicity, mice were immunized with a strong adjuvant (Complete Freund’s Adjuvant) to induce robust CD4+ T-cell responses. The HLA-transgenic mice were immunized with either WT EcAII or 3.1.E2, and T-cell responses were measured in draining lymph node cells by cytokine ELISPOT assays for IFN-γ levels following recall with either the initial enzyme itself or with overlapping 20-mer synthetic peptides corresponding to the sequence of the enzyme used in the initial immunization (Fig. 3A). For WT EcAII, the highest level of T-cell activation was observed in response to 20-mer WTp211-230, which contained the predicted core region I216VYNYANAS. Deimmunization resulted in a significant decrease in T-cell activation by peptides containing the mutated sequences relative to I216VYNYANAS and V304LLQLALTYQ in the parental enzyme. In contrast, although mutagenesis of the M115RPSTSMSA region resulted in a sequence with improved CPR score with respect to binding DRB1*0401, no statistical difference in cytokine stimulation could be observed for the mutant peptide. This result was probably a consequence of the complex relationship between antigen processing, MHC-II binding, and TCR recognition and signaling (47). Interestingly, although the cytokine responses induced by the 17 N-terminal overlapping peptides were essentially indistinguishable (p = 0.182) regardless of whether the mice had been immunized with WT or mutant enzyme, a significantly reduced response (p < 0.0001) was observed for the 3.1.E2-immunized population across the 15 C-terminal overlapping peptides. One possible explanation for this result is that the mutations in 3.1.E2 may have affected antigen processing, thus altering MHC-II loading (48). Importantly, mice immunized with 3.1.E2 also displayed a statistically significant (p = 0.02) 10-fold reduction in anti-EcAII IgG titer relative to mice receiving the WT enzyme (Fig. 3B). Given that the activation of CD4+ T cells is in most cases required for the longevity and proliferation of B cells and for antibody isotype switching (49), this result strongly implicated that the removal of EcAII T-cell epitopes in 3.1.E2 resulted in reduced T-cell help, and thus led to lower antibody titers relative to the WT enzyme.
Fig. 3.
T-cell activation and antibody responses in HLA-DRB1*0401 transgenic mice: (A) IFN-γ production by lymph node T cells from HLA-DRB1*0401 transgenic mice immunized with either EcAII (n = 10 mice) or 3.1.E2 (n = 8 mice) and challenged with overlapping 20-mer peptides as shown. For each mouse, cytokine signals to each peptide were normalized using the cytokine signal generated to whole antigen in order to account for response variability across each sample population. Error bars shown are SEM. *, p = 0.182; **, p < 0.0001; paired Student’s t test, two-tailed, comparing recall responses. (B) Relative anti-EcAII serum IgG titers induced by EcAII (n = 6 mice) and 3.1.E2 (n = 6 mice) in HLA-DRB1*0401 transgenic mice. Error bars shown are SEM. *, p = 0.02; unpaired Student’s t test, two-tailed, comparing antibody titers.
Discussion
Human or humanized protein deimmunization has so far relied on the introduction of one or at most, a very limited number of conservative amino acid substitutions that attempt to remove immunogenic epitopes without perturbing therapeutic function. However, more drastic reengineering of the polypeptide sequence is often required for the deimmunization of heterologous enzymes that have not undergone tolerance induction. Introducing substantial changes in the primary sequence of enzymes without affecting stability and function poses a significant challenge. We demonstrated that the use of combinatorial mutagenesis and neutral drift screens that directly interrogate protein function can be exploited to take large leaps in sequence space and thus generate variant polypeptides with reduced propensity to bind to MHC-II and elicit T-dependent antibody responses. The EcAII 3.1.E2 mutant contained eight amino acid substitutions, three of which are not observed in any of the nearly 500 bacterial type II asparaginases in the database, yet retained near WT catalytic efficiency and stability. EcAII 3.1.E2 exhibited substantially reduced immunogenicity in HLA-transgenic mice and thus constitutes a very promising candidate for alleviating adverse responses in the treatment of childhood ALL. Further, the development of an asparaginase displaying reduced immunogenicity could prove critical for longer-term treatment in adult ALL or for relapsing patients.
The neutral drift screening strategy we developed may be readily applied for the combinatorial deimmunization of a number of other heterologous therapeutic enzymes used in cancer treatment that function by systemic amino acid depletion, such as L-methioninase or arginine deiminase. Likewise, different neutral drift screens may be readily designed for the deimmunization of heterologous binding proteins, e.g., enzyme inhibitors (50), using established methods such as phage or microbial display. In general, the effect of mutations on protein expression may be accounted for by constructing a fusion of the target protein to a fluorescent protein that emits at a different wavelength [e.g., red fluorescent protein, which incidentally can be secreted in the bacterial periplasm in a fluorescent form (51)]. The use of fluorescent protein fusions to monitor expression and folding in vivo is well established (52), and thus two-color sorting could be used to select for both expression and activity simultaneously. Ultimately, however, the stability of the isolated mutants would likely need to be determined in medium throughput assays (e.g., 96-well plates). Finally, although in this work we employed a computational approach for monitoring immunogenicity, experimental methods for identifying T-cell epitopes can also be applied to monitor the immunogenic propensity of mutant proteins.
Methods
FACS Screening.
M9 medium supplemented with 0.4% glucose, 3.5 μg/mL thiamine, 1 mM MgSO4, .1 mM CaCl2, 160 μg/mL of the amino acids L-Asp and L-Tyr, 80 μg/mL of the 18 remaining amino acids, 30 μg/mL kanamycin, and 200 μg/mL ampicillin was inoculated with a frozen aliquot of E. coli JC1 transformed with pQE80L-GFP (11.3.3) (53) and either a library or a single mutant. Cultures were grown at 37 °C to an A600 = 0.9–1.1, harvested by centrifugation (6,000 × g, 4 °C, 6 min), and washed twice with cold 0.9% NaCl. The cell pellets were resuspended in supplemented M9 medium containing 19 amino acids (no L-Asp, Tyr at 160 μg/mL, remaining amino acids at 80 μg/mL). GFP expression was induced following the media shift by addition of IPTG to a final concentration of 1 mM. After 1 h of induction at 37 °C, the cells were harvested by centrifugation (6,000 × g, 4 °C, 6 min), washed twice with PBS, and resuspended in PBS to a final A600 ∼ 0.05–0.1 for flow cytometric analysis and cell sorting.
Flow cytometric analyses were performed with a FACSAria (BD Biosciences) using a 488-nm solid-state laser for excitation and a 530/30 band pass filter for detection. The throughput rate of cells was adjusted to 4,000–5,000 events per second, and ∼107 cells were sorted each round in single cell mode except for the initial sort of each library, which was done in purity mode. A gate in the fluorescence channel was set to recover the 4–5% most highly fluorescent cells while additional gates were set based on both the forward- and side-scatter channels to exclude sorting nonsingle cell events. The sorted cells were collected in 0.5 mL of 2xYT medium and then plated onto 2xYT medium supplemented with 30 μg/mL kanamycin and 200 μg/mL ampicillin. Following overnight growth at 30 °C, the clones were pooled and stored in 15% glycerol at -80 °C in aliquots.
Transgenic Mice.
HLA-DR4 (DRB1*0401) transgenic mice were generated as described previously (54) and bred under specific pathogen-free conditions at the University of Texas at San Antonio. Transgenic mice were injected at 6–10 wk of age with the antigen in complete Freund’s adjuvant (CFA). WT EcAII and variant 3.1.E2 were purified and treated for endotoxin reduction (see SI Methods). CFA was prepared by mixing Mycobacterium tuberculosis H37RA (Difco Laboratories) at 5 mg/mL into Incomplete Freund’s Adjuvant. Antigens were mixed with the adjuvant to yield a 2 mg protein/mL emulsion, of which 50 μL was injected subcutaneously as specified. Ten days later, popliteal and inguinal lymph nodes were removed and single cell suspensions were adjusted to 5 × 106 cells/mL in HL-1 media (BioWhittaker). Serum was obtained by terminal cardiac puncture. All animal care and experimental procedures were conducted according to guidelines of the Institutional Care and Use Committee at the University of Texas at San Antonio.
Methods protocols for strain engineering, library construction, transgenic mouse studies, and biochemical characterization are described in SI Methods.
Supplementary Material
Acknowledgments.
We thank Sai Reddy for many helpful discussions. This work was supported by National Institutes of Health Grant CA139059 (to G.G.) and Grant GM065551 (to G.G.) and from Grant RG3701 from the National Multiple Sclerosis Society (to T.G.F.). J.R.C. also acknowledges the US Department of Homeland Security (DHS) for a Graduate Fellowship under the DHS Scholarship and Fellowship Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014739108/-/DCSupplemental.
References
- 1.Leader B, Baca QJ, Golan DE. Protein therapeutics: A summary and pharmacological classification. Nat Rev Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 2.Kim W, et al. Trends in enzyme therapy for phenylketonuria. Mol Ther. 2004;10:220–224. doi: 10.1016/j.ymthe.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Chohan S, Becker MA. Update on emerging urate-lowering therapies. Curr Opin Rheumatol. 2009;21:143–149. doi: 10.1097/BOR.0b013e328325bd94. [DOI] [PubMed] [Google Scholar]
- 4.Ni Y, Schwaneberg U, Sun ZH. Arginine deiminase, a potential anti-tumor drug. Cancer Lett. 2008;261:1–11. doi: 10.1016/j.canlet.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 5.Tan Y, Xu M, Hoffman RM. Broad selective efficacy of recombinant methioninase and polyethylene glycol-modified recombinant methioninase on cancer cells in vitro. Anticancer Res. 2010;30:1041–1046. [PubMed] [Google Scholar]
- 6.Cooney DA, Handschumacher RE. L-asparaginase and L-asparagine metabolism. Annu Rev Pharmacol. 1970;10:421–440. doi: 10.1146/annurev.pa.10.040170.002225. [DOI] [PubMed] [Google Scholar]
- 7.Haskell CM, Canellos GP, Cooney DA, Hansen HH. Biocmical and pharmacologic effects of L-asparaginase in man. J Lab Clin Med. 1970;75:763–770. [PubMed] [Google Scholar]
- 8.Rytting M. Peg-asparaginase for acute lymphoblastic leukemia. Expert Opin Biol Th. 2010;10:833–839. doi: 10.1517/14712591003769808. [DOI] [PubMed] [Google Scholar]
- 9.Schellekens H. Immunogenicity of therapeutic proteins: Clinical implications and future prospects. Clin Ther. 2002;24:1720–1740. doi: 10.1016/s0149-2918(02)80075-3. [DOI] [PubMed] [Google Scholar]
- 10.Jevsevar S, Kunstelj M, Porekar VG. PEGylation of therapeutic proteins. Biotechnol J. 2010;5:113–128. doi: 10.1002/biot.200900218. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong JK, et al. Antibody against poly(ethylene glycol) adversely affects PEG-asparaginase therapy in acute lymphoblastic leukemia patients. Cancer. 2007;110:103–111. doi: 10.1002/cncr.22739. [DOI] [PubMed] [Google Scholar]
- 12.Sherman MR, Saifer MGP, Perez-Ruiz F. PEG-uricase in the management Of treatment-resistant gout and hyperuricemia. Adv Drug Deliver Rev. 2008;60:59–68. doi: 10.1016/j.addr.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Yeung VP, et al. Elimination of an immunodominant CD4+ T cell epitope in human IFN-beta does not result in an in vivo response directed at the subdominant epitope. J Immunol. 2004;172:6658–6665. doi: 10.4049/jimmunol.172.11.6658. [DOI] [PubMed] [Google Scholar]
- 14.Harding FA, et al. A beta-lactamase with reduced immunogenicity for the targeted delivery of chemotherapeutics using antibody-directed enzyme prodrug therapy. Mol Cancer Ther. 2005;4:1791–1800. doi: 10.1158/1535-7163.MCT-05-0189. [DOI] [PubMed] [Google Scholar]
- 15.Jones EY, Fugger L, Strominger JL, Siebold C. MHC class II proteins and disease: A structural perspective. Nat Rev Immunol. 2006;6:271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 16.Wang P, et al. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bander NH, et al. Phase I trial of 177lutetium-labeled J591, a monoclonal antibody to prostate-specific membrane antigen, in patients with androgen-independent prostate cancer. J Clin Oncol. 2005;23:4591–4601. doi: 10.1200/JCO.2005.05.160. [DOI] [PubMed] [Google Scholar]
- 18.Macfarlane DJ, et al. Safety, pharmacokinetic and dosimetry evaluation of the proposed thrombus imaging agent 99 mTc-DI-DD-3B6/22-80B3 Fab’. Eur J Nucl Med Mol I. 2006;33:648–656. doi: 10.1007/s00259-005-0025-y. [DOI] [PubMed] [Google Scholar]
- 19.Holgate RG, Baker MP. Circumventing immunogenicity in the development of therapeutic antibodies. IDrugs. 2009;12:233–237. [PubMed] [Google Scholar]
- 20.Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 21.Warmerdam PA, et al. Elimination of a human T-cell region in staphylokinase by T-cell screening and computer modeling. Thromb Haemostasis. 2002;87:666–673. [PubMed] [Google Scholar]
- 22.Amitai G, Gupta RD, Tawfik DS. Latent evolutionary potentials under the neutral mutational drift of an enzyme. HFSP J. 2007;1:67–78. doi: 10.2976/1.2739115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloom JD, Romero PA, Lu Z, Arnold FH. Neutral genetic drift can alter promiscuous protein functions, potentially aiding functional evolution. Biol Direct. 2007;2:17. doi: 10.1186/1745-6150-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bershtein S, Tawfik DS. Advances in laboratory evolution of enzymes. Curr Opin Chem Biol. 2008;12:151–158. doi: 10.1016/j.cbpa.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 25.Gupta RD, Tawfik DS. Directed enzyme evolution via small and effective neutral drift libraries. Nat Methods. 2008;5:939–942. doi: 10.1038/nmeth.1262. [DOI] [PubMed] [Google Scholar]
- 26.Link AJ, Jeong KJ, Georgiou G. Beyond toothpicks: New methods for isolating mutant bacteria. Nat Rev Microbiol. 2007;5:680–688. doi: 10.1038/nrmicro1715. [DOI] [PubMed] [Google Scholar]
- 27.Breaker RR, Banerji A, Joyce GF. Continuous in-vitro evolution of bacteriophage RNA-polymerase promoters. Biochemistry. 1994;33:11980–11986. doi: 10.1021/bi00205a037. [DOI] [PubMed] [Google Scholar]
- 28.Gates CM, Stemmer WP, Kaptein R, Schatz PJ. Affinity selective isolation of ligands from peptide libraries through display on a lac repressor “headpiece dimer”. J Mol Biol. 1996;255:373–386. doi: 10.1006/jmbi.1996.0031. [DOI] [PubMed] [Google Scholar]
- 29.Yano T, Kagamiyama H. Directed evolution of ampicillin-resistant activity from a functionally unrelated DNA fragment: A laboratory model of molecular evolution. Proc Natl Acad Sci USA. 2001;98:903–907. doi: 10.1073/pnas.031442298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broome JD. Evidence that the L-asparaginase of guinea pig serum is responsible for its antilymphoma effects. II. Lymphoma 6C3HED cells cultured in a medium devoid of L-asparagine lose their susceptibility to the effects of guinea pig serum in vivo. J Exp Med. 1963;118:121–148. doi: 10.1084/jem.118.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asselin BL, et al. In vitro and in vivo killing of acute lymphoblastic leukemia cells by L-asparaginase. Cancer Res. 1989;49:4363–4368. [PubMed] [Google Scholar]
- 32.Avramis VI, et al. A randomized comparison of native Escherichia coli asparaginase and polyethylene glycol conjugated asparaginase for treatment of children with newly diagnosed standard-risk acute lymphoblastic leukemia: A Children’s Cancer Group study. Blood. 2002;99:1986–1994. doi: 10.1182/blood.v99.6.1986. [DOI] [PubMed] [Google Scholar]
- 33.Pui C-H, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 34.Horowitz B, et al. Asparagine synthetase activity of mouse leukemias. Science. 1968;160:533–535. doi: 10.1126/science.160.3827.533. [DOI] [PubMed] [Google Scholar]
- 35.Ho DH, Whitecar JP, Jr, Luce JK, Frei E., 3rd L-asparagine requirement and the effect of L-asparaginase on the normal and leukemic human bone marrow. Cancer Res. 1970;30:466–472. [PubMed] [Google Scholar]
- 36.Capizzi RL, et al. L-asparaginase: Clinical, biochemical, pharmacological, and immunological studies. Ann Intern Med. 1971;74:893–901. doi: 10.7326/0003-4819-74-6-893. [DOI] [PubMed] [Google Scholar]
- 37.Story MD, Voehringer DW, Stephens LC, Meyn RE. L-asparaginase kills lymphoma cells by apoptosis. Cancer Chemother Pharmacol. 1993;32:129–133. doi: 10.1007/BF00685615. [DOI] [PubMed] [Google Scholar]
- 38.Ueno T, et al. Cell cycle arrest and apoptosis of leukemia cells induced by L-asparaginase. Leukemia. 1997;11:1858–1861. doi: 10.1038/sj.leu.2400834. [DOI] [PubMed] [Google Scholar]
- 39.Zeidan A, Wang ES, Wetzler M. Pegasparaginase: Where do we stand? Expert Opin Biol Th. 2009;9:111–119. doi: 10.1517/14712590802586058. [DOI] [PubMed] [Google Scholar]
- 40.Avramis VI, Tiwari PN. Asparaginase (native ASNase or pegylated ASNase) in the treatment of acute lymphoblastic leukemia. Int J Nanomed. 2006;1:241–254. [PMC free article] [PubMed] [Google Scholar]
- 41.Duval M, et al. Comparison of Escherichia coli-asparaginase with Erwinia-asparaginase in the treatment of childhood lymphoid malignancies: Results of a randomized European Organisation for Research and Treatment of Cancer-Children’s Leukemia Group phase 3 trial. Blood. 2002;99:2734–2739. doi: 10.1182/blood.v99.8.2734. [DOI] [PubMed] [Google Scholar]
- 42.Derst C, Henseling J, Röhm KH. Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase activity by amino acid replacements at position 248. Protein Sci. 2000;9:2009–2017. doi: 10.1110/ps.9.10.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asselin BL, et al. Measurement of serum L-asparagine in the presence of L-asparaginase requires the presence of an L-asparaginase inhibitor. Cancer Res. 1991;51:6568–6573. [PubMed] [Google Scholar]
- 44.Dorak MT, et al. Unravelling an HLA-DR association in childhood acute lymphoblastic leukemia. Blood. 1999;94:694–700. [PubMed] [Google Scholar]
- 45.Southwood S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 46.Stecher AL, de Deus PM, Polikarpov I, Abrahão-Neto J. Stability of L-asparaginase: An enzyme used in leukemia treatment. Pharm Acta Helv. 1999;74:1–9. doi: 10.1016/s0031-6865(99)00009-6. [DOI] [PubMed] [Google Scholar]
- 47.Huang J, et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 2010;464:932–936. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- 49.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 50.Stoop AA, Craik CS. Engineering of a macromolecular scaffold to develop specific protease inhibitors. Nat Biotechnol. 2003;21:1063–1068. doi: 10.1038/nbt860. [DOI] [PubMed] [Google Scholar]
- 51.Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 52.Link AJ, Skretas G, Strauch EM, Chari NS, Georgiou G. Efficient production of membrane-integrated and detergent-soluble G protein-coupled receptors in Escherichia coli. Protein Sci. 2008;17:1857–1863. doi: 10.1110/ps.035980.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoo TH, Link AJ, Tirrell DA. Evolution of a fluorinated green fluorescent protein. Proc Natl Acad Sci USA. 2007;104:13887–13890. doi: 10.1073/pnas.0701904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ito K, et al. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. J Exp Med. 1996;183:2635–2644. doi: 10.1084/jem.183.6.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeLano DW. The PyMol Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.