Abstract
The major obstacle in cancer treatment is the resistance of cancer cells to therapies. Nrf2 is a transcription factor that regulates a cellular defense response and is ubiquitously expressed at low basal levels in normal tissues due to Keap1-dependent ubiquitination and proteasomal degradation. Recently, Nrf2 has emerged as an important contributor to chemoresistance. High constitutive expression of Nrf2 was found in many types of cancers, creating an environment conducive for cancer cell survival. Here, we report the identification of brusatol as a unique inhibitor of the Nrf2 pathway that sensitizes a broad spectrum of cancer cells and A549 xenografts to cisplatin and other chemotherapeutic drugs. Mechanistically, brusatol selectively reduces the protein level of Nrf2 through enhanced ubiquitination and degradation of Nrf2. Consequently, expression of Nrf2-downstream genes is reduced and the Nrf2-dependent protective response is suppressed. In A549 xenografts, brusatol and cisplatin cotreatment induced apoptosis, reduced cell proliferation, and inhibited tumor growth more substantially when compared with cisplatin treatment alone. Additionally, A549-K xenografts, in which Nrf2 is expressed at very low levels due to ectopic expression of Keap1, do not respond to brusatol treatment, demonstrating that brusatol-mediated sensitization to cisplatin is Nrf2 dependent. Moreover, a decrease in drug detoxification and impairment in drug removal may be the primary mechanisms by which brusatol enhances the efficacy of chemotherapeutic drugs. Taken together, these results clearly demonstrate the effectiveness of using brusatol to combat chemoresistance and suggest that brusatol can be developed into an adjuvant chemotherapeutic drug.
Keywords: chemosensitization, reactive oxygen species, antioxidant response, natural compounds
Cancer is the second leading cause of death in developed countries. Lung cancer is one of the most commonly diagnosed cancers and comprises 15–30% of the total number of cancer cases. Despite the fact that many new chemotherapeutic drugs have been developed and many aggressive treatments are available, cancer death rates continue to rise. The major obstacle in cancer treatment is the resistance to therapies, including chemotherapies. Two types of resistance are observed. (i) Intrinsic resistance: cancer cells are inherently drug resistant and do not respond. (ii) Acquired resistance: cancer cells acquire resistance to chemotherapy following an initial response, which is reflected in the recurrence of cancer in many patients following chemotherapy. Mechanistically, the resistance phenomena may be explained by (i) mutation or overexpression of drug target proteins, and/or (ii) inactivation of drugs by a reduction in uptake or enhanced detoxification and removal of drugs. Currently, radiation and platinum-based drugs are the standard treatment for lung cancer (1). However, the toxicity profiles and high rate of relapse with platinum compounds limit their use and effectiveness. Therefore, there is an urgent need to develop new adjuvants that enhance the efficacy of platinum-based treatments and circumvent chemoresistance.
Nrf2 is an important transcription factor that regulates the antioxidant response by inducing the expression of genes bearing an antioxidant response element (ARE) in their regulatory regions. Activation of the Nrf2 pathway composes a cellular protective system that promotes cell survival under detrimental environments (2–4). Interestingly, many Nrf2 target genes, including drug metabolizing enzymes, conjugating enzymes, and drug transporters, play a crucial role in determining drug response and resistance. Nrf2 is ubiquitously expressed in all human organs at low constitutive levels due to tight regulation by Keap1, a substrate adaptor protein for a Cullin3-based E3 ubiquitin ligase (5–8). Under basal conditions, Nrf2 is constantly targeted for Keap1-mediated ubiquitination and subsequent proteasomal degradation to maintain low Nrf2 protein levels. Upon activation, the enzymatic activity of the Keap1-Cullin3 E3 ubiquitin ligase is compromised, resulting in stabilization of Nrf2 and activation of Nrf2 downstream genes.
Oxidative stress is implicated in the initiation and progression of cancer. Under oxidative stress, Nrf2 induces the transcription of cellular protective genes to combat carcinogenic reactive intermediates. Therefore, activation of the Nrf2 pathway is important in chemoprevention as demonstrated by these findings: (i) many chemopreventive compounds have been identified as Nrf2 activators (4, 9–12), and (ii) Nrf2-null mice are highly susceptible to chemical carcinogens and are no longer protected by chemopreventive compounds (3, 13, 14). Paradoxically, recent findings point to the “dark side” of Nrf2, as many studies have shown that constitutively high levels of Nrf2 promote cancer formation and contribute to chemoresistance (15–18). When Yamamoto's team revealed the structure of the Nrf2–Keap1 interaction, they demonstrated that two different mutations in the Kelch domain of Keap1 in lung cancer disrupted the interaction between Nrf2 and Keap1, leading to hyperactivation of the Nrf2-mediated protective response (19). Additionally, Biswal's group identified somatic mutations that likely disrupt the repressor function of Keap1 at a frequency of 50% (6/12) or 19% (10/54) in cancer cell lines or cancer samples, respectively. Furthermore, loss of heterozygosity at 19p13.2, where Keap1 is located, was observed at a frequency of 61 and 41% in non-small cell lung carcinoma (NSCLC)-derived cell lines (72 in total) and tumor tissues (39 samples in total), respectively (20). In another study, somatic mutations in Keap1 were found in 5 out of 65 (8%) patients who had adenocarcinoma (ADC), squamous cell carcinoma (SCC), or large cell carcinoma (LCC) (21). Furthermore, nuclear expression of Nrf2 or low expression of Keap1 was found at a frequency of 26 or 56%, respectively, in 304 NSCLC tissue samples studied (22). In addition to lung cancer, overexpression of Nrf2 has been shown in several other cancers. For example, Nrf2 expression was increased in 91.5% of cancer tissues from head and neck squamous cell carcinoma (HNSCC) (23), and the frequency of Keap1 mutation was 30.7% (4 out of 13) in gallbladder cancer samples (24). By the same token, mutations in Nrf2 can disrupt Keap1-dependent repression and result in overexpression of Nrf2. Somatic mutations in the coding region of Nrf2 that allow Nrf2 to escape repression by the Keap1-Cullin3 E3 ubiquitin ligase were found in lung (11/103; 10.7%) and head and neck (3/12, 10.7%) tumors (25). In a similar study, Nrf2 mutations were found in esophagus (8/70; 11.4%), skin (1/17; 6.3%), lung (10/125; 8.0%), and larynx (3/23; 13.0%) tumors (26). More recently, we have reported a study on Nrf2 expression in endometrial cancer patients (117 cases). There was no detectable Nrf2 expression in complex hyperplasia. However, Nrf2 was highly expressed in endometrial endometrioid carcinoma (type I) (28%) or endometrial serous carcinoma (type II) (89%) (27), which is the most malignant and recurrent carcinoma among female genital malignancies.
Recent studies with cultured cells and mouse xenograft models strongly support the notion that Nrf2 is a great target to overcome chemoresistance. For instance, suppression of endogenous Nrf2, either by transfecting Nrf2-siRNA or overexpressing Keap1, sensitized cancer cells to chemotherapeutic drugs. Conversely, overexpression of Nrf2 in cancer cells that have low basal levels of Nrf2, enhanced resistance in a variety of cancer cells including neuroblastoma, breast, ovarian, prostate, lung, and pancreatic cancer cells (28–32). Combined use of Nrf2-siRNA and carboplatin inhibited the growth of A549 xenografts in mice (33). In addition, we have demonstrated that suppression of Nrf2 by Keap1 overexpression sensitized SPEC-2 cells, which are derived from type II endometrial cancer, and SPEC-2 xenografts to cisplatin (27). In support of a role for Nrf2 in chemoresistance, expression of Nrf2 in cancer cells increased during acquisition of drug resistance (34, 35). Collectively, these results demonstrate that Nrf2 contributes to chemoresistance observed in many types of cancers originating from different organs. Further, this illustrates the urgent need to identify compounds that suppress the Nrf2 pathway and develop them into druggable compounds to enhance the effectiveness of cancer treatments.
Results
Brusatol Selectively Inhibited the Nrf2 Pathway.
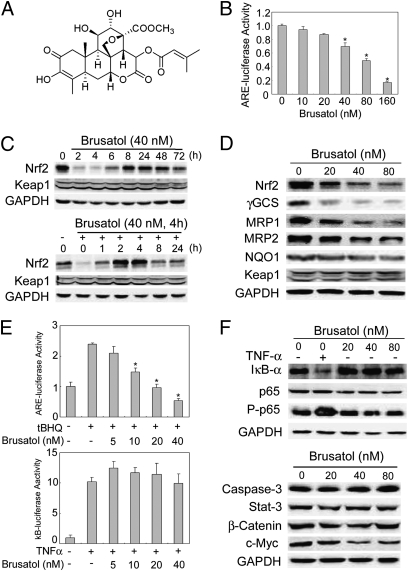
To combat Nrf2-mediated chemoresistance, we searched for compounds that suppress the Nrf2 pathway by screening a large number of natural products for their ability to inhibit ARE-luciferase activity using a stable cell line, MDA-MB-231-ARE-Luc (36). A plant extract from Brucea javanica (L) Merr. (Simaroubaceae), an evergreen shrub grown in Southeast Asia and Northern Australia, was found to inhibit ARE-luciferase activity and the protein levels of Nrf2. Subsequently, the plant extract was further fractionated and purified compounds were tested for their ability to inhibit the Nrf2 pathway, which resulted in the identification of brusatol, a quassinoid (Fig. 1A). The purification procedure and the identification of brusatol are described (Fig. S1). Brusatol inhibits ARE-luciferase activity in a dose-dependent manner in the MDA-MB-231-ARE-Luc stable cell line (Fig. 1B). Furthermore, 40 nM of brusatol significantly decreased the protein level of Nrf2 after just 2–4 h of treatment and was able to maintain reduced Nrf2 protein levels compared with control for up to 72 h (Fig. 1C, Upper). Interestingly, brusatol but not brucein C, another quassinoid with a similar chemical structure, reduced Nrf2 in a dose-dependent manner (Fig. 1D, Nrf2, and Fig. S2 A and B). Additionally, a dose-dependent reduction of Nrf2 protein levels by brusatol was observed in all cell lines tested, including HeLa, MDA-MB-231, Ishikawa, and SPEC-2 (Fig. S3). To demonstrate whether the effects of brusatol are reversible, we tested the ability of Nrf2 to recover after removal of brusatol. Again, brusatol decreased Nrf2 protein levels when compared with untreated cells (Fig. 1C, Lower). Moreover, following the removal of brusatol, Nrf2 protein level quickly recovered within 1 h and surpassed those of basal levels at 2–4 h. Subsequently, Nrf2 protein levels reached equilibrium 8–24 h after the removal of brusatol (Fig. 1C, Lower). In addition to Nrf2, the protein level of Nrf2-target genes, including γGCS, MRP1, and MRP2, was also reduced in a dose-dependent manner, whereas only a slight reduction in NQO1 was observed (Fig. 1D).
Fig. 1.
Brusatol selectively inhibited the Nrf2 pathway. (A) Structure of brusatol. The structure was determined by NMR and HRMS. (B) Brusatol inhibited ARE-dependent luciferase activity. MDA-MB-231-ARE-Luc cells were treated with several doses of brusatol for 16 h. (C) Brusatol-mediated reduction of Nrf2 was reversible. (Upper) A549 cells were treated with 40 nM of brusatol for the indicated time points. (Lower) A549 cells were pretreated with 40 nM of brusatol for 4 h and then brusatol was removed and cells were further cultured for the indicated periods. (D) Brusatol reduced the protein levels of Nrf2 and its downstream genes. A549 cells were treated with the indicated doses of brusatol for 16 h. (E) Brusatol inhibited ARE-dependent, but not κB-dependent luciferase activity. A549 cells were transfected with ARE-dependent firefly luciferase (Upper) or κB-dependent firefly luciferase (Lower), along with TK renilla. At 32 h posttransfection, cells were treated with 40 μM of tBHQ (Upper) or 10 ng/mL TNFα (Lower), along with the indicated doses of brusatol for 16 h. Relative luciferase activity was measured by normalizing firefly luciferase activity to renilla luciferase activity. (F) Brusatol had no effect on the NFκB pathway or other proteins. A549 cells were treated with the indicated doses of brusatol for 4 h.
Conversely, the protein level of Keap1 was not affected by brusatol (Fig.1 C and D). In contrast to a previous report indicating that brusatol activated the NF-κB pathway (37), brusatol had no effect on κB-luciferase activity, whereas it strongly suppressed ARE-luciferase activity (Fig. 1E), demonstrating that brusatol selectively inhibits the Nrf2 pathway. This notion was further supported by similar protein levels of IκBα and phosphorylated p65 in response to different doses of brusatol, whereas TNFα decreased IκBα and increased phosphorylated p65 (Fig. 1F, Upper). In addition, brusatol had no effect on caspase-3, a key modulator of the apoptotic pathway, Stat-3, or β-catenin; however, it moderately reduced c-Myc expression (Fig. 1F, Lower). Collectively, these results demonstrate that brusatol specifically inhibits the Nrf2 pathway by reducing the protein level of Nrf2.
Brusatol Inhibited the Nrf2 Pathway Through Enhanced Ubiquitination and Degradation of Nrf2.
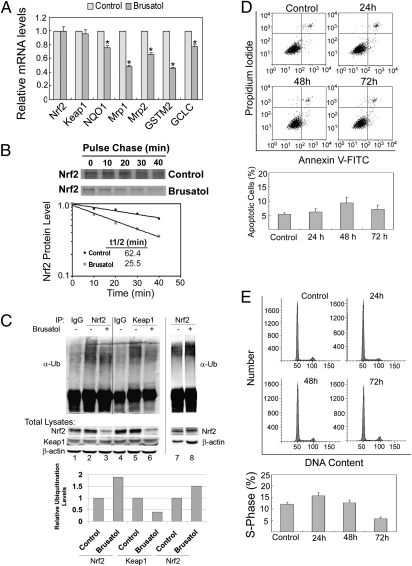
To test the possibility that brusatol reduces Nrf2 through a posttranslational mechanism, mRNA expression of Nrf2, Keap1, and Nrf2-target genes were measured by real-time RT-PCR. Brusatol did not change the mRNA level of Nrf2 or Keap1, but decreased the level of NQO1, MRP1, MRP2, GSTm2, and GCLC (Fig. 2A). Next, half-life was measured using a pulse-chase assay. Brusatol reduced the half-life of Nrf2 from 62.4 min to 25.5 min (Fig. 2B). In accordance with this notion, in vivo ubiquitination analysis indicated that ubiquitination of Nrf2 (Fig. 2C, ubiquitin-conjugated Nrf2 normalized to the amount of Nrf2, lanes 2 and 3; or by using equal amounts of Nrf2 for the ubiquitination assay, lanes 7 and 8) was enhanced 1.5- to 2-fold by brusatol, whereas ubiquitination of Keap1 was decreased (Fig. 2C, for quantification see bar graph below). Interestingly, this brusatol-mediated switch in ubiquitination from Keap1 to Nrf2 is opposite of what we reported previously with the Nrf2 activator tert-butylhydroquinone (tBHQ), which resulted in reduced ubiquitination of Nrf2, but enhanced ubiquitination of Keap1 (38). Collectively, these results imply that brusatol likely modulates the ubiquitin-mediated protein degradation system to enhance Nrf2 degradation. Additionally, the effect of brusatol on cell growth and death was also investigated. Brusatol did not cause any significant cell death up to 72 h after treatment as measured by Annexin V and propidium iodide (PI) staining (Fig. 2D), whereas it slightly arrested cell cycle at the S phase at 72 h (Fig. 2E).
Fig. 2.
Brusatol reduced Nrf2 through enhanced ubiquitination and degradation of Nrf2. (A) Brusatol reduced the mRNA level of Nrf2 target genes. A549 cells were treated with 40 nM of brusatol for 16 h and subjected to qRT-PCR. (B) Brusatol decreased the half-life of Nrf2. A549 cells were left untreated or treated with 40 nM of brusatol for 4 h. A pulse-chase experiment was carried out to measure half-life of Nrf2. (C) Brusatol enhanced ubiquitination of Nrf2, but decreased ubiquitination of Keap1. A549 cells were left untreated or treated with 40 nM of brusatol for 4 h and cells were subjected to in vivo ubiquitination analysis for Nrf2 and Keap1 (lanes 1–6). The amount of Nrf2 and Keap1 in the total lysate was detected and used for normalization. In another ubiquitination assay, cells were treated with 40 nM of brusatol for 2 h and equal amounts of Nrf2 protein was used for ubiquitination analysis (lanes 7 and 8). (D) Brusatol treatment did not cause any obvious cell death. A549 cells were treated with 40 nM of brusatol for the indicated time points before measurement of apoptosis using Annexin V and PI. (E) Brusatol slightly arrested cell cycle at the S phase. The treated cells were subjected to cell cycle analysis by PI staining.
Brusatol Sensitized Cancer Cells and Xenografts to Chemotherapeutic Drugs.
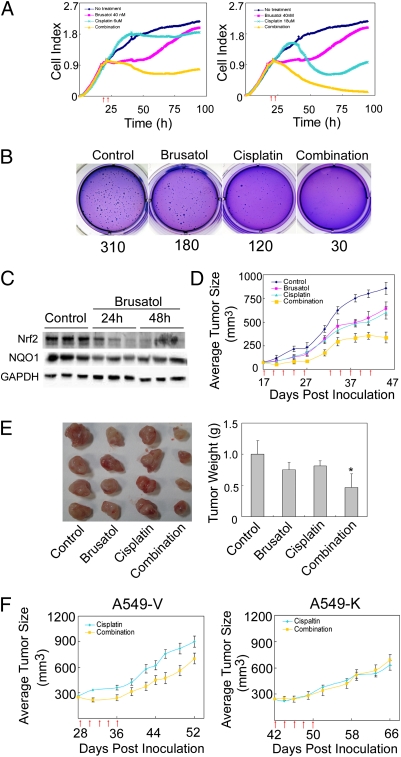
Because Nrf2 was found to be highly expressed in cancer tissues and contributes to chemoresistance, a reduction of Nrf2 should sensitize cancer cells to chemotherapeutic drugs. Next, the effects of brusatol on cytotoxicity were measured using the xCELLigence system (Roche). A549 lung cancer cells were treated with DMSO, brusatol, cisplatin, or in combination, and cell growth was monitored (Fig. 3A). A total of 6 μM of cisplatin displayed a minor reduction in cell number, whereas treatment with brusatol alone resulted in transient growth arrest followed by a quick growth period (Fig. 3A, Left). However, cotreatment of brusatol with cisplatin dramatically reduced cell number (Fig. 3A, Left). Using a higher dose of cisplatin (18 μM) alone, we saw substantial cell death up to 60 h posttreatment. However, an increase in cell growth was observed afterward, indicating the onset of drug resistance. In contrast, when cells were cotreated with brusatol, the number of viable cells decreased dramatically after 60 h and there were no viable cells at 96 h (Fig. 3A, Right). Brucein C, which was unable to reduce Nrf2 protein levels, did not enhance cisplatin-mediated cell death (Fig. S2C). Furthermore, brusatol sensitized A549 cells to other chemotherapeutic drugs such as carboplatin, 5-fluorouracil, etoposide, and pacilitaxel (Fig. S4). In addition, similar experiments were performed in several other cancer cell lines including HeLa and MDA-MB-231 cells, and the brusatol-mediated sensitization to chemotherapeutic drugs was observed in both cancer cell lines (Fig. S5). As another way to measure toxicity, a colony formation assay was conducted and our results demonstrated that brusatol or cisplatin alone reduced the number of colonies formed; however, combined treatment dramatically reduced colony formation (Fig. 3B). These results indicate that brusatol enhanced cytotoxicity induced by chemotherapeutics drugs.
Fig. 3.
Brusatol sensitized cancer cells and xenografts to chemotherapeutic drugs in an Nrf2-dependent manner. (A) Brusatol sensitized A549 cells to cisplatin treatment. A549 cells were pretreated with 40 nM of brusatol for 4 h (first arrow) before the addition of DMSO or cisplatin (second arrow) (6 μM in Left panel and 18 μM in Right panel). (B) Combination treatment markedly inhibited colony formation. A549 cells were treated with PBS, 40 nM of brusatol, 3 μM of cisplatin, or 40 nM of brusatol and 3 μM of cisplatin in combination for 4 wk. The average number of colonies formed per well is shown below the image. (C) Brusatol reduced Nrf2 in xenografts in vivo up to 48 h postinjection. Nude mice (three per group) were injected with A549 cells. Once tumor size reached 80 mm3, mice were injected with brusatol (2 mg/kg) and killed at 24 h and 48 h postinjection. Tumors were isolated and subjected to immunoblot analysis. (D) Brusatol sensitized xenografts to cisplatin treatment. Once tumor size reached 80 mm3, mice (10 per group) were treated with DMSO, cisplatin (2 mg/kg), brusatol (2 mg/kg), or in combination through i.p. every other day for a total of five times (five-time cisplatin treatment regimen); then treatment stopped. At day 33, the same five-time cisplatin treatment regimen was repeated. Small arrows under the x-axis represent the day of injection. (E) Combination treatment significantly reduced tumor weight. Tumors were excised and weighed at the end of the experiment (46 d). (F) A reduction in Nrf2 was required for brusatol-mediated sensitization to cisplatin. Twenty nude mice were injected with either A549-V or A549-K (1 × 107 cells). When the average tumor size reached 280 mm3 (28 d for A549-V–derived tumors; 42 d for A549-K–derived tumors), mice (10 per group) were treated with cisplatin or in combination every other day for a total of five times. Tumor size was measured.
To explore the anticancer effect of brusatol in vivo, we used A549 xenografts grown in nude mice as a model. Nude mice were injected with A549 cells to induce tumor growth, followed by a single i.p. injection of 2 mg/kg brusatol. Tumors were isolated 24 h or 48 h postinjection. We found that Nrf2 protein levels were significantly decreased at 24 h or 48 h postinjection, indicating that brusatol is able to reach the tumor tissue and inhibit the Nrf2 pathway (Fig. 3C). To measure tumor growth, two different experiments were performed. In the first experiment, once the tumor size reached an average of 230 mm3, DMSO, brusatol (2 mg/kg), cisplatin (2 mg/kg), or cisplatin (2 mg/kg) and brusatol (2 mg/kg) combined treatment was i.p. injected every other day for a total of five times. This will be referred to as the “five-time cisplatin treatment regimen.” Cisplatin or brusatol alone did not inhibit tumor growth significantly, whereas in the combination group, tumor size was significantly reduced (Fig. S6A, Left). No significant weight loss was observed in any group (Fig. S6A, Right). These results indicate that brusatol is able to combat intrinsic resistance. Because many patients develop acquired resistance after several treatments with cisplatin, we wanted to mimic this in our mouse model. In the second experiment, once the tumor size reached an average of 80 mm3 (day 18) the mice were treated with the five-time cisplatin treatment regimen. One week after the end of the first treatment regimen, a second five-time cisplatin treatment regimen was used on day 33. Brusatol or cisplatin alone delayed tumor growth (Fig. 3D). However, cotreatment of brusatol with cisplatin significantly reduced tumor sizes (Fig. 3D). The sensitizing effect of brusatol was more substantial after the second regimen (Fig. 3D), although mice in the combination group lost weight slightly after the second regimen (Fig. S6B). The experiment was terminated at 46 d after the injection of A549 cells and tumors were isolated. The reduction of tumor size and weight in the combination group was apparent (Fig. 3E). Next, tumor tissues were analyzed for apoptosis and cell proliferation by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) and immunohistochemistry (IHC) with Ki67, respectively. Brusatol alone did not induce visible apoptosis, whereas cisplatin and combined treatment significantly induced apoptotic cells (Fig. S6C, Upper). Furthermore, cisplatin treatment inhibited cell proliferation slightly, whereas there was a significant decrease in cell proliferation when cells were cotreated (Fig. S6C, Lower). Taken together, these results clearly demonstrate the effectiveness of using brusatol to combat both intrinsic and acquired chemoresistance.
Brusatol-Mediated Sensitization to Cisplatin Treatment Was Nrf2 Dependent.
To test whether brusatol-mediated sensitization to chemotherapeutic drugs relies on its ability to decrease Nrf2 protein expression, two A549-derived stable cell lines established in our laboratory were used (28). A549-V and A549-K are A549 cells stably transfected with an empty vector or a vector containing Keap1-CBD, respectively. Stable knockdown of Nrf2 in A549-K and enhanced susceptibility of this cell line to chemotherapeutic drugs, compared with A549-V, has been reported by our group previously (28). Nude mice were injected with either A549-V or A549-K cells. The five-time cisplatin treatment regimen started once the average tumor size reached 280 mm3. It is interesting to note that A549-K cells took much longer than A549-V to reach the same tumor size (Fig. 3F, 28 d for A549-V and 42 d for A549-K), further confirming the notion that high constitutive levels of Nrf2 promote tumor formation. Remarkably, xenografts derived from A549-V cells had a significant reduction in tumor size in the combination group compared with that in the cisplatin group (Fig. 3F, Left). In contrast, xenografts derived from A549-K cells responded to cisplatin and combined treatment equally (Fig. 3F, Right). No weight loss was observed in this experiment (Fig. S7A). These results indicate that a reduction in Nrf2 protein levels is necessary for brusatol-mediated sensitization to cisplatin and that brusatol can mimic Keap1 in reducing the protein level of Nrf2, thus sensitizing cancer cells to chemotherapy.
Next, xenografts derived from A549-V and A549-K after 52 and 66 d injection, respectively, were examined for Nrf2 expression by immunoblot analysis. Interestingly, the xenografts derived from A549-K cells still expressed Keap1-CBD and had reduced Nrf2 expression even after 66 d without puromycin selection (Fig. S7B). Lower Nrf2 expression in A549-K–derived xenografts was confirmed by IHC staining with an Nrf2 antibody (Fig. S7C, Top). Furthermore, A549-K–derived xenografts had more TUNEL positive cells than A549-V–derived xenografts in the cisplatin-treated groups (Fig. S7C, Middle), indicating reduced Nrf2 expression renders cells more sensitive to cisplatin. Cotreatment with brusatol increased the number of apoptotic cells only in A549-V–derived xenografts, not in A549-K–derived xenografts, which already had a high level of apoptosis in response to cisplatin alone (Fig. S7C, Middle). Moreover, the combined treatment of brusatol with cisplatin further reduced cell proliferation when compared with cisplatin-treated tumors (Fig. S7C, Bottom). Collectively, these results show that brusatol is able to enhance chemotherapeutic efficacy through specific inhibition of the Nrf2-dependent cellular defense mechanism.
Brusatol Treatment Decreased Glutathione Levels and Increased the Intracellular Concentration of Cisplatin.
It was hypothesized that reduced detoxification due to a decrease in the formation of cisplatin–glutathione conjugates and impaired export of intracellular cisplatin due to limited expression of exporters, such as MRP1 and MRP2, may be the underlying mechanisms for brusatol-mediated sensitization to cisplatin. Indeed, brusatol decreased intracellular glutathione levels in a dose-dependent manner (Fig. S7D). Furthermore, brusatol treatment enhanced the intracellular concentration of cisplatin by a fold of 1.62 (Fig. S7E).
Discussion
Using a rationally targeted approach, we have identified brusatol as a unique inhibitor of the Nrf2 pathway. We have demonstrated that brusatol sensitized cancer cells and xenografts to cisplatin and other chemotherapeutic drugs. Brusatol-mediated sensitization to cancer therapy relies on its ability to suppress the Nrf2 pathway through reduced Nrf2 protein expression. No obvious toxicity was observed at the doses that were sufficient to sensitize cancer cells to chemotherapeutic treatments. Furthermore, the inhibitory effect of brusatol on the Nrf2 pathway is reversible as Nrf2 quickly recovers to basal levels after removal of brusatol. Because brusatol is capable of inhibiting Nrf2 in many cell types, it can be used to enhance the efficacy of a wide variety of chemotherapeutic drugs to treat many types of cancers. Our study demonstrates the potential therapeutic use of brusatol to combat chemoresistance.
A group at the University of North Carolina investigated the use of brusatol as an antitumor agent for leukemia cancer in the 1970s and 80s. This group reported that bruceantin, brucein D, brucein E, bruceoside A, and brusatol inhibited DNA, RNA, and protein synthesis as well as oxidative phosphorylation in p-388 lymphocytic leukemia cells (39, 40). Inhibition of overall protein synthesis was observed in rabbit reticulocytes, as well as in cell lines derived from lymphocytic leukemia, Ehrlich carcinoma, hepatoma, and lymphoid leukemia (41–44). It should be noted that nanomolar concentrations of brusatol were used in our study, whereas micromolar concentrations were used in the in vitro studies conducted by the aforementioned groups (42, 43). In our study, brusatol (20–80 nM) specifically inhibited the protein level of Nrf2 in a dose-dependent manner (Fig. 1D). Despite a moderate reduction of c-Myc, no effect on caspase-3, Stat-3, β-catenin, p65, or IκB-α was observed (Fig. 1F), further demonstrating that brusatol is not a universal protein synthesis inhibitor at the concentration used in our study. The controversial results between our study and theirs are most likely due to the difference in either the concentrations of brusatol or cell types used. Moreover, we show that despite the structural similarity between brucein C and brusatol, brucein C had no effect on the protein level of Nrf2 (Fig. S2B). These results further demonstrate that brusatol-mediated inhibition of the Nrf2 pathway has a high degree of specificity. Between 2001 and 2004, Pezzuto's group found that brusatol was able to induce G1 arrest and terminal differentiation in myeloid leukemia cell lines such as HL-60, K562, Kasumi-1, NB4, U937, and BV173, through inhibition of c-Myc expression (45, 46). We did observe a moderate reduction in c-Myc expression following brusatol treatment (Fig. 1F, Lower); however, we did not detect any significant changes in the number of cells in G1 phase in A549 cells treated with 40 nM of brusatol for 24 h, 48 h, or 72 h (Fig. 2E). Currently, it is not clear whether a reduction in c-Myc expression contributes to the growth inhibition observed in response to brusatol treatment alone in our in vitro and in vivo experiments (Fig. 3 A and D). Nevertheless, a reduction in Nrf2 protein expression certainly reduced tumor growth because A549-K xenografts took much longer than A549-V xenografts to reach 280 mm3 (42 d vs. 28 d) (Fig. 3F). It is also unclear whether inhibition of both Nrf2 and c-Myc by brusatol is independent of each other or whether it is a result of cross-talk between the Nrf2 and c-Myc pathways. In addition, the same group demonstrated activation of the NFκB pathway by brusatol in HL-60 cells undergoing cell differentiation (37). However, we failed to detect any effect of brusatol on the NFκB pathway in A549 cells, as demonstrated by both κB-luciferase activity and the protein level of IκB-α and P-p65 (Fig. 1 E, Lower and F, Upper).
Brusatol inhibits the Nrf2 signaling pathway by reducing the protein level of Nrf2. The reduction in Nrf2 in response to brusatol was observed in all cell lines tested, including A549, HeLa, MDA-MB-231, Ishikawa, and SPEC-2, regardless of the status of Keap1 or Nrf2 being wild type or mutated. A549 cells bear a G-T mutation at amino acid position 333 in the first Kelch domain of Keap1, which gives rise to a constitutively high basal level of Nrf2 (20). Ishikawa and Spec-2 cells were isolated from type I and type II endometrial cancer patients, respectively, and we did not find any mutations in the coding region of Keap1 or Nrf2 in either cell line. However, Spec-2 has higher basal and inducible levels of Nrf2 than Ishikawa (27). MDA-MB-231 has very low basal levels of Nrf2 and the Nrf2-Keap1 pathway is highly inducible. Moreover, no mutation was found in either Nrf2 or Keap1 in HeLa cells (7, 25). It will be interesting to know whether brusatol-mediated enhancement of ubiquitination and degradation is Keap1 dependent. Because the cell lines tested had different statuses of Keap1, brusatol-mediated Nrf2 degradation may be independent of Keap1. However, in A549 cells, despite the point mutation in Keap1, an induction of Nrf2 by sulforaphane was observed. This suggests that the function of Keap1 is not completely disrupted and leads us to believe that brusatol enhances Nrf2 degradation in a Keap1-dependent manner. The detailed mechanism by which brusatol enhances Nrf2 ubiquitination warrants further investigation.
Materials and Methods
See SI Materials and Methods for greater detail.
Animal Treatment.
Athymic nude mice were purchased from Harlan Laboratories. Mice 4–6 wk old were injected with A549 cells. Once the tumors reached 80 mm3 (for the two times five-time cisplatin treatment regimen in Fig. 3D) or 280 mm3 (for the single five-time cisplatin treatment regimen in Fig. 3F and Fig. S6), mice were randomly allocated into four groups and treated i.p. with DMSO, cisplatin (2 mg/kg), brusatol (2 mg/kg), or in combination every other day for a total of five times. In Fig. 3D, after the initial five-time cisplatin treatment regimen, treatment stopped for 1 wk to allow mice to recover before the second five-time cisplatin treatment regimen was repeated.
Reporter Gene Assay, in Vivo Ubiquitination, and Pulse-Chase Analysis.
MDA-MB-231-ARE-Luc cells were used to measure luciferase activity. For the dual luciferase reporter gene assay, A549 cells were transfected with all of the necessary vectors and firefly and renilla luciferase activity was measured using the Promega dual-luciferase reporter gene assay system. In vivo ubiquitination analysis was conducted as reported previously (47). The half-life of Nrf2 was measured by pulse-chase analysis.
Cell Viability Assay, Colony Formation Assay, Apoptotic Cell Death, and Cell Cycle Analysis.
Cell viability was measured by the xCELLigence system (Roche). Colony formation was performed using a standard protocol. An in situ cell death detection kit (Roche) was used for detecting apoptotic cell death in tumor tissue and analyzed under a fluorescence microscope (Zeiss Observer Z1, Marianas digital microscopy workstation). Apoptotic cells in cultured cells were detected using Annexin V-FITC apoptosis detection kit (Sigma) and analyzed by flow cytometry. For cell cycle analysis, 1 × 106 cells were incubated with RNase A and PI before analysis using flow cytometry.
Supplementary Material
Acknowledgments
This study was supported by the following grants: RSG-07-154 (American Cancer Society) and RO1ES015010 (National Institute of Environmental Health Sciences), which were awarded to D.D.Z., and ES006694 (National Institutes of Health), a center grant.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014275108/-/DCSupplemental.
References
- 1.Schiller JH. Current standards of care in small-cell and non-small-cell lung cancer. Oncology. 2001;61(Suppl 1):3–13. doi: 10.1159/000055386. [DOI] [PubMed] [Google Scholar]
- 2.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 3.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 4.Jeong WS, Jun M, Kong AN. Nrf2: a potential molecular target for cancer chemoprevention by natural compounds. Antioxid Redox Signal. 2006;8:99–106. doi: 10.1089/ars.2006.8.99. [DOI] [PubMed] [Google Scholar]
- 5.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes JD, Kelleher MO, Eggleston IM. The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur J Nutr. 2008;47(Suppl 2):73–88. doi: 10.1007/s00394-008-2009-8. [DOI] [PubMed] [Google Scholar]
- 10.Kong AN, et al. Signal transduction events elicited by cancer prevention compounds. Mutat Res. 2001;480-481:231–241. doi: 10.1016/s0027-5107(01)00182-8. [DOI] [PubMed] [Google Scholar]
- 11.Itoh K, Mimura J, Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: A historical overview. Antioxid Redox Signal. 2010;13:1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 12.Dinkova-Kostova AT, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramos-Gomez M, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci USA. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khor TO, et al. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res (Phila) 2008;1:187–191. doi: 10.1158/1940-6207.CAPR-08-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: Permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Hayes JD, McMahon M. The double-edged sword of Nrf2: Subversion of redox homeostasis during the evolution of cancer. Mol Cell. 2006;21:732–734. doi: 10.1016/j.molcel.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Kensler TW, Wakabayashi N. Nrf2: Friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padmanabhan B, et al. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Singh A, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohta T, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 22.Solis LM, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stacy DR, et al. Increased expression of nuclear factor E2 p45-related factor 2 (NRF2) in head and neck squamous cell carcinomas. Head Neck. 2006;28:813–818. doi: 10.1002/hed.20430. [DOI] [PubMed] [Google Scholar]
- 24.Shibata T, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135(4):1358–1368. doi: 10.1053/j.gastro.2008.06.082. 1368.e1–4. [DOI] [PubMed] [Google Scholar]
- 25.Shibata T, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YR, et al. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol. 2010;220:446–451. doi: 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- 27.Jiang T, et al. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res. 2010;70:5486–5496. doi: 10.1158/0008-5472.CAN-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang XJ, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P, et al. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Homma S, et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res. 2009;15:3423–3432. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- 31.Cho JM, Manandhar S, Lee HR, Park HM, Kwak MK. Role of the Nrf2-antioxidant system in cytotoxicity mediated by anticancer cisplatin: Implication to cancer cell resistance. Cancer Lett. 2008;260:96–108. doi: 10.1016/j.canlet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 32.Hong YB, et al. Nuclear factor (erythroid-derived 2)-like 2 regulates drug resistance in pancreatic cancer cells. Pancreas. 2010;39:463–472. doi: 10.1097/MPA.0b013e3181c31314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh A, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shim GS, Manandhar S, Shin DH, Kim TH, Kwak MK. Acquisition of doxorubicin resistance in ovarian carcinoma cells accompanies activation of the NRF2 pathway. Free Radic Biol Med. 2009;47:1619–1631. doi: 10.1016/j.freeradbiomed.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Kim SK, et al. Increased expression of Nrf2/ARE-dependent anti-oxidant proteins in tamoxifen-resistant breast cancer cells. Free Radic Biol Med. 2008;45:537–546. doi: 10.1016/j.freeradbiomed.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 36.Du Y, et al. Oridonin confers protection against arsenic-induced toxicity through activation of the Nrf2-mediated defensive response. Environ Health Perspect. 2008;116:1154–1161. doi: 10.1289/ehp.11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuendet M, Gills JJ, Pezzuto JM. Brusatol-induced HL-60 cell differentiation involves NF-kappaB activation. Cancer Lett. 2004;206:43–50. doi: 10.1016/j.canlet.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 38.Zhang DD, et al. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 39.Hall IH, et al. Antitumor agents. XXXIV: Mechanism of action of bruceoside A and brusatol on nucleic acid metabolism of P-388 lymphocytic leukemia cells. J Pharm Sci. 1979;68:883–887. doi: 10.1002/jps.2600680726. [DOI] [PubMed] [Google Scholar]
- 40.Eigebaly SA, et al. Antitumor agents. XXXV: Effects of brusatol, bruceoside A, and bruceantin on P-388 lymphocytic leukemia cell respiration. J Pharm Sci. 1979;68:887–890. doi: 10.1002/jps.2600680727. [DOI] [PubMed] [Google Scholar]
- 41.Willingham W, Jr, et al. Mechanism of eukaryotic protein synthesis inhibition by brusatol. Biochim Biophys Acta. 1981;654:169–174. doi: 10.1016/0005-2787(81)90168-4. [DOI] [PubMed] [Google Scholar]
- 42.Liou YF, Hall IH, Okano M, Lee KH, Chaney SG. Antitumor agents XLVIII: Structure-activity relationships of quassinoids as in vitro protein synthesis inhibitors of P-388 lymphocytic leukemia tumor cell metabolism. J Pharm Sci. 1982;71:430–435. doi: 10.1002/jps.2600710414. [DOI] [PubMed] [Google Scholar]
- 43.Hall IH, Liou YF, Lee KH, Chaney SG, Willingham W., Jr Antitumor agents LIX: Effects of quassinoids on protein synthesis of a number of murine tumors and normal cells. J Pharm Sci. 1983;72:626–630. doi: 10.1002/jps.2600720612. [DOI] [PubMed] [Google Scholar]
- 44.Lee KH, Okano M, Hall IH, Brent DA, Soltmann B. Antitumor agents XLV: Bisbrusatolyl and brusatolyl esters and related compounds as novel potent antileukemic agents. J Pharm Sci. 1982;71:338–345. doi: 10.1002/jps.2600710320. [DOI] [PubMed] [Google Scholar]
- 45.Mata-Greenwood E, et al. Novel esters of glaucarubolone as inducers of terminal differentiation of promyelocytic HL-60 cells and inhibitors of 7,12-dimethylbenz[a]anthracene-induced preneoplastic lesion formation in mouse mammary organ culture. J Nat Prod. 2001;64:1509–1513. doi: 10.1021/np010212p. [DOI] [PubMed] [Google Scholar]
- 46.Mata-Greenwood E, et al. Brusatol-mediated induction of leukemic cell differentiation and G(1) arrest is associated with down-regulation of c-myc. Leukemia. 2002;16:2275–2284. doi: 10.1038/sj.leu.2402696. [DOI] [PubMed] [Google Scholar]
- 47.Chen W, et al. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.