Abstract
Formation of many dsDNA viruses begins with the assembly of a procapsid, containing scaffolding proteins and a multisubunit portal but lacking DNA, which matures into an infectious virion. This process, conserved among dsDNA viruses such as herpes viruses and bacteriophages, is key to forming infectious virions. Bacteriophage P22 has served as a model system for this study in the past several decades. However, how capsid assembly is initiated, where and how scaffolding proteins bind to coat proteins in the procapsid, and the conformational changes upon capsid maturation still remain elusive. Here, we report Cα backbone models for the P22 procapsid and infectious virion derived from electron cryomicroscopy density maps determined at 3.8- and 4.0-Å resolution, respectively, and the first procapsid structure at subnanometer resolution without imposing symmetry. The procapsid structures show the scaffolding protein interacting electrostatically with the N terminus (N arm) of the coat protein through its C-terminal helix-loop-helix motif, as well as unexpected interactions between 10 scaffolding proteins and the 12-fold portal located at a unique vertex. These suggest a critical role for the scaffolding proteins both in initiating the capsid assembly at the portal vertex and propagating its growth on a T = 7 icosahedral lattice. Comparison of the procapsid and the virion backbone models reveals coordinated and complex conformational changes. These structural observations allow us to propose a more detailed molecular mechanism for the scaffolding-mediated capsid assembly initiation including portal incorporation, release of scaffolding proteins upon DNA packaging, and maturation into infectious virions.
Keywords: cryo-EM, asymmetric reconstruction, P22 phage, asymmetric procapsid, portal protein
DsDNA viruses infecting both prokaryotes and eukaryotes share a common assembly pathway proceeding from a precursor (procapsid) to an infectious virion (1–4). In addition to the coat proteins, the procapsid requires scaffolding proteins, absent from the virion, for proper assembly, and a portal for DNA packaging and subsequent DNA ejection. However, despite a half-century of research on icosahedral viruses, it remains unclear how initially identical subunits adopt both hexameric and pentameric conformations in the virus and select the correct locations needed to form closed shells of the proper size (5). Packaging of DNA through the portal is accompanied by the exit of scaffolding proteins from the procapsid and conformational changes in the coat proteins as the capsid matures (2, 6).
Understanding the molecular mechanisms of dsDNA virus assembly and maturation requires knowledge of the interactions among the coat, scaffolding, and portal proteins, all of which are essential for these processes. X-ray crystallography (7–9) and electron cryomicroscopy (cryo-EM) (10–12) have yielded near-atomic to atomic resolution models of several dsDNA icosahedral viruses and provided a structural framework of interactions among their coat proteins. However, the structural details of procapsid portal incorporation, scaffolding protein binding and release, and capsid shell expansion during maturation remain largely unknown. In particular, the scaffolding-dependent association of the coat protein with the portal ring at a single fivefold capsid vertex has been difficult to characterize. The structures reported here of the P22 virions and procapsids reveal the conformational transition upon maturation, as well as the contacts among the portal, scaffolding subunits, and the coat subunits. These contacts offer a mechanism by which icosahedral shell assembly is initiated and controlled in P22 and possibly in some other dsDNA viruses.
Results and Discussion
Cryo-EM Structure of P22 Procapsid at 3.8-Å Resolution.
The bacteriophage P22 procapsid is a well-characterized morphogenetic precursor of the mature phage, consisting of hundreds of copies of the gene 5 (gp5) coat and gene 8 (gp8) scaffolding proteins, multiple copies of three ejection proteins (gp7, gp16, gp20, also known as pilot proteins), and a unique multisubunit gene 1 (gp1) portal (13). We used single-particle cryo-EM to determine its structure at 3.8-Å resolution, enforcing icosahedral symmetry with approximately 23,400 particle images (Fig. 1A and Fig. S1 A and B). The procapsids were isolated from cells infected with nonsense mutants of the terminase complex defective in DNA packaging and represent the physiological precursor prior to DNA packaging and capsid maturation.
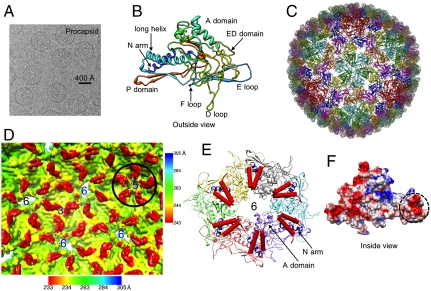
Fig. 1.
The icosahedral structure of the P22 procapsid at 3.8-Å resolution and the interactions between the scaffolding protein and the coat protein. (A) A typical cryo-EM image of the P22 procapsid. (B) A Cα backbone model from residues 10–425 with eight annotated domains for one gp5 subunit derived from the density. The model is colored from blue (N terminus) to red (C terminus). (C) The Cα model for the entire procapsid. (D) The internal surface view of a portion of procapsid density map along an icosahedral threefold axis (labeled “3”), showing the V-shaped densities in red around hexamers (labeled “6”) and pentamers (labeled “5”). The V-shaped densities at pentamers, shown in a black circle, are displayed at a lower contour level than the rest of the map. The density map was low-pass filtered from 3.8 to 7.0 Å, and was radially colored according to the color scheme shown at the bottom except the inset of a pentamer in the black circle using different color scheme shown on the right. (E) Interactions between the coat protein (colored ribbons, N arm colored in blue) and the C-terminal helix-loop-helix motif of the scaffolding protein (red cylinders) in a hexamer. (F) The charge distribution of one procapsid subunit model. The scaffolding protein-binding region (dashed circle) is negatively charged.
From our map, Cα backbone models for each of the seven structurally similar but nonidentical copies of the coat protein (gp5) in the asymmetric unit of the T = 7 procapsid were built with a hybrid de novo modeling method using Gorgon (14) and Rosetta (15) (Fig. 1 B and C and Fig. S2 A, C and E). The coat proteins are organized as pentamers and skewed hexamers as previously reported for the GuHCl treated procapsid (16–20). Each of the gp5 models consists of eight distinct domains: A domain, extra-density (ED) domain, E loop, D loop, F loop, P domain, N arm, and a long helix (Fig. 1B). No significant sequence homology to other proteins is found for the entire coat protein. Part of the gp5 has similar secondary structure elements and a characteristic coat protein fold, which is found in HK97 and other phages (7, 8, 11, 19, 21, 22). However, the fold of the ED domain is different from the previous model based on a lower resolution cryo-EM map (20).
Bacteriophages T4 (23), Lambda (24), and ϵ15 (10) utilize additional coat proteins that bind to the outside of the major coat protein, which stabilizes the virion. In bacteriophage HK97 (7), no such additional proteins are present in the mature capsid. Whereas P22 has only one coat protein, the coat protein contains the ED domain at the capsid surface (Fig. 1B). This domain has a unique fold, composed primarily of β-strands, a short α-helix, and a protruding loop (the “D loop”) (Fig. 1B and Fig. S2 A and C). The D loop is located at the intersubunit interface between neighboring capsomeres, and provides strong interactions across the local twofold axes (Fig. S3A). Out of 21 temperature-sensitive mutations that affect the coat protein monomer folding and/or capsid assembly (25–27), 14 are found in this domain, suggesting it plays a key role in stabilizing the monomeric coat protein and/or the procapsid shell (Fig. S3C). Previous intermediate-resolution studies proposed that this domain stabilizes the coat protein monomer (20). Our model suggests that more extensive interactions in the D loop and ED domain are involved in stabilizing the entire procapsid (see Fig. 4 below) in lieu of the covalent modifications exhibited by HK97 phage or the additional coat proteins in T4, Lambda, and ϵ15 phages.
Scaffolding Protein and Its Interactions with Coat Protein.
V-shaped densities at the hexamers (Fig. 1D), which cannot be accounted for by the coat protein model, are found on the inner surface of the coat protein shell (Movie S1). These densities are not observed in a separate 7.0-Å reconstruction of the scaffolding protein-deficient P22 procapsid (empty procapsid) (Fig. S4A), suggesting that these V-shaped densities are part of the scaffolding proteins. Each V-shaped density agrees in size and shape with the NMR structure of the P22 scaffolding protein C-terminal region (residues 238–303, Protein Data Bank ID 2GP8) consisting of a helix-loop-helix motif (28) (Fig. S4B). Although all of the V-shaped densities within an asymmetric unit are similar, the interface between each scaffolding protein density and the coat protein varies in the skewed hexamer.
Earlier studies suggested that there are two distinct scaffolding binding sites on the coat protein (29, 30). Based on our model, one of the two scaffolding binding sites occurs at the negatively charged residues on the N arm of the coat protein (Fig. 1 E and F), which interact with the positively charged C-terminal domain of the scaffolding protein (28). These electrostatic interactions are consistent with previous biochemical studies (31, 32). Note that the interactions between the scaffolding protein and coat protein have never been visualized in such fine detail and modeled in any authentic bacteriophage procapsid, although previous attempts at lower resolutions were made in herpes virus capsid (33) and P22 (17).
The second scaffolding protein binding site is located at the tip of the A domain of the coat protein, which is bent toward the interior of the procapsid shell (Fig. 1E and Fig. S4B). In four (S1, S2, S4, S5) of the six positions at each skewed hexamer, the scaffolding protein spans two coat proteins, binding to the N arm of one coat protein and the A-domain tip of an adjacent coat protein. In the other two positions (S3, S6) at each hexamer, the scaffolding protein does not contact either its own or the adjacent coat protein’s A-domain tip; the scaffolding protein is only bound to one coat protein at its N arm. At the pentamers, the scaffolding density appears considerably weaker with the base of the V shape of the scaffolding protein situated below the A-domain tips of two adjacent coat proteins (Fig. 1D).
Based on our cryo-EM structures of P22 procapsids, which were averaged from approximately 23,400 particle images, every coat protein is associated with one scaffolding protein. As the scaffolding protein densities at the pentamers appear at a lower isosurface value, it may be possible that there is only partial occupancy of scaffolding protein at the pentamers. Regardless, this number of scaffolding proteins is likely higher than 250–300 copies as previously deduced from biochemical data (34, 35).
Interactions Between Scaffolding Proteins and the Unique Portal in Asymmetric Procapsid Structure.
To investigate the interactions between the scaffolding proteins and the portal vertex, the procapsid was reconstructed to 8.7-Å resolution without imposing icosahedral symmetry (Fig. 2 A–C, Fig. S5, and Movie S2). This map shows the density features and locations of the coat protein and the scaffolding proteins to be the same as those in the icosahedral map (Fig. 1D), thus validating the proposed interaction sites between the scaffolding and coat proteins (Fig. 1E). The map also reveals a unique portal at one of the twelve vertices, as well as its interactions with the capsid shell. Similar to the biochemically isolated portal (36, 37), the in situ portal of the procapsid has 12-fold symmetry and can be divided into the clip, stem, wing, and crown domains (36, 38) (Fig. S5C). An extra barrel-shaped domain extending next to the crown domain in the portal toward the interior of the capsid has not been previously reported (Fig. S5C), and may correspond to the C-terminal portion of the portal protein as in P-SSP7 phage (39). The clip domain is loosely connected to the coat proteins by the P domains and F loops from the surrounding five hexamers (Fig. S5 B–F).
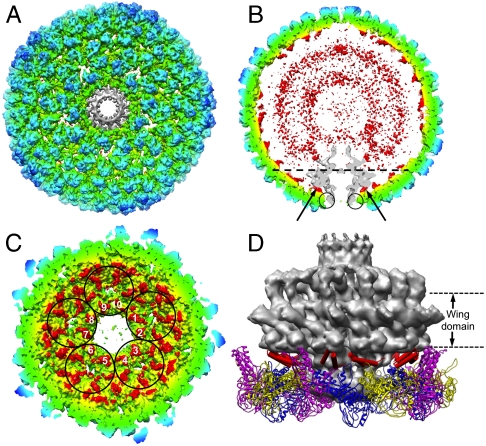
Fig. 2.
Asymmetric reconstruction of the P22 procapsid. (A) The asymmetric procapsid map at 8.7-Å resolution. (B) A central slice of the asymmetric reconstruction. The inner two disordered concentric shells (red) are composed mainly of scaffolding proteins. The portal (gray) interacts with the scaffolding proteins at positions pointed by black arrows and with the coat proteins at the positions marked with circles. In (A), the portal density (gray) was segmented out and 12-fold averaged. In both A and B, the density map was color-coded using the same scheme as in Fig. 1D, except the portal is in gray. (C) The C termini of 10 scaffolding proteins are labeled from 1 to 10, from five hexamers (circled) surrounding the portal vertex. The view direction is at the level of, and normal to, the dashed line in B and the portal has been removed for clarity. (D) Side view of the interaction model among the 10 scaffolding protein C termini (red cylinders), coat proteins (ribbons), and the 12-fold averaged portal density (gray). A difference map between the procapsid asymmetric structure and its icosahedrally averaged structure showed no significant difference for the bound scaffolding proteins and capsid shell; the only difference was at the portal. The red cylinders were fit into the scaffolding protein densities of icosahedrally averaged structure.
As in the icosahedral reconstruction, the C termini of the scaffolding proteins are also observed on the inner surface of capsid shell in the asymmetric map (Fig. 2C). Again, their densities are nonequivalent in the skewed hexamer, truly representing unique interactions between the coat and scaffolding proteins as shown above (Fig. 1 D and E). Unexpectedly, a set of scaffolding subunits is interposed between and interacts with the portal and coat subunits (Fig. 2 C and D). The interaction of the scaffolding protein with the portal is actually much more extensive than that of the coat protein with the portal, because the “loop” in the helix-loop-helix motif of the scaffolding protein is in close contact with the wing domain of the portal (Fig. 2D and Movie S2). The presence of the scaffolding proteins here explains why the portal is not incorporated into procapsids in the absence of scaffolding proteins (40) and the mutation (G287E) in the loop, proximal to the portal, of the helix-loop-helix motif in the C-terminal portion of scaffolding protein influences the portal incorporation into procapsids (41).
Our asymmetric map shows that the C termini of 10 scaffolding proteins (Fig. 2C) interact with the portal in P22, which is different from a recent model for ϕ29 phage (42). Nonidentical interactions between the wing domain of the portal and the surrounding scaffolding proteins may be a structural mechanism to accommodate the symmetry mismatch between the 12 portal proteins and the 10 scaffolding proteins at the portal vertex. It should be emphasized that the 10 numbered scaffolding proteins (Fig. 2C) are not associated with a pentamer (there is no pentamer at this vertex). Rather, each of the five surrounding hexamers contributes two of its associated scaffolding proteins to the interaction with the portal. Each of these two scaffolding proteins has different orientations with respect to the portal, in addition to the symmetry mismatch mentioned above (Movie S2).
In contrast to the well-ordered C termini bound to the coat proteins, the rest of the scaffolding proteins appear as two disordered concentric shells in the icosahedral and asymmetric (Fig. 2B) reconstructions. These rings presumably arise from the hundreds of elongated scaffolding subunits with their 10∶1 axial ratio (43, 44). The inability to visualize the conformations for the rest of the scaffolding proteins suggests that the N-terminal regions of the scaffolding subunits are not ordered or vary from particle to particle. This region of the scaffolding protein may function to exclude cytoplasmic proteins from entering the growing shell, in addition to interacting with the ejection and portal proteins and self-associating during procapsid assembly.
Virion Structure at 4-Å Resolution and Conformational Changes upon Capsid Maturation.
In contrast to the spherical procapsid, the P22 virion appears more angular and has the characteristic DNA fingerprint (Fig. 3A). A 4.0-Å icosahedral reconstruction was obtained from approximately 18,300 virion particle images (Fig. S1 C and D). The skewed hexamers in the procapsid become more symmetric in the virion, with smaller openings at the center of the hexamers (Fig. 1C, Fig. 3B, and Fig. S3 A and B). These changes are coordinated with the increase in diameter from ∼610 to ∼710 Å and thinning of the shell wall in the virion (Fig. S1 A and C and Movie S3). Cα backbone models were again built for the seven coat proteins using our hybrid de novo method (Fig. 3B and Fig. S2 B, D, and F). Our models of the procapsid and virion differ from earlier models based on lower resolution maps of empty and expanded procapsids (20) and reveal structural changes not previously seen.
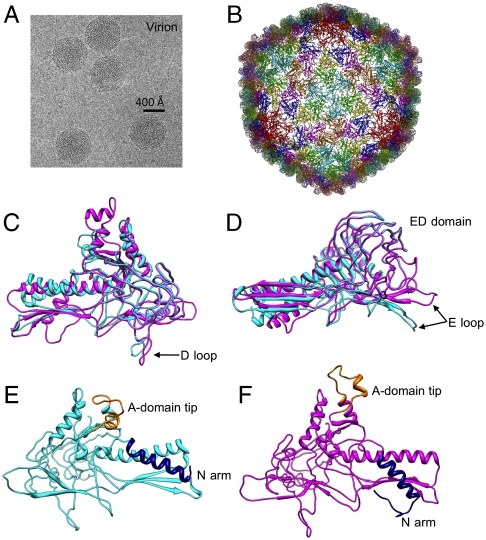
Fig. 3.
The icosahedral structure of the P22 virion at 4.0-Å resolution and conformational changes upon maturation. (A) A typical cryo-EM image of P22 virion. (B) The Cα backbone model of gp5 (residues 1–425) for the virion. Outside (C) and tangential (D) views of a subunit model from the virion (magenta) and procapsid (cyan) superimposed, showing conformational changes at the long helix, D loop, and E loop. An interior view of a procapsid subunit (E) and of a virion subunit (F). The movement of A-domain tip (orange) and the N arm (navy) are apparent in the virus maturation.
Despite having similar folds, conformational differences are observed in the various domains of the coat protein in the procapsid and virion (Fig. 3 C and D and Movie S3). A set of conformational changes in the coat protein occurs in the two previously mentioned regions that interact with the scaffolding protein. The A-domain tip (Fig. 3 E and F) flexes toward the centers of the capsomeres during maturation, narrowing the capsomere openings and resulting in a more symmetric hexamer (Fig. S3 A and B). The N arm, another site for scaffolding protein binding, is rotated approximately 20° clockwise in the virion, placing it below the β-sheet region of the P domain (Fig. 3 E and F). In addition, the long helix is “unkinked.”
Conformational changes in regions of the coat protein not directly interacting with the scaffolding protein also occur during capsid expansion and maturation. In the procapsid, the tip of the E loop (G60–A67) interacts with the tip of a P domain (G396–S402) of a coat protein in an adjacent asymmetric unit across the icosahedral threefold axes via a salt bridge between K66 and D397. In the virion, the E loop is shifted approximately 20° outward (Fig. 3D), resting on the large β-sheet region of the P domain in a neighboring subunit within the same capsomere (Fig. S3B), while the tips of the three P domains remain in the same position at the threefold axes.
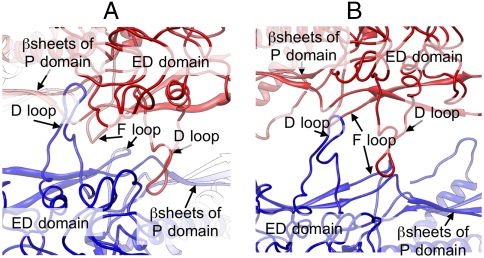
In the procapsid, several charged residues at the tip of the D loop extend into a pocket of complementary charges formed by part of the ED domain and β-sheet portion of the P domain of the neighboring coat protein subunit in an adjacent capsomere along the local twofold symmetry axes (Fig. 4A). Seven temperature-sensitive mutations that affect coat protein monomer folding and/or capsid assembly (25–27) are located within this binding pocket or the loop itself (Fig. S3C). Along with the adjacent F loops, these interactions stabilize neighboring capsomeres in the procapsid (Fig. 4A). In the virion, these interactions do not exist; rather the D loops are shifted out of their pockets in the adjacent asymmetric unit. To maintain stability in the virion, this shift of the D loops forms new electrostatic interactions between the two D loops of the subunits across the capsomeres (Fig. 4B). Thus, intercapsomere interfaces in the procapsid are composed of the D and E loops interacting with the P domains as well as the F loops, whereas only interactions between the D loops seem to be involved in intercapsomere interfaces in the virion.
Fig. 4.
Structural difference at the capsomere interface. (A and B) Zoom-in view of local twofold interactions in the procapsid (A) and the virion (B) models.
During capsid expansion, intracapsomere interactions compensate for the loss of intercapsomere interactions in the virion (Fig. S3 A and B). Helix 2 and helix 3 in the A domains (Fig. S2 C and D) of neighboring subunits form strong lateral electrostatic interactions that vary considerably between the procapsid and virion. In the procapsid, the interacting surface area at this interface is similar for the pentameric subunits and four of the six hexameric subunits (labeled as C1, C2, C4, and C5 in Fig. S3A). In the other two hexameric procapsid subunits (labeled as C3 and C6 in Fig. S3A), there is a marked reduction (∼10%) in total interaction surface area at this interface. Helix 2 and helix 3 in these two subunits, due to the skewing, are shifted such that the electrostatic interactions between the neighboring subunits are out-of-register and do not participate in any interactions. As the skewed hexamers become more equivalent in the virion, these two helices move into register in all hexameric positions. The interacting surface area is nearly identical at all hexameric and pentameric positions, forming the core intracapsomere interactions. Along with the E loop/P domain interactions in the virion (Fig. S3 A and B), these interactions likely account for the increased stability of the virion but, at the same time, still accommodate the skewing of the procapsid hexamers.
Molecular Mechanism for Capsid Assembly, Scaffolding Protein Release, and Capsid Maturation.
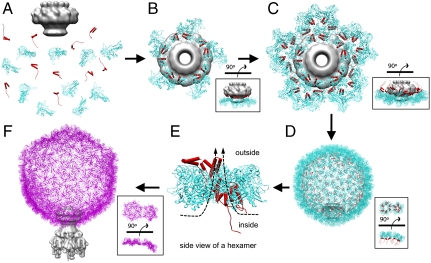
Based on our cryo-EM structures and their associated models, we can propose a detailed molecular mechanism for P22 procapsid assembly, including portal incorporation, scaffolding protein release, and capsid maturation (Fig. 5). Beyond P22, this model also may help elucidate the conformational switching involved in forming hexamers and pentamers, as well as the cascade of events associated with packaging the viral genome in icosahedral viruses.
Fig. 5.
Pathway for capsid assembly, scaffolding protein release and capsid maturation. (A and B) The portal (gray) associates with scaffolding (red) and coat (cyan) proteins to initiate procapsid assembly. The assembly continues with the addition of scaffolding and coat subunits to the growing shell (C) until the full procapsid is assembled (D). DNA is then packaged into the procapsid shell through the channel of the portal by the terminase motor. The scaffolding proteins are released by the electrostatic forces from DNA being packaged and exit through the central large openings of hexamers (E). During the release of scaffolding proteins, conformational changes associated with the maturation transition occur. After capsid expansion and DNA packaging, the tail hub, needle, and tail spikes are attached to the portal to form an infectious virion (magenta) (F). In B and C, the insets show the side views. In D and F, the insets show one hexamer and the adjacent pentamer rotated from the side view to the end-on view.
To form a functional P22 procapsid, at least four types of proteins are required: coat, scaffolding, ejection, and portal proteins, in which the portal proteins form a unique 12-fold portal complex at a fivefold vertex. The lack of scaffolding proteins results in the failure to incorporate the portal and can lead to incomplete particles (29, 32, 40, 45). Based on our density maps, we propose that the formation of a unique portal complex with the requisite scaffolding and coat proteins is likely the key for initiating proper procapsid assembly (Fig. 5 A and B). Scaffolding proteins may play a critical role during the capsid assembly nucleation because the portal would not be incorporated into the procapsid when the scaffolding proteins are absent. Note that the scaffolding protein in bacteriophage HK97 is a domain at the N terminus of the coat protein, and hexamers and pentamers are present in solution in vitro (46). In contrast, in bacteriophage P22 the scaffolding protein is a separate protein from the coat protein and no evidence exists for preformed hexamers or pentamers in solution (43, 44, 47, 48), suggesting that capsomeres form during P22 procapsid assembly. Once nucleated, procapsid assembly proceeds by the addition of scaffolding and coat subunits to the growing shell until the full procapsid is assembled with the proper size and shape as directed by the scaffolding proteins (Fig. 5 C and D). Coat proteins are likely added as monomers or dimers, with the mediation of scaffolding proteins, because our structures show that each scaffolding protein’s C-terminal helix-loop-helix motif interacts with the N arm of the corresponding coat protein. Though the exact timing is not known, ejection proteins are also incorporated and possibly interact with the scaffolding and portal proteins before procapsid assembly is completed (29, 49). The ejection proteins are likely located close to the portal but remain to be identified in our asymmetric procapsid map.
The control of conformational switching involved in forming hexamers and pentamers at the correct locations in icosahedral viruses remains poorly defined. In P22, the fact that four of the scaffolding subunits bind to two different coat protein subunits in a skewed hexamer (Fig. S4B) may explain how scaffolding proteins control the ratio between hexamers and pentamers during icosahedral shell assembly. In the absence of scaffolding proteins, the coat proteins sometimes polymerize into smaller T = 4 shells (45). Both the T = 4 and T = 7 particles have 12 pentamers, though the T = 4 particles have 30 skewed hexamers compared to 60 in T = 7 particles. In the presence of both proteins, the binding of scaffolding proteins to coat proteins shifts the equilibrium towards hexamers, generating more authentic T = 7 particles. Presumably, the skewing of the hexamers in the procapsid is also a result of those two subunits being tilted out of the plane of the capsomere (labeled as C3 and C6 in Fig. S3A). This observation, along with the differences in the coat-scaffolding protein interactions in a procapsid asymmetric unit, may dictate the capsid geometry and the interfaces between capsomeres. Although scaffolding proteins mediate the assembly of the procapsid shell, they are not required for maintaining the capsid shell, as evident in our empty procapsid structure (Fig. S4A) and the T = 4 capsid structure (45), both of which show the skewed conformation for hexamers.
Once the capsid shell forms (Fig. 5D), DNA packaging can begin. In the early stages of DNA packaging, the terminase complex (gp2 and gp3) docks against the portal and hydrolyzes ATP to drive DNA into the procapsid shell through the portal. Upon entry into the procapsid shell via the portal channel, DNA then encounters the scaffolding proteins, which have negatively charged residues at their N termini (35). We propose that, as more negatively charged DNA enters, the scaffolding subunits are expelled out of the shell through the large central openings (∼18 × 42 Å) of the skewed hexamers (Fig. 5E) (the openings at pentamers in the procapsid seem to be closed because they are similar in size to the ones in pentamers and hexamers of the virion) and the capsid is triggered to expand by ∼100 Å. The fold of the coat subunit remains basically unchanged, though numerous structural changes, notably the unkinking of the long helix, the N arm rotation, the flexing of the A-domain tip, and movements of D loop and E loop, occur (Fig. 3 C–F). Capsid expansion does not appear to be directed by the scaffolding protein, as the icosahedral structures of the procapsid shell with and without scaffolding protein bound appear to be virtually identical (Fig. S4A). Rather, the coat protein itself may exist in several metastable states (including those found in the procapsid and virion) and, depending on the state of the capsid shell, may be able to switch conformations to best accommodate local interactions necessary for maintaining the integrity of the capsid. The forces behind maturation, i.e., the dsDNA packaging which results in the release of the scaffolding proteins, likely drives the coat protein into alternative contacts with neighboring coat proteins and necessitates small shifts in the coat protein structure. In the P22 procapsid, the portal may also be involved in controlling capsid expansion, as seen in phage T4 procapsids (50). In our procapsid asymmetric structure, the scaffolding proteins were observed to extensively connect to the portal. Once the scaffolding proteins are expelled by the packaging DNA, the removal of these connections between the portal and the scaffolding proteins could possibly trigger the rearrangement of coat proteins during DNA packaging. To accommodate such rearrangements while still encapsulating the viral genome and providing a stable structure, secondary coat proteins and/or additional domains are required to maintain capsid stability and potentially propagate structural transitions as suggested previously (20, 36, 37). Only after the capsid has fully expanded and packaged its DNA, the tail hub proteins (gene 4 and gene 10), along with the needle (gene 26) and tail spikes (gene 9) are attached to form an infectious virion (Fig. 5F).
In short, our cryo-EM studies of P22, in two morphogenetic states, provide structural details and support a more developed and detailed mechanism for capsid nucleation, scaffolding-mediated assembly, scaffolding protein release, and DNA-triggered maturation. As there is conservation among dsDNA viruses, a similar mechanism is likely to be found in other dsDNA viruses, including those that infect animals, such as adenoviruses and herpesviruses.
Materials and Methods
P22 procapsids and infectious virions were purified from Salmonella typhimurium using an established procedure (47, 51). The samples were applied to Quantifoil grids, flash-frozen using a Vitrobot and imaged. Data for the icosahedral and asymmetric reconstructions were collected on photographic film on a 300-kV JEM-3000SFF with a specimen temperature of ∼4 K. Additional images for the asymmetric procapsid reconstruction were captured on a Gatan 10 × 10 k CCD (US10000XP, model 990) with 2× binning on a 300-kV JEM-3200FSC with energy slit of 20 eV in the in-column energy filter. Data processing for the icosahedral procapsid and virion structures from approximately 23,400 and 18,300 particle images, respectively, was performed using EMAN (52) with newly implemented programs for global optimization and defocus compensation. The asymmetric procapsid reconstruction was obtained with approximately 43,850 particle images as previously described (39). Resolutions for the icosahedral structures were assessed using the 0.5 criterion of the Fourier shell correlation (53) from two independent half datasets. The resolution for the asymmetric procapsid reconstruction was assessed based on the comparison with the 3.8-Å procapsid icosahedral reconstruction. Segmentation of the coat protein subunits was done using Chimera (54). De novo Cα backbone modeling for each of the seven subunits in an asymmetric units in both procapsid and virion was performed using a hybrid of modeling tools and cryo-EM constrained refinement available in Gorgon (14) and Rosetta (15, 55), respectively. Iterative model refinement in Coot (56) and Gorgon was used to optimize the models and remove clashes of subunits within and across capsomeres.
Supplementary Material
Acknowledgments.
We thank Xiangan Liu for his advice on the asymmetric reconstruction and Juan Chang and Frazer Rixon for discussions and assistance in the manuscript preparation. This research was supported by the National Institutes of Health (R01AI0175208, P41RR002250, R01GM079429, and PN2EY016525), the National Science Foundation (NSF IIS-0705644 and IIS-0705474), and The Robert Welch Foundation (Q1242). This work was supported in part by the access to the computing resources from Rice University Computational Research Cluster supported by NSF (CNS-0421109), National Energy Research Scientific Center supported by the Department of Energy (DE-AC02-05CH11232), and the Texas Advanced Computing Center at the University of Texas at Austin (MCB100112).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The icosahedral cryo-EM density maps of bacteriophage P22 procapsid, empty procapsid, and virion have been deposited in the Electron Microscopy data bank, European Bioinformatics Institute, http://www.ebi.ac.uk/ (accession nos. EMD-1824, EMD-1825, and EMD-1826, respectively). The asymmetric density map of bacteriophage P22 procapsid and the 12-fold averaged portal have been deposited in the Electron Microscopy data bank (accession nos. EMD-1827 and EMD-1828, respectively). The Cα models for the asymmetric units of the bacteriophage P22 procapsid and virion have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2XYY and 2XYZ, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015739108/-/DCSupplemental.
References
- 1.Earnshaw WC, Hendrix RW, King J. Structural studies of bacteriophage lambda heads and proheads by small angle X-ray diffraction. J Mol Biol. 1979;134:575–594. doi: 10.1016/0022-2836(79)90368-1. [DOI] [PubMed] [Google Scholar]
- 2.King J, Chiu W. The procapsid-to-capsid transition in double-stranded DNA bacteriophages. In: Chiu W, Burnett RM, Garcea RL, editors. Structural Biology of Viruses. New York: Oxford University Press; 1997. pp. 288–311. [Google Scholar]
- 3.Heymann JB, et al. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- 4.Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 6.Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 7.Wikoff WR, et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 8.Gertsman I, et al. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. Crystal structure of human adenovirus at 3.5 A resolution. Science. 2010;329:1071–1075. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang W, et al. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, et al. Cryo-EM structures of a marine podovirus infecting Prochlorococcus elucidate the viral DNA release mechanism. Nat Struct Mol Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King J, et al. Structure and assembly of the capsid of bacteriophage P22. Philos Trans R Soc, B. 1976;276:37–49. doi: 10.1098/rstb.1976.0096. [DOI] [PubMed] [Google Scholar]
- 14.Baker ML, Zhang J, Ludtke SJ, Chiu W. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nat Protoc. 2010;5:1697–1708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 16.Prasad BV, et al. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- 17.Thuman-Commike PA, et al. Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids. Biophys J. 1999;76:3267–3277. doi: 10.1016/S0006-3495(99)77479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, et al. Visualization of the maturation transition in bacteriophage P22 by electron cryomicroscopy. J Mol Biol. 2000;297:615–626. doi: 10.1006/jmbi.2000.3601. [DOI] [PubMed] [Google Scholar]
- 19.Jiang W, et al. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- 20.Parent KN, et al. P22 coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical crosslinks. Structure. 2010;18:390–401. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morais MC, et al. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Fokine A, et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci USA. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwasaki K, et al. Molecular architecture of bacteriophage T4 capsid: Vertex structure and bimodal binding of the stabilizing accessory protein, Soc. Virology. 2000;271:321–333. doi: 10.1006/viro.2000.0321. [DOI] [PubMed] [Google Scholar]
- 24.Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage lambda capsids. J Mol Biol. 1993;233:682–694. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- 25.Gordon CL, King J. Temperature-sensitive mutations in the phage P22 coat protein which interfere with polypeptide chain folding. J Biol Chem. 1993;268:9358–9368. [PubMed] [Google Scholar]
- 26.Teschke CM, King J. In vitro folding of phage P22 coat protein with amino acid substitutions that confer in vivo temperature sensitivity. Biochemistry. 1995;34:6815–6826. doi: 10.1021/bi00020a028. [DOI] [PubMed] [Google Scholar]
- 27.Teschke CM, Parent KN. ‘Let the phage do the work’: Using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology. 2010;401:119–130. doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, et al. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J Mol Biol. 2000;297:1195–1202. doi: 10.1006/jmbi.2000.3620. [DOI] [PubMed] [Google Scholar]
- 29.Greene B, King J. Binding of scaffolding subunits within the P22 procapsid lattice. Virology. 1994;205:188–197. doi: 10.1006/viro.1994.1634. [DOI] [PubMed] [Google Scholar]
- 30.Parker MH, Brouillette CG, Prevelige PE., Jr Kinetic and calorimetric evidence for two distinct scaffolding protein binding populations within the bacteriophage P22 procapsid. Biochemistry. 2001;40:8962–8970. doi: 10.1021/bi0026167. [DOI] [PubMed] [Google Scholar]
- 31.Parker MH, Casjens S, Prevelige PE., Jr Functional domains of bacteriophage P22 scaffolding protein. J Mol Biol. 1998;281:69–79. doi: 10.1006/jmbi.1998.1917. [DOI] [PubMed] [Google Scholar]
- 32.Weigele PR, Sampson L, Winn-Stapley D, Casjens SR. Molecular genetics of bacteriophage P22 scaffolding protein’s functional domains. J Mol Biol. 2005;348:831–844. doi: 10.1016/j.jmb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Zhou ZH, et al. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc Natl Acad Sci USA. 1998;95:2778–2783. doi: 10.1073/pnas.95.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King J, Casjens S. Catalytic head assembling protein in virus morphogenesis. Nature. 1974;251:112–119. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- 35.Eppler K, Wyckoff E, Goates J, Parr R, Casjens S. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology. 1991;183:519–538. doi: 10.1016/0042-6822(91)90981-g. [DOI] [PubMed] [Google Scholar]
- 36.Lander GC, et al. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure. 2009;17:789–799. doi: 10.1016/j.str.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng H, et al. A conformational switch in bacteriophage p22 portal protein primes genome injection. Mol Cell. 2008;29:376–383. doi: 10.1016/j.molcel.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lebedev AA, et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Rochat RH, Chiu W. Reconstructing Cyano-bacteriophage P-SSP7 structure without imposing symmetry. Protoc Exch. 2010 10.1038/nprot.2010.96. [Google Scholar]
- 40.Earnshaw W, King J. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J Mol Biol. 1978;126:721–747. doi: 10.1016/0022-2836(78)90017-7. [DOI] [PubMed] [Google Scholar]
- 41.Moore SD, Prevelige PE., Jr A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein. J Virol. 2002;76:10245–10255. doi: 10.1128/JVI.76.20.10245-10255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu CY, et al. A docking model based on mass spectrometric and biochemical data describes phage packaging motor incorporation. Mol Cell Proteomics. 2010;9:1764–1773. doi: 10.1074/mcp.M900625-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuller MT, King J. Assembly in vitro of bacteriophage P22 procapsids from purified coat and scaffolding subunits. J Mol Biol. 1982;156:633–665. doi: 10.1016/0022-2836(82)90270-4. [DOI] [PubMed] [Google Scholar]
- 44.Parker MH, Stafford WF, 3rd, Prevelige PE., Jr Bacteriophage P22 scaffolding protein forms oligomers in solution. J Mol Biol. 1997;268:655–665. doi: 10.1006/jmbi.1997.0995. [DOI] [PubMed] [Google Scholar]
- 45.Thuman-Commike PA, Greene B, Malinski JA, King J, Chiu W. Role of the scaffolding protein in P22 procapsid size determination suggested by T = 4 and T = 7 procapsid structures. Biophys J . 1998;74:559–568. doi: 10.1016/S0006-3495(98)77814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie Z, Hendrix RW. Assembly in vitro of bacteriophage HK97 proheads. J Mol Biol. 1995;253:74–85. doi: 10.1006/jmbi.1995.0537. [DOI] [PubMed] [Google Scholar]
- 47.Fuller MT, King J. Purification of the coat and scaffolding proteins from procapsids of bacteriophage P22. Virology. 1981;112:529–547. doi: 10.1016/0042-6822(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 48.Prevelige PE, Jr, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas D, Prevelige P., Jr A pilot protein participates in the initiation of P22 procapsid assembly. Virology. 1991;182:673–681. doi: 10.1016/0042-6822(91)90608-e. [DOI] [PubMed] [Google Scholar]
- 50.Ray K, Oram M, Ma J, Black LW. Portal control of viral prohead expansion and DNA packaging. Virology. 2009;391:44–50. doi: 10.1016/j.virol.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prevelige PE, Jr, Thomas D, King J. Scaffolding protein regulates the polymerization of P22 coat subunits into icosahedral shells in vitro. J Mol Biol. 1988;202:743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- 52.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 53.Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- 54.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.