Abstract
Primary cilia are required for proper Sonic Hedgehog (Shh) signaling in mammals. However, their role in the signal transduction process remains unclear. We have identified sister of open brain (sopb), a null allele of mouse Intraflagellar transport protein 122 (Ift122). IFT122 negatively regulates the Shh pathway in the cilium at a step downstream of the Shh ligand and the transmembrane protein Smoothened, but upstream of the Gli2 transcription factor. Ift122sopb mutants generate primary cilia, but they show features of defective retrograde intraflagellar transport. IFT122 controls the ciliary localization of Shh pathway regulators in different ways. Disruption of IFT122 leads to accumulation of Gli2 and Gli3 at cilia tips while blocking the ciliary localization of the antagonist TULP3. Suppressor of Fused and Smoothened localize to the cilium through an IFT122-independent mechanism. We propose that the balance between positive and negative regulators of the Shh pathway at the cilium tip controls the output of the pathway and that Shh signaling regulates this balance through intraflagellar transport.
Keywords: neural patterning, axoneme, kinesin 2, cytoplasmic dynein 2
Cilia and flagella act as sensory organelles, allowing cells to perceive extracellular signals and execute signal transduction (1). In mammals, primary cilia are required for Hedgehog (Hh) signaling, but how cilia participate in the pathway remains unclear (2–8).
An important mechanism for ciliary/flagellar assembly is intraflagellar transport (IFT). IFT is the microtubule-based transport of cargo within cilia and flagella (9). Cargo is loaded onto IFT particles and transported in an anterograde direction from the base of the cilium toward the cilium tip by kinesin 2 motors and, in a retrograde direction, back to the cilium base by cytoplasmic dynein 2 (10). IFT particles are composed of two complexes, A and B, which contain at least 6 and 10 IFT proteins, respectively (9, 11). On the basis of mutant phenotypes and measurement of transport rates, IFT proteins of the A and B complexes have been suggested to act in retrograde and anterograde transport, respectively (9, 12–14), although this strict classification of IFT A and B complex function remains equivocal (15–17).
The morphogen Sonic Hedgehog (Shh) controls the patterning of many tissues during mammalian embryonic development (18). In the context of neural-tube patterning, Shh is produced ventrally by the notochord and floor plate, forming a ventral-to-dorsal gradient. Cells along the dorsal-ventral axis adopt different fates depending on the concentration and duration of the signal to which they are exposed (19). Shh activates the pathway by binding to a negative regulator, Patched1 (Ptch1), thereby releasing Smoothened (Smo) from inhibition. Smo controls gene expression via the Gli family of transcription factors. Of the three mammalian Gli proteins (Glis 1–3), Gli2 and Gli3 are the primary regulators of the pathway in development (20). Gli2 acts primarily as a transcriptional activator, whereas Gli3 can function as either a transcriptional activator or a repressor, depending on its proteolytic processing (21, 22). The steps linking Smo activity to that of Gli proteins remain unclear (18, 23).
The discovery of the role of primary cilia in the vertebrate Shh pathway has shed light on this process (2–8). Several major regulators of the pathway localize to primary cilia. Smo and Ptch1 localize to the cilium through a mechanism gated by Shh. In the absence of ligand, Ptch1 and Smo localize to the cilium and cytoplasm, respectively, whereas the pattern is reversed upon Shh exposure (6, 24). All three Gli proteins and the pathway regulators Sufu, TULP3, and Kif7 localize to cilia tips (7, 25–27). Moreover, accumulation of Gli proteins and Kif7 at cilia tips is induced by Shh. Mouse mutations affecting ciliary assembly and structure have distinct Shh signaling phenotypes. Mutations that prevent ciliogenesis, such as those disrupting Kif3a, Ift172, and Ift88, cause dampened expression of Shh target genes (2, 4, 28). In such mutants, both Gli activator function and formation of the Gli3 repressor are impaired. Mutations in genes such as Thm1, Dync2h1, Arl13b, and bromi allow cilia to form, yet perturb ciliary structure and have varied effects on the pathway (3, 29–31). For example, both the IFT139 homolog THM1 and the cytoplasmic dynein 2 subunit DYNC2H1 function in retrograde IFT, yet loss of THM1 causes ligand-independent activation of the pathway, whereas disruption of DYNC2H1 results in dampened Shh responses (3, 28, 29).
Although IFT proteins are essential for Shh signaling, it is unknown whether they control Shh signaling by providing a permissive environment for the pathway or whether they play more direct roles in regulating the pathway. By their nature, IFT proteins could control the ciliary localization of Shh pathway components, but support for this possibility has been lacking. Here we show that intraflagellar transport protein 122 (IFT122) is a potent negative regulator of Shh signaling acting at a step between Smo and Gli2. Importantly, we find that IFT122 controls the ciliary localization of a subset of Shh pathway components, suggesting a more direct role for intraflagellar transport in Shh signal transduction. We propose that IFT122 controls the activity of the pathway through regulating the balance between Shh pathway activators and repressors at the tips of primary cilia.
Results
sister of open brain Mutants Show Indications of Hyperactive Shh Signaling.
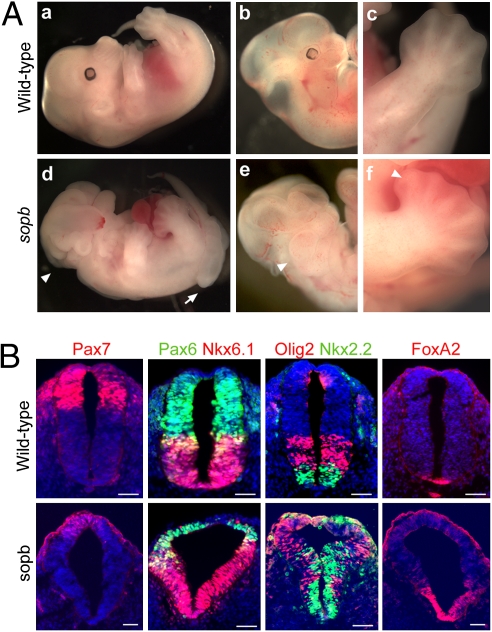
In a screen for mutations disrupting embryonic development in mice (32), we identified a mouse line exhibiting a recessive phenotype similar to that of Rab23/open brain (opb) mutants (33), leading us to name this mutation sister of open brain (sopb). sopb homozygotes died around embryonic day 13.5 (e13.5) with neural tube defects, preaxial polydactyly, enlarged branchial arches, and eye defects (Fig. 1A). We hypothesized that sopb, like Rab23/opb, disrupts an antagonist of the Shh pathway.
Fig. 1.
sopb mutants display features indicative of increased Shh signaling. (A) Wild-type (a–c) and sopb mutant (d–f) embryos shown at e13.5. sopb mutants exhibit exencephaly (arrowhead in d) and an open caudal neural tube (arrow in d), enlarged branchial arches (arrowhead in e), and preaxial polydactyly (arrowhead in f). (B) Neural patterning in e10.5 wild-type and sopb mutants in the neural tube at hindlimb levels. In the mutant neural tube, markers for dorsal and lateral cell identities (Pax7 and Pax6) are expressed in reduced, dorsally restricted domains, whereas markers for ventral identities (FoxA2, Nkx2.2, Olig2, and Nkx6.1) are expressed in dorsally expanded domains. Scale bars: 50 μm.
Shh promotes specification of different classes of ventral progenitors and inhibits that of dorsal cell types in the neural tube. To investigate Shh signaling in sopb mutants, we analyzed neural patterning using markers for progenitor subtypes. The e10.5 sopb mutant neural tube showed a ventralized phenotype at the level of the hindlimbs (Figs. 1B; Fig. S1). In wild-type embryos, Pax7 and Pax6 are expressed at high levels in dorsal and lateral neural progenitors, respectively, and they are repressed ventrally by Shh signaling. In the triangular-shaped neural tube of sopb mutants, Pax7 expression was absent or dorsally restricted, and Pax6 expression was shifted dorsally (Fig. 1B). A similar pattern was observed for other markers inhibited by Shh (Fig. S1). Concomitantly, markers for ventral fates induced by Shh, such as Nkx2.2, Nkx6.1, Olig2, and FoxA2, were expressed in dorsally expanded or shifted domains in sopb mutant neural tubes (Fig. 1B). Direct targets of the Shh pathway Ptch1 and Gli1 were also expressed in ectopic dorsal domains in the sopb neural tube (Fig. S1). Despite the ventralization of cell fates at hindlimb levels, neural patterning was largely normal at forelimb levels (Fig. S2).
Increased activity of the Shh pathway was also observed in the branchial arches and limb buds of sopb mutants (Fig. S3). Gene expression patterns in the mutant limb buds were expanded or shifted anteriorly, consistent with increased Shh signaling. Collectively, these data indicate that sopb is required to restrict the activity of the Shh pathway.
sopb Encodes an Antagonist of the Shh Pathway.
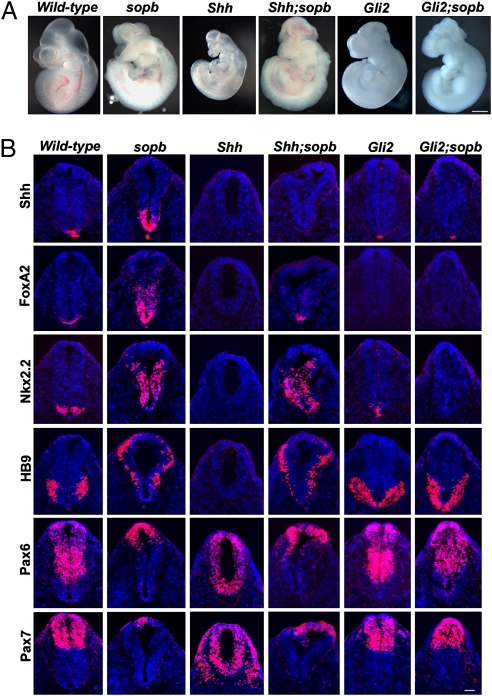
The ventralization of neural fate in sopb mutants could be due to increased production of Shh as the size of the Shh expression domain was expanded in sopb mutant neural tubes (Fig. S1). Alternatively, the defect could be due to ligand-independent effects in signal-receiving cells. To distinguish between these possibilities, we analyzed the neural patterning phenotype of Shh;sopb double mutants. Shh single-mutant embryos are small and exhibit holoprosencephaly (34), whereas sopb mutants are normal in size and exencephalic. Morphologically, Shh;sopb double mutants resembled sopb single mutants (Fig. 2A). Moreover, dorsal-ventral neural patterning was similar in sopb and Shh;sopb mutants (Fig. 2B). In Shh mutants, ventral cell types such as FoxA2+ floor plate, Nkx2.2+ V3 interneuron progenitors, and HB9+ motor neurons are not specified, and the expression of Pax6 and Pax7 expands ventrally. In Shh;sopb double mutants, as in sopb single mutants, expression of Nkx2.2 and HB9 was expanded dorsally and Pax6 and Pax7 expression was inhibited ventrally. Expression of FoxA2, requiring the highest level of Shh signaling, was rescued in the double-mutant neural tube, indicating that the ectopic pathway activity occurs without the ligand in sopb mutants. We note that FoxA2 expression was not expanded in double mutants as in sopb single mutants, suggesting that sopb mutant cells retain some ability to respond to the ligand.
Fig. 2.
The sopb phenotype is Shh-independent and Gli2-dependent. (A) Whole-mount images of wild-type, single-mutant (sopb, Shh, and Gli2), and double-mutant (Shh;sopb and Gli2;sopb) embryos at e10.5. The growth retardation phenotype of Shh mutants was suppressed in Shh;sopb mutants. (B) Neural patterning in the e10.5 caudal neural tubes assayed by Shh, FoxA2, Nkx2.2, HB9, Pax6, and Pax7 expression in single and double mutants. Patterning in Shh;sopb mutants resembled that of sopb single mutants. The specification of FoxA2+, Nkx2.2+, and HB9+ ventral fates, which lack Shh single mutants, was rescued in Shh;sopb mutants. The ventralization of cell fate and the triangular morphology of the sopb neural tube were suppressed in Gli2;sopb mutants, whose patterning phenotype resembled that of Gli2 single mutants. Scale bars: 1 mm in A and 50 μm in B.
Next we asked whether the ectopic activation of Shh pathway in sopb mutants relies on Gli function. As Gli2 is the major transcriptional activator of the pathway in neural patterning, we generated and analyzed Gli2;sopb double mutants. Gli2 mutants show a dorsalized phenotype; ventral neural cell types either are not specified or are found in ventrally displaced domains (35) in contrast to the ventralized sopb mutant phenotype. Patterning and morphology of the Gli2;sopb double-mutant neural tube resembled those of Gli2 single mutants (Fig. 2B). These findings suggest that the sopb gene product represses the Shh pathway through restricting Gli2 activity.
sopb Encodes the Mouse Homolog of IFT122.
The sopb locus was mapped to a 0.9-Mb interval on chromosome 6 (Fig. S4A). This interval contains the gene encoding IFT122, whose homologs in other organisms control ciliogenesis (17, 36–38). This was a promising candidate given the relationship between Shh signaling and cilia. Sequencing revealed a T-to-C transition mutation in the start codon of Ift122 in sopb mutants (Fig. S4B). Consistent with a block in translation, we did not detect the 135-kDa IFT122 protein in sopb embryos with antiserum against the C terminus (Fig. S4C). These data indicate that sopb is a null allele of Ift122.
IFT122 was originally identified in Chlamydomonas as a component of IFT particles orchestrating flagellar assembly. Mutations in Ift122 homologs in Caenorhabditis elegans, Drosophila, and Tetrahymena (36, 38, 39) do not block ciliogenesis, yet they result in profound morphological ciliary defects. Recently, IFT122 missense mutations were found in a subset of cranioectodermal dysplasia (CED) patients whose fibroblasts exhibit ciliary defects (40). Some CED phenotypic features, such as shortening of the limbs and digits, may result from defects in Hedgehog signaling.
A targeted mutation (Ift122Med1Δ1–3) disrupting both Ift122 and Med1/Mbd4 in mice has been described (41). Curiously, in these mutants ventral neural fates requiring high levels of Shh signaling were not specified, whereas motorneurons requiring lower levels showed dorsal expansion and ciliogenesis appeared largely disrupted. This phenotype differs from that of sopb mutants (see Discussion). As two genes are disrupted in the Ift122Med1Δ1–3 allele, the sopb allele provides an opportunity to investigate IFT122 function on its own.
Ift122sopb Mutants Generate Swollen Cilia with Defective Retrograde IFT.
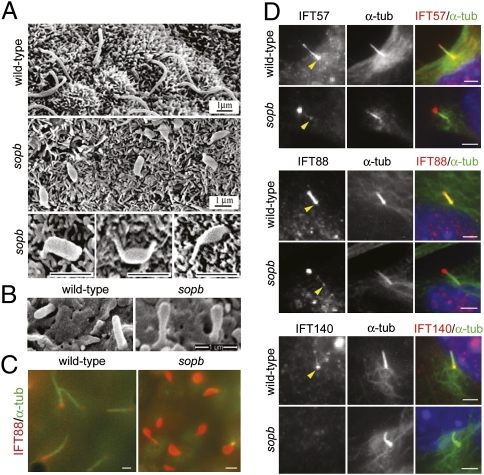
We found that EGFP-tagged mouse IFT122 concentrated at the base and tips of cilia, in addition to cytoplasmic localization, in serum-starved primary mouse embryonic fibroblasts (pMEFs; Fig. S4D). Cilia in the Ift122sopb mutant node were generated, but they had an abnormal swollen morphology (Fig. 3A). In the neural tube at lumbar and brachial levels, the mutant cilia showed swelling at their distal ends (Figs. 3B; Fig. S5C). Neural tube cilia were generated at near normal frequency, but cilia frequency was somewhat reduced in Ift122sopb pMEFs (Fig. S5A). The nodal cilia defect is likely to disrupt left-right patterning, as the leftward fluid flow generated by the nodal cilia rotation is crucial for this process (42). Indeed, Ift122sopb mutants showed left-right patterning defects such as randomization of heart looping direction and bilateral expression of Nodal in the lateral plate mesoderm (Fig. S6).
Fig. 3.
Ciliary defects in Ift122sopb mutants. (A) Scanning electron microscopy (SEM) images of cilia in the wild-type and mutant node at head-fold stages. Nodal cilia in the mutant were short, with pronounced swelling along their lengths or at their tips. (B) SEM images of cilia from the e10.5 wild-type and Ift122sopb neuroepithelium at lumbar levels. (C) Mutant nodal cilia showed dramatic accumulation of IFT88 (red). Axonemes were labeled with acetylated α-tubulin (green). (D) Ciliary localization of the complex B IFT proteins IFT57 and IFT88 and the complex A IFT protein IFT140 in wild-type and Ift122sopb pMEFs. IFT57 and IFT88 (red) localize to the base (yellow arrowheads) and along the axoneme (marked by acetylated α-tubulin staining in green) of cilia from wild-type pMEFs, whereas these proteins localized primarily to the tips of the mutant cilia. IFT140 localized to the base and tips of wild-type cilia, but failed to localize to Ift122sopb cilia. Scale bars: 1 μm in A–C and 2 μm in D.
IFT122, along with other IFT proteins such as IFT139 and IFT140, is part of the A complex of IFT particles (9). Disruption of complex A IFT proteins reduces retrograde IFT rates in Chlamydomonas (13, 14), and in a variety of systems loss of such proteins results in swollen cilia or flagella filled with complex B IFT proteins (13, 29, 36–38). These data suggest that complex A IFT proteins are primarily required for retrograde transport. On the basis of the swollen morphology of sopb mutant cilia, we hypothesized that mouse IFT122 also functions in retrograde IFT.
We examined the ciliary localization of complex B proteins IFT88 and IFT57 and the complex A protein IFT140 in sopb pMEFs. In wild-type cells, IFT88 and IFT57 localized primarily near the basal body and along the axonemes (Fig. 3D). In contrast, these proteins concentrated at the tips of the mutant cilia with little or no staining at their base. Double labeling with the centrosomal marker γ-tubulin confirmed that this accumulation was at the cilium tip (Fig. S5D). IFT140 was undetectable within the cilia of Ift122sopb cells, although it localized to cilia of wild-type cells (Fig. 3D). In vivo, we observed a similar phenotype as the cilia of the mutant node, mesenchyme, and neuroepithelium showed accumulation of IFT88 (Figs. 3C; Fig. S5E). These data suggest that mouse IFT122 functions in retrograde IFT and facilitates complex A stability.
IFT122 Suppresses Shh Signaling in the Cilium at a Step Downstream of Smoothened.
We next asked whether the roles for IFT122 in Shh signaling and IFT are linked at the level of the cilium. To test this, we disrupted ciliogenesis in Ift122sopb mutants by introducing a mutation in Kif3a, which encodes a kinesin 2 subunit mediating anterograde IFT. Kif3a mutants lack cilia and show suppression of Shh pathway activity (2, 43). Kif3a;Ift122sopb double mutants lacked primary cilia and resembled Kif3a single mutants in their morphology and neural tube patterning (Fig. S7 A–C). These findings suggest that ciliogenesis is required for the ectopic activation of the Shh pathway observed in Ift122sopb mutants.
Recent studies suggest that, in the absence of Shh, Smo is transported into cilia but quickly exits by retrograde IFT, preventing its ciliary accumulation (28, 44). This prompted us to ask whether the constitutive activation phenotype in Ift122sopb mutants is due to defects in Smo transport. We tested this through epistasis experiments and by examining the ciliary localization of Smo. If IFT122 regulates the Shh pathway through Smo, activity of the pathway in Ift122sopb mutants should be blocked by loss of Smo. However, Smo;Ift122sopb double mutants at e9.5 were morphologically similar to Ift122sopb single mutants, and the pathway remained ectopically active in the double mutant neural tube (Fig. S7 A and B). In addition, Smo localized properly to Ift122sopb mutant cilia. In wild-type and Ift122sopb cells, Smo localized to cilia upon stimulation of the pathway with Smoothened agonist (SAG), but not in its absence (Fig. 4A). These findings indicate that transport of Smo into and out of cilia does not require IFT122, although its ciliary exit requires the retrograde transport motor dynein (28).
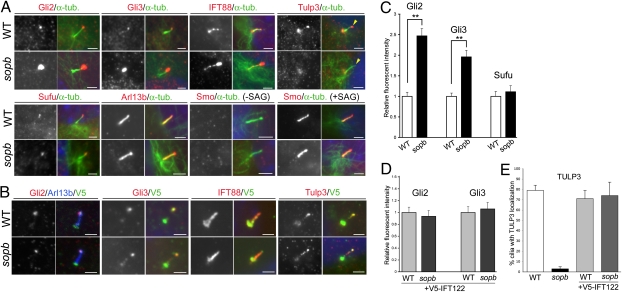
Fig. 4.
Localization of Shh pathway components in Ift122sopb cilia. (A) Localization of Gli2, Gli3, IFT88, TULP3, Sufu, Arl13b, and Smo in the cilia of serum-starved wild-type (WT) and Ift122sopb pMEFs. These markers are shown alone (Left panels in each set of four) and with acetylated α-tubulin staining in green (Right panels in each set). Smo localization was assayed with and without stimulation with SAG. TULP3 was largely absent from the mutant cilia tips (yellow arrowheads). Staining at the base of cilia is nonspecific (26). (B) Wild-type and sopb mutant cells transfected with V5-tagged IFT122 (green) were stained for Gli2, Gli3, IFT88, and TULP3 (red) and Arl13b (blue) in merged panels (Right panels in each set). Exogenous IFT122 rescued the Gli2, Gli3, and IFT88 accumulation defect and restored TULP3 localization to the cilium tip in sopb mutant cells. (C) Fluorescent intensity of Gli2, Gli3, and Sufu at cilia tips in wild-type and Ift122sopb cells (relative to wild-type average). (D) Fluorescent intensity of Gli2 and Gli3 in V5-IFT122–transfected cells. (E) Percentage of cilia showing TULP3 staining at cilia tips in wild-type and Ift122sopb cells with or without the V5-IFT122 construct. Error bars reflect SEM; **P < 0.01. Scale bars: 2 μm.
IFT122 Controls the Localization of Shh Pathway Activators and Repressors Within the Cilium.
The genetic experiments suggest that Gli proteins are regulated directly or indirectly by IFT122. Determining how these transcription factors are affected in Ift122sopb mutants may provide insight into how IFT122 functions to antagonize the Shh pathway. Mouse mutations that perturb IFT, such as Ift172, Ift88, Kif3a, Thm1/aln, and Dync2h1, increase the ratio between of the full-length form of Gli3 (Gli3FL) and the processed repressor form (Gli3Rep) (3, 25, 29). The Gli3FL/Gli3Rep ratio is also increased in Ift122Med1Δ1–3 mutants (41). In Ift122sopb whole embryos, the levels of Gli3FL, Gli3Rep, and Gli2 proteins did not appear altered (Fig. S8 A and B). However, in mutant limb buds, there was a 4.2-fold increase in the Gli3FL/Gli3Rep ratio (Fig S8 C and E), and Gli2 levels in the mutant limb buds appeared somewhat increased (Fig. S8 C and D). Thus, loss of IFT122 seems to affect Gli activator and repressor levels in specific developmental contexts.
We next monitored the ciliary localization of endogenous Gli2, Gli3, Sufu, Smo, Arl13b, and TULP3 by immunofluorescence in wild-type and Ift122sopb pMEFs (Fig. 4A). Although the total levels of Gli2 and Gli3 proteins were not significantly affected in sopb mutant pMEF extracts (Fig S9E), we found that Gli2 and Gli3 immunofluorescence at the tips of Ift122sopb mutant cilia was significantly increased relative to wild-type cilia (2.47 ± 0.18- and 1.96 + 0.16-fold, respectively; Fig. 4C), paralleled by an increase in the frequency of Gli2+ and Gli3+ cilia in the mutants (Fig. S9C). Consistent with a role for IFT122 downstream of Smo, treatment with a Smo antagonist (cyclopamine) did not suppress this phenotype (Fig. S9A). Treatment with the Smoothened agonist SAG had little effect on ciliary Gli2 and Gli3 in sopb mutant cells, yet it increased the intensity and frequency of Gli2 and Gli3 immunostaining in wild-type cilia (Fig. S9 A and C).
Not all Shh pathway components accumulated in the mutant cilia. Sufu was detected on the tips of wild-type and mutant cilia, but there was no significant difference in staining intensity between genotypes (Fig. 4 A and C). Similarly, the localization of Smoothened and Arl13b to cilia appeared normal in Ift122sopb mutant cells, although the frequency of Arl13b-expressing cilia was somewhat reduced. In contrast, TULP3, a repressor of Shh pathway (26), was largely absent from the tips of mutant cilia (4.2% positive, n = 71), whereas it localized to cilia tips in the majority of wild-type cells (81%, n = 63; Fig. 4 A and E). This is likely to be a defect in transport as steady-state TULP3 protein levels were normal in Ift122sopb mutants (Fig. S8C and Fig. S9E), and because TULP3 localized to the tips of Dync2h1lln mutant cilia (Fig. S9F), the defect in sopb mutant cells is unlikely to result secondarily from retrograde IFT defects. The accumulation of Gli2 and Gli3 at the cilium tip was suppressed and the localization of TULP3 was restored to normal by expression of V5-tagged wild-type IFT122 in sopb mutant cells (Fig. 4 B, D, and E). Along with the changes in Gli2, Gli3, and TULP3 localization, sopb mutant pMEFS exhibited increased activity of the Shh pathway (as assayed by expression of Ptch1) in the presence and absence of cyclopamine, yet the mutant cells retained responsiveness to SAG (Fig. S9D). Collectively, our data suggest that IFT122 regulates Shh pathway activity by differentially controlling the ciliary localization of pathway regulators.
Discussion
We find that mouse IFT22 is a negative regulator of the Shh pathway, acting at a step downstream of Shh and Smo and upstream of Gli2. The cilia of Ift122sopb mutants adopt a swollen morphology with complex B IFT proteins accumulating at their tips, hallmarks of defective retrograde IFT. A role for mouse IFT122 in retrograde IFT is consistent with that postulated for its homologs in other organisms (17, 36–38). Furthermore, our analysis of Shh pathway components indicates that their ciliary localization occurs by IFT122-dependent and -independent mechanisms: Ift122sopb mutant cilia show accumulation of Gli2 and Gli3 and loss of TULP3 localization at their tips, whereas the localization of Smo and Sufu is unaffected.
IFT122 and Intraflagellar Transport.
It is commonly thought that complex A and B IFT proteins play important roles in retrograde and anterograde IFT, respectively. However, some findings suggest that proteins of each complex participate in aspects of both anterograde and retrograde transport (15–17). For example, ciliary localization of the TRPV channel subunit IAV in Drosophila requires the IFT A protein IFT140, but not IFT dynein, suggesting an anterograde IFT role for IFT140 (17). Our results suggest that IFT122 is important for retrograde transport of some ciliary proteins, including Gli2, Gli3, IFT57, and IFT88. However, the loss of TULP3 localization to the tips of the mutant cilia suggests that IFT122 plays additional roles in ciliogenesis, such as anterograde transport of some proteins or loading of cargo onto IFT particles before anterograde transport.
Multiple roles for IFT122 in ciliogenesis may explain why our results differ from those described for another allele, Ift122Med1Δ1–3, which also disrupts the DNA repair gene Med1/Mdb4 (41). Ciliogenesis appears to be largely blocked in Ift122Med1Δ1–3 mutants, which may explain the loss of some ventral neural cell fates, as observed in other mutants lacking cilia (3, 4). In contrast, Ift122sopb mutants retain cilia with a swollen morphology and exhibit widespread ventralization of cell fates in the posterior neural tube. As both Ift122sopb and Ift122Med1Δ1–3 alleles appear to be null, the differences may be explained by the strain backgrounds on which they were studied (C3Heb/FeJ and C57BL/6, respectively). Alternatively, the disruption of Med1/Mdb4 in the Ift122Med1Δ1–3 allele could modify the ciliogenesis phenotype caused by loss of IFT122, leading to a strong defect in anterograde IFT.
Regulation of the Sonic Hedgehog Pathway by IFT122.
Our findings underscore the complex relationship between the primary cilium and Hedgehog signaling in mice. Genetic studies suggest that the dynein 2 subunit DYNC2H1, the IFT139-related protein THM1/aln, and IFT122 all function in retrograde IFT. However, their precise roles with respect to the Shh pathway may differ. In contrast to Ift122sopb mutants, disruption of Dync2h1 represses Shh signaling. Although Gli proteins accumulate at the tips of cilia in both mutants, Smo constitutively localizes to Dync2h1lln, but not Ift122sopb, mutant cilia in the absence of Shh signaling (28, 44). Moreover, TULP3 localizes normally to Dync2h1lln mutant cilia, but this localization is disrupted in Ift122sopb mutants. Therefore, complex A IFT proteins and the retrograde motor dynein have overlapping, yet distinct, functions in transporting Shh pathway components. The functions of individual complex A IFT proteins in Hedgehog signaling also appear to be distinct. Recent findings suggest that mammalian IFT122, WDR19, and IFT140 represent core IFT A proteins required for TULP3 association, whereas THM1 and WDR35 are not required for this association (45). It is unclear whether Gli proteins accumulate abnormally in Thm1aln mutant cilia as they do in Ift122sopb mutants. Exogenously expressed Gli2 and Gli3 localized to Thm1aln mutant cilia tips, but the overexpression of proteins hinders a proper quantitative analysis. Distinct roles for IFT122 and THM1 in Gli protein and TULP3 transport are likely to impact Shh pathway activity differently. Indeed, disruption of Thm1/aln leads to ectopic activation of the Shh pathway, but to a lesser extent than in Ift122sopb mutants on the basis of neural patterning and genetic suppression data (29; this study). Thus, the roles of individual components of the IFT machinery in controlling the Hedgehog pathway are likely to differ depending on their precise functions in the transport process.
The cilium tip may be a cellular compartment where Gli activity is regulated. Several regulators of the Shh pathway localize here, and stimulation with Shh ligands causes Gli proteins to accumulate at this site (7, 26, 27, 46). Importantly, a similar phenomenon is observed in Ift122sopb mutant cilia without Shh stimulation. Because disruption of IFT122 bypasses the requirement for Shh ligands for both pathway activity and ciliary accumulation of Gli proteins, we suggest that Shh signaling shifts the balance between activators of the pathway (Gli2 and Gli3) and inhibitors (Sufu and TULP3) at the cilium tip by regulating their association with the intraflagellar transport machinery. Shifting this balance in favor of the Gli proteins may protect them from inhibition, allowing them to activate target genes.
Materials and Methods
Mouse Strains.
sopb is an N-ethyl-nitrosourea–induced recessive mutation obtained from a phenotype-based screen for abnormal embryos at e9.5 (32). The mutation was induced on a C57BL/6J chromosome and backcrossed to a C3Heb/FeJ background. The sopb mutant phenotype was analyzed after more than five generations of backcrossing to C3Heb/FeJ. All double mutants were obtained from intercrosses between double heterozygous animals on a C3Heb/FeJ background, and wild-type littermates served as controls. Analysis of Nodal-LacZ expression was performed on sopb and wild-type embryos heterozygous for the Nodaltm1Rob allele. Shhtm1Chg, Kif3Atm1Gsn, Smotm1Amc, Gli2tm1Al,j, and Nodaltm1Rob mutant alleles were genotyped from yolk sac and ear punch samples as described (34, 43, 47–49). Three to five double mutants were analyzed for each epistasis experiment. Animal work was carried out under Institutional Animal Care and Use Committee-approved protocols at Princeton University.
Immunostaining of Sections.
Immunofluorescence analysis of tissue was performed as described (50). Frozen sections (10 μm) were stained with primary antibodies, followed by detection with Cy2- or Cy3-conjugated secondary antibodies (Jackson ImmunoResearch) and counterstained with DAPI (Sigma-Aldrich). Images were obtained using a Nikon E800 compound microscope with a CCD camera (Princeton Instruments) and MetaMorph software (Universal Imaging). Photoshop (Adobe) was used for image processing. Mouse monoclonal antibodies against Shh, FoxA2, Nkx2.2, HB9, Nkx6.1, Pax6, and Pax7 were obtained from the Developmental Studies Hybridoma Bank. Rabbit α-Pax6 and goat α-Olig2 polyclonal antibodies were purchased from Covance and R&D Systems, respectively.
Immunostaining of Embryonic Fibroblasts.
Cells grown on coverslips were fixed for 10 min in 4% paraformaldehyde or ice-cold acetone (for anti-TULP3 staining) and immunostainined as described (26). Samples were imaged using conventional (Nikon E800) and confocal (Leica SP5) fluorescent microscopes. Primary antibodies used were mouse α-acetylated α-tubulin (Sigma-Aldrich); mouse α-γ–tubulin (Sigma-Aldrich); rabbit α-Arl13b (gift from T. Caspary, Emory University, Atlanta); rabbit α-mGli3 (51); rabbit α-Sufu (Santa Cruz Biotechnology); rabbit α-Smo (gift from K. Anderson, Sloan Kettering Institute, New York); rabbit α-IFT88, α-IFT57, and α-IFT140 (gifts from G. Pazour, University of Massachusetts Medical School, Worcester, MA); rabbit α-TULP3 (26); and guinea pig α-Gli2 (52). Quantitation of immunofluorescence intensity was performed using National Institutes of Health ImageJ software. Details of the quantitation methods as well as a detailed account of additional experimental procedures can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Kathryn Anderson (Sloan Kettering Institute, New York), Tamara Caspary (Emory University, Atlanta), Gregory Pazour (University of Massachusetts Medical School, Worcester, MA), Philip Beachy (Stanford University, Stanford, CA), Alexandra Joyner (Sloan Kettering Institute, New York), Elizabeth Robertson (University of Oxford, Oxford), and Andrew McMahon (Harvard University, Cambridge, MA) for their generous gifts of mouse strains, polyclonal antibodies, and cell lines; Cathy Vu, Margaret Bisher, and Joseph Goodhouse (Princeton University) for help with sequencing, scanning electron microscopy, and confocal microscopy, respectively; and Rebecca Burdine (Princeton University) and members of the Eggenschwiler laboratory for comments on the manuscript. Monoclonal antibodies against neural patterning markers were from the Developmental Studies Hybridoma Bank. This work was supported by National Institutes of Health Grant R01 HD050761 (to J.T.E.) and by National Institutes of Health National Research Service Award 5 F32 NS050480 (to J.Q.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011410108/-/DCSupplemental.
References
- 1.Bloodgood RA. Sensory reception is an attribute of both primary cilia and motile cilia. J Cell Sci. 2010;123:505–509. doi: 10.1242/jcs.066308. [DOI] [PubMed] [Google Scholar]
- 2.Huangfu D, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 3.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 5.Rohatgi R, Milenkovic L, Corcoran RB, Scott MP. Hedgehog signal transduction by Smoothened: Pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci USA. 2009;106:3196–3201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 7.Wen X, et al. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol Cell Biol. 2010;30:1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 10.Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole DG, et al. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blacque OE, et al. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol Biol Cell. 2006;17:5053–5062. doi: 10.1091/mbc.E06-06-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piperno G, et al. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J Cell Biol. 1998;143:1591–1601. doi: 10.1083/jcb.143.6.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iomini C, Babaev-Khaimov V, Sassaroli M, Piperno G. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J Cell Biol. 2001;153:13–24. doi: 10.1083/jcb.153.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedersen LB, et al. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr Biol. 2005;15:262–266. doi: 10.1016/j.cub.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 16.Iomini C, Li L, Esparza JM, Dutcher SK. Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetic interactions in Chlamydomonas reinhardtii. Genetics. 2009;183:885–896. doi: 10.1534/genetics.109.101915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee E, Sivan-Loukianova E, Eberl DF, Kernan MJ. An IFT-A protein is required to delimit functionally distinct zones in mechanosensory cilia. Curr Biol. 2008;18:1899–1906. doi: 10.1016/j.cub.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varjosalo M, Taipale J. Hedgehog: Functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 19.Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: A sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 20.Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 21.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 22.Motoyama J, et al. Differential requirement for Gli2 and Gli3 in ventral neural cell fate specification. Dev Biol. 2003;259:150–161. doi: 10.1016/s0012-1606(03)00159-3. [DOI] [PubMed] [Google Scholar]
- 23.Østerlund T, Kogerman P. Hedgehog signalling: How to get from Smo to Ci and Gli. Trends Cell Biol. 2006;16:176–180. doi: 10.1016/j.tcb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 25.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norman RX, et al. Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Hum Mol Genet. 2009;18:1740–1754. doi: 10.1093/hmg/ddp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liem KF, Jr, He M, Ocbina PJ, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci USA. 2009;106:13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ocbina PJ, Anderson KV. Intraflagellar transport, cilia, and mammalian Hedgehog signaling: Analysis in mouse embryonic fibroblasts. Dev Dyn. 2008;237:2030–2038. doi: 10.1002/dvdy.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran PV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Ko HW, et al. Broad-minded links cell cycle-related kinase to cilia assembly and hedgehog signal transduction. Dev Cell. 2010;18:237–247. doi: 10.1016/j.devcel.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García-García MJ, et al. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc Natl Acad Sci USA. 2005;102:5913–5919. doi: 10.1073/pnas.0501071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- 34.Chiang C, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 35.Matise MP, Epstein DJ, Park HL, Platt KA, Joyner AL. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- 36.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 37.Absalon S, et al. Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol Biol Cell. 2008;19:929–944. doi: 10.1091/mbc.E07-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsao CC, Gorovsky MA. Tetrahymena IFT122A is not essential for cilia assembly but plays a role in returning IFT proteins from the ciliary tip to the cell body. J Cell Sci. 2008;121:428–436. doi: 10.1242/jcs.015826. [DOI] [PubMed] [Google Scholar]
- 39.Avidor-Reiss T, et al. Decoding cilia function: Defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- 40.Walczak-Sztulpa J, et al. Cranioectodermal dysplasia, Sensenbrenner syndrome, is a ciliopathy caused by mutations in the IFT122 gene. Am J Hum Genet. 2010;86:949–956. doi: 10.1016/j.ajhg.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cortellino S, et al. Defective ciliogenesis, embryonic lethality and severe impairment of the Sonic Hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene Ift122/Wdr10, partially overlapping with the DNA repair gene Med1/Mbd4. Dev Biol. 2009;325:225–237. doi: 10.1016/j.ydbio.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiratori H, Hamada H. The left-right axis in the mouse: From origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- 43.Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci USA. 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukhopadhyay S, et al. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 2010;24:2180–2193. doi: 10.1101/gad.1966210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen MH, et al. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001;106:781–792. [PubMed] [Google Scholar]
- 48.Mo R, et al. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- 49.Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- 50.Eggenschwiler JT, Anderson KV. Dorsal and lateral fates in the mouse neural tube require the cell-autonomous activity of the open brain gene. Dev Biol. 2000;227:648–660. doi: 10.1006/dbio.2000.9918. [DOI] [PubMed] [Google Scholar]
- 51.Bulgakov OV, Eggenschwiler JT, Hong DH, Anderson KV, Li T. FKBP8 is a negative regulator of mouse sonic hedgehog signaling in neural tissues. Development. 2004;131:2149–2159. doi: 10.1242/dev.01122. [DOI] [PubMed] [Google Scholar]
- 52.Cho A, Ko HW, Eggenschwiler JT. FKBP8 cell-autonomously controls neural tube patterning through a Gli2- and Kif3a-dependent mechanism. Dev Biol. 2008;321:27–39. doi: 10.1016/j.ydbio.2008.05.558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.