Abstract
Duplicated genes provide an important raw material for adaptive evolution. However, the relationship between gene duplication and the emergence of new biochemical functions is complicated, and it has been difficult to quantify the likelihood of evolving novelty in any systematic manner. Here, we describe a comprehensive search for artificially amplified genes that are able to impart new phenotypes on Escherichia coli, provided their expression is up-regulated. We used a high-throughput, library-on-library strategy to screen for resistance to antibiotics and toxins. Cells containing a complete E. coli ORF library were exposed to 237 toxin-containing environments. From 86 of these environments, we identified a total of 115 cases where overexpressed ORFs imparted improved growth. Of the overexpressed ORFs that we tested, most conferred small but reproducible increases in minimum inhibitory concentration (≤16-fold) for their corresponding antibiotics. In many cases, proteins were acting promiscuously to impart resistance. In the absence of toxins, most strains bore no fitness cost associated with ORF overexpression. Our results show that even the genome of a nonpathogenic bacterium harbors a substantial reservoir of resistance genes, which can be readily accessed through overexpression mutations. During the growth of a population under selection, these mutations are most likely to be gene amplifications. Therefore, our work provides validation and biochemical insight into the innovation, amplification, and divergence model of gene evolution under continuous selection [Bergthorsson U, Andersson DI, Roth JR (2007) Proc Natl Acad Sci USA 104:17004–17009], and also illustrates the high frequency at which novel traits can evolve in bacterial populations.
Keywords: antibiotic resistance, evolutionary innovation, molecular evolution, protein promiscuity, phenotype microarray
All species must adapt to survive in changing environments. At the molecular level, the presence of novel nutrients and toxins can drive the evolution of proteins that recognize or metabolize them (1, 2). Duplicated copies of preexisting genes provide the primary genetic source for functional innovation (3, 4). However, the relationship between duplication and the emergence of new biochemical functions is complicated because gene duplications are generally expected to be either selectively neutral (5) or deleterious (6). Therefore, it has been difficult to systematically assess the likelihood of evolving novel traits. In the present study, we have sought to address this gap in the Darwinian paradigm.
Numerous hypotheses have been proposed to explain the fates of duplicated genes within populations (7–9). A particularly appealing model identifies a mechanism by which selection can act continuously to favor both an increase in gene dosage, and divergence of one copy from the parental gene. This model, termed adaptive radiation (10) or the innovation, amplification, and divergence (IAD) model (11), is rooted in the notion that many existing proteins display broad specificity and secondary activities in addition to the function that they evolved to carry out (12). In primordial times, this “promiscuity” would have allowed many metabolic functions to be carried out by a minimal number of multitasking proteins (13). In more specialized, modern-day proteins, promiscuity is likely to be a product of contingency: active sites typically contain a variety of reactive groups (proton donors and acceptors, metal ions, and so forth) that could (by chance) play roles in secondary reactions (14). These secondary activities are assumed to provide the necessary reservoir of selectable functions.
In the IAD model, selection is imposed on a weak, promiscuous activity. To increase fitness, duplications (and higher-order amplifications) of the promiscuous gene are selected and maintained in the population. In turn, this increase in copy number improves the likelihood of point mutations that enhance the promiscuous activity, by increasing the number of mutational targets yet still allowing at least one copy to retain the parental activity (11).
Two central tenets of the IAD model are: (i) that gene amplification events are common; and (ii) that promiscuous activities are widespread, as well as being biochemically and mechanistically diverse. Experiments with unselected bacterial populations have shown the first point to be accurate (15), with cells that bear duplications at any given locus reaching steady-state frequencies of ∼10−3 (16). We are interested in cataloguing the promiscuous activities within an entire proteome to understand the protein biochemistry that underlies the evolution of new phenotypes.
Multicopy suppression experiments are a powerful and tractable way of demonstrating that adaptation can begin with the increased expression of promiscuous proteins. In these experiments, conditionally auxotrophic bacterial strains are transformed with plasmid-encoded libraries of ORFs. The libraries are screened for genes that can suppress the phenotype of the original mutation, provided they are present in high copy number (17–19) or overexpressed (20, 21). Previously, we used the multicopy suppression approach in an attempt to assess the prevalence of promiscuity, on a proteome-wide scale. We made use of the ASKA (A Complete Set of Escherichia coli K-12 ORF Archive) library, which comprises every E. coli ORF cloned into the expression vector pCA24N (22). The plasmids of the library were pooled and used to transform 104 conditionally auxotrophic, single-gene deletion strains. Twenty-one of the auxotrophies were specifically suppressed by the overexpression of noncognate E. coli genes (23). The implication was that many proteins are multifunctional; however, the ability of proteome-wide promiscuity to drive an organism's adaptation to new environments remained unknown. The next logical step was to survey the entire E. coli proteome for its latent ability to confer genuinely new phenotypes (rather than to recreate old ones).
Here, we describe a comprehensive search for promiscuous proteins that can impart new phenotypes on E. coli. We provide experimental evidence that the overexpression of preexisting E. coli proteins can provide resistance to >80 antibiotics and toxins. Our results suggest that the evolution of novel traits is surprisingly likely, and that even the genomes of well-characterized bacteria harbor substantial reservoirs of latent resistance determinants.
Results
Library-on-Library Screen.
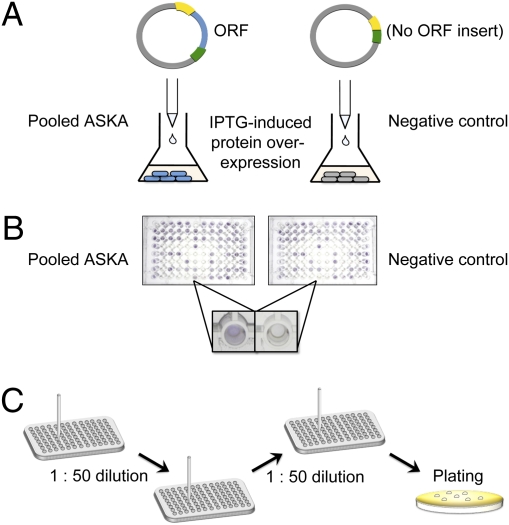
We have conducted a global survey to uncover latent resistance genes in the E. coli genome. E. coli cells were transformed with the pooled plasmids of the ASKA ORF library and then used to inoculate every well of 10 phenotype microarray (PM) plates (Fig. 1 A and B). In total, these 10 PM plates contained 237 toxins (including antibacterial agents), with each toxin present at four concentrations (24). Growth in each toxin-containing well could be monitored by purple color development because of the presence of a tetrazolium indicator dye (25). A negative control (E. coli harboring the empty ASKA vector, pCA24N-NoIns) was used to inoculate a second set of PM plates in parallel. By comparing the growth rates of the ASKA pool and the control, we were able to screen for examples in which the isopropyl-β-d-thiogalactopyranoside (IPTG)-induced overexpression of ASKA-encoded genes conferred increased fitness (i.e., an improved capacity for survival and reproduction) in the presence of a toxin.
Fig. 1.
The library-on-library screen and subsequent isolation of ASKA-encoded resistance genes. (A) E. coli cells harboring the pooled plasmids of the ASKA ORF collection (blue) and the negative control clone (gray) were grown to midlog phase in parallel, and protein overexpression was induced by the addition of IPTG (50 μM). (B) Both cultures were used to inoculate every well of two sets of PM plates. A well was scored positive when the ASKA pool out-grew the negative control, as shown by more rapid tetrazolium color development (Inset). (C) For each positive PM well, the fittest ASKA clone (or clones) was isolated by two rounds of serial transfer before an aliquot of the enriched culture was plated on nonselective medium to allow identification of the ASKA ORF (or ORFs).
The ASKA expression vector, pCA24N, has the same modified pMB1 replication origin as pQE30 (Qiagen), which gives it a copy number of 300 to 400 per cell. The pCA24N backbone also encodes the lacIq repressor, for tightly controlling the expression of each ASKA ORF. Therefore, we were specifically seeking examples where protein overexpression was essential for increased fitness. However, induction with a high concentration of IPTG (1 mM) severely inhibits the growth of 51% of ASKA strains (including almost all that overexpress membrane proteins), and in addition, another 28% show moderate growth inhibition (22). To avoid this source of bias, we used a significantly lower concentration of IPTG (50 μM) to induce protein expression. This amount is sufficient for derepression of the pCA24N T5-lacO promoter, but minimizes fitness differences associated with overexpression of different ORFs (20, 23).
In duplicate screens, the ASKA pool out-grew the negative control in the presence of 99 toxins (42% of those tested). In the majority of cases, these improved growth phenotypes manifested as small but reproducible changes in the intensity of tetrazolium color development within the first 48 h of incubation (Dataset S1). The ASKA pool showed enhanced growth in 50 of the 78 antibacterial agents (64%) contained within the PM plates. We also noted increased growth rates in the presence of 49 of the other 159 toxins that were tested (31%), which included antifungal and antiparasitic agents, inorganic salts, and dyes (Dataset S1).
Identification of Novel Resistance Genes.
Next we sought to deconvolute the results of our library-on-library screen by isolating individual ASKA clones that were responsible for the observed improvements in growth rates. In each of the 99 conditions identified above, we used serial transfer to enrich for the fittest clones within the ASKA pool (Fig. 1C). After two transfers, cells from the enriched population were plated under nonselective conditions. PCR and DNA sequencing revealed the plasmid-encoded genes that had imparted improved growth.
An ASKA ORF was scored as a hit if it occurred at least twice among the eight clones sequenced from a serially passaged population. Given the large size of the ASKA pool (5,272 clones), it was highly unlikely (P ≈ 0.005) that any two of the eight randomly chosen colonies would contain the same ASKA plasmid, unless that clone had been positively selected during serial transfer. Our approach yielded hits from 86 of the 99 conditions tested, including representatives of all of the compound classes that were tested (Table 1). For example, the PM plates contained 22 β-lactam antibiotics, and we isolated ASKA clones that improved growth in 11 of them (Table 1, row 1). Our serial enrichment strategy yielded a single hit from 60 of the 86 conditions. In 38 of these, the ASKA clone was enriched to apparent homogeneity (i.e., eight of eight sequenced clones were identical). However, serial transfer gave two different hits in 23 conditions, and three hits in another three cases (the β-lactam, aztreonam; the antiseptic, benzethonium chloride; and the toxic alkaloid, sanguinarine). Overall, therefore, we identified 115 positively selected hits from 86 toxin-containing environments (Table 1 and Dataset S1).
Table 1.
Summary of hits obtained from the library-on-library screen
| Class of toxic compound | Number of positives* | Number of ASKA hits† |
| Antibacterials | ||
| β-Lactams | 11 (of 22) | 14 |
| Aminoglycosides | 4 (of 14) | 4 |
| Anti-folates | 7 (of 9) | 10 |
| Quinolones | 4 (of 9) | 6 |
| Tetracyclines | 3 (of 8) | 4 |
| MLS antibiotics | 6 (of 7) | 10 |
| Glycopeptides | 1 (of 3) | 1 |
| Nitrofurans | 3 (of 3) | 4 |
| Rifamycins | 1 (of 2) | 2 |
| Steroid antibacterial | 1 (of 1) | 1 |
| Other toxins | 45 (of 159) | 59 |
| Total | 86 (of 237) | 115 |
*The number of toxic compounds for which screening and serial enrichment yielded at least one ASKA-encoded resistance gene. The total number of toxins that were screened in each class is listed in parentheses.
†Total number of ASKA ORFs that were isolated from PM wells containing toxins of each compound class.
In 13 cases, serial transfer failed to enrich any individual ASKA clone to a frequency of two or more in the eight sequenced clones (Dataset S1). One explanation is that there were many plasmid-encoded ORFs that all imparted fitness advantages (of similar magnitudes) in those conditions. It is also possible that chromosomal mutations were responsible for improved growth, although this is unlikely when the experimental design (two independent ASKA pools, both required to out-grow negative control populations) and the small size of each input population (∼50,000 cells) are considered. Alternatively, further transfers may have been required to identify the fastest-growing clones in the ASKA pool. By imparting stringent criteria for identifying novel resistance genes, we minimized the probability of isolating false-positives.
Diverse Mechanisms of Resistance.
Our initial focus was on identifying proteins—and particularly enzymes—that possessed weak secondary activities. However, it rapidly became apparent that our library-on-library screen had uncovered many different classes of protein that were contributing to resistance. Therefore, we used the EcoCyc (26) and UniProt (27) databases, in addition to the references cited in Dataset S1, to assign proposed modes of action for each of the 115 cases that we had identified (Table 2).
Table 2.
Proposed modes of action for 115 cases of resistance imparted by ASKA ORFs
| Mechanism of resistance | Frequency |
| Efflux pump/transporter | 22 |
| Regulatory effect | 19 |
| Catalytic promiscuity and/or substrate ambiguity | 15 |
| Prophage gene | 14 |
| Stress response | 13 |
| Envelope/capsule synthesis | 8 |
| Biofilm formation and regulation | 5 |
| Overexpression of the drug's target | 1 |
| Maintenance of metabolic flux | 1 |
| Unknown | 17 |
| Total | 115 |
Eighteen genes occurred multiple times and were responsible for improving growth in a total of 72 toxin-containing environments (Table S1), suggesting broad and nonspecific mechanisms. Consistent with previous results (28–31), the expression of efflux pumps and transporters was common among this subset of compound nonspecific resistance determinants. For example, expression of the Bcr multidrug transporter increased growth in the presence of three antifolate antibiotics (sulfachloropyridazine, sulfathiazole, and sulfisoxazole), two tetracyclines (oxytetracycline and rolitetracycline), and one β-lactam (cefoperazone). An uncharacterized outer-membrane β-barrel, YcbS, improved growth in the presence of four toxic compounds: a mutagen, 9-aminoacridine; an uncoupling agent, FCCP; a herbicide, methyl viologen; and an alkaloid, sanguinarine. Similarly, the MdtM and Cmr efflux pumps improved growth in the presence of five and three toxins, respectively (Table S1). The selection of these membrane proteins confirmed that we had minimized any bias associated with IPTG-induced overexpression.
We also discovered regulators, stress-response proteins, and proteins from capsule biosynthesis that each improved fitness in the presence of multiple toxins (Table S1). The only readily predictable example was the transcriptional activator MarA, which we isolated from environments containing two quinolones (ciprofloxacin and ofloxacin), a tetracycline (penimepicycline), a macrolide (tylosin), and a mutagen (proflavine). MarA is known to mediate multidrug resistance by up-regulating the efflux system and down-regulating membrane permeability (32, 33). In other cases, we found genes that had not previously been associated with resistance, including the biofilm modulation protein, Bdm, the small stress response protein, YcgZ, and the global regulator of transcription, CpdA (cAMP phosphodiesterase). The likely pleiotropic effects of these in altering the expression of downstream resistance determinants can be rationalized. Another prominent hit was the UDP-glucose 4-epimerase (GalE) ASKA strain, which was isolated from seven PM wells. GalE is involved in a variety of galactose metabolic processes, including glycolysis and the biosynthesis of lipopolysaccharide and capsule components. The host strain, E. coli DH5α-E, is gal−, although the exact mutations responsible for this phenotype are not known. It is possible that the fitness effects seen when GalE was overexpressed were simply a result of complementation of the gal− mutation. However, if this was the case, we would have expected the GalE-expressing ASKA strain to out-grow the negative control in many more than seven of the 237 environments that we tested. Instead, GalE expression appears to mediate a suite of pleiotropic effects that are relevant for the evolution of resistance. Finally, other multiple-occurring ORFs hint at the functions of previously uncharacterized proteins (Table S1). For example, the YejG-expressing clone was isolated from PM wells containing apramycin, sisomicin, and tobramycin, suggesting that it may possess broad enzymatic activity toward these aminoglycoside antibiotics.
In 43 cases, an ASKA-encoded gene improved growth in the presence of a single toxin (Dataset S2). These compound-specific responses included 12 cases in which metabolic enzymes appeared to display catalytic promiscuity or substrate ambiguity. Examples included the carbonic anhydrase, CynT, evincing growth in the presence of enoxacin [reminiscent of the human enzyme's promiscuous esterase activity (34)] and the purine nucleotidase, YrfG, increasing fitness in the presence of a toxic analog (6-mercaptopurine). Finally, 13 of the 17 instances in which we were unable to assign a mechanism of action (Table 2) involved uncharacterized ASKA ORFs that were isolated from a single environment (Dataset S2).
Interplay of Fitness and Resistance.
Mutations that confer antibiotic resistance can have a variety of effects on the fitness of the cell in the absence of drug (35). Resistance mutations often bear fitness costs, although examples of cost-free resistance (36) and mutual compensation (i.e., simultaneous increases in resistance and fitness) have also been described (37). Our library-on-library screen revealed cases where ASKA clones were responsible for enhancing fitness in the presence of a toxin. It was unclear from our screen whether the plasmid-encoded ORFs were improving the general vigor of the host or increasing resistance. To investigate this further, we measured the fitness value (W) for each of 25 strains relative to the negative-control strain (i.e., E. coli DH5α-E with pCA24N-NoIns). Fitness tests were conducted in the absence of toxin, but the presence of 50 μM IPTG. Using the same set of 25 strains, we also determined the minimum inhibitory concentrations (MICs) for 29 toxin/strain combinations. We chose to focus on anti-bacterial compounds and to test examples of each mechanistic class (as defined in Table 2).
In the absence of toxin, only two of the 25 ASKA strains showed significantly reduced fitness (i.e., W < 1; P < 0.01) compared with the negative control (Table 3). Nine ASKA strains were not significantly different from the control (P > 0.01), and 14 of the strains out-competed it (W > 1; P < 0.01). In contrast, almost all of MICs determined for ASKA strains were increased compared with those that were measured for the negative-control strain (Table 3).
Table 3.
Relative fitness and drug resistance data for E. coli cells overexpressing ORFs from the ASKA library
| ASKA ORF | W ± SE* | Compound | MIC (μg/mL) with ASKA ORF | MIC (μg/mL) with pCA24N-NoIns | Fold-increase in MIC |
| β-Lactams | |||||
| bdm | 1.47 ± 0.04† | Cephalothin | 16 | 8 | 2.0 |
| bfd | 1.16 ± 0.03† | Penicillin G | 48 | 32 | 1.5 |
| rbsR | 1.40 ± 0.04† | Aztreonam | 0.19 | 0.064 | 3.0 |
| yeaD | 1.20 ± 0.03† | Aztreonam | 0.094 | 0.064 | 1.5 |
| ycgZ | 1.72 ± 0.05† | Aztreonam | 0.38 | 0.064 | 5.9 |
| ycgZ | See above | Cefuroxime | 16 | 6 | 2.7 |
| Aminoglycosides | |||||
| yejG | 0.94 ± 0.04 | Sisomicin | 0.19 | 0.125 | 1.5 |
| yejG | See above | Tobramycin | 1.5 | 0.5 | 3.0 |
| Anti-folates | |||||
| folA | 0.84 ± 0.03† | Trimethoprim | >32 | 0.064 | >500 |
| luxS | 1.11 ± 0.02† | Sulfadiazine | 24 | 16 | 1.5 |
| nudB | 0.68 ± 0.04† | Sulfadiazine | 32 | 16 | 2.0 |
| Quinolones | |||||
| cynT | 1.17 ± 0.04† | Enoxacin | 0.25 | 0.19 | 1.3 |
| marA | 1.08 ± 0.02 | Ciprofloxacin | 0.125 | 0.032 | 3.9 |
| galE | 1.16 ± 0.01† | Nalidixic acid | 96 | 32 | 3.0 |
| yfdO | 1.03 ± 0.02 | Nalidixic acid | 48 | 32 | 1.5 |
| Tetracyclines | |||||
| bcr | 1.03 ± 0.04 | Oxytetracycline | 6 | 2 | 3.0 |
| Macrolide-Lincosamide-Streptogramin | |||||
| cmr | 1.45 ± 0.02† | Lincomycin | 1,536 | 192 | 8.0 |
| rbsR | See above | Lincomycin | 384 | 192 | 2.0 |
| ydfW | 1.51 ± 0.12† | Spiramycin | 36 | 24 | 1.5 |
| ydfW | See above | Erythromycin | 96 | 32 | 3.0 |
| ydaC | 1.33 ± 0.03† | Erythromycin | 128 | 32 | 4.0 |
| Glycopeptides | |||||
| mzrA | 1.23 ± 0.07 | Vancomycin | 256 | 128 | 2.0 |
| Nitrofurans | |||||
| tfaX | 1.18 ± 0.02† | Nitrofurantoin | 0.094 | 0.19 | 0.5 |
| Rifamycins | |||||
| feoC | 0.78 ± 0.07 | Rifampicin | 6 | 4 | 1.5 |
| tusE | 0.87 ± 0.04 | Rifampicin | 6 | 4 | 1.5 |
| Other toxins | |||||
| mdtM | 1.07 ± 0.03 | Puromycin | 128 | 8 | 16.0 |
| puuD | 1.12 ± 0.03† | Benzethonium chloride | 6 | 6 | 1.0 |
| ybcD | 1.02 ± 0.02 | Benzethonium chloride | 8 | 6 | 1.3 |
| yhiK | 1.19 ± 0.01† | Benzethonium chloride | 12 | 6 | 2.0 |
*Fitness (W) of the ASKA strain, in the absence of toxin, relative to a neutrally-marked negative control that harbors pCA24N-NoIns. Values are expressed as mean ± SE (n = 8).
†Significance level of P < 0.01 for a two-tailed test with the null hypothesis that W = 1, calculated using the t distribution and 7 df.
By far the largest increase in MIC was for trimethoprim, where overexpression of its intracellular target, dihydrofolate reductase (folA), effected very high levels of resistance (at least 500-fold greater than the negative control strain). The FolA-expressing strain was also one of the least fit in the absence of antibiotic (W = 0.84). Expression of the multidrug transporters MdtM and Cmr led to 16-fold and 8-fold increases in the MICs for puromycin and lincomycin, respectively, without imparting fitness costs in a toxin-free environment. The remainder of the selected ASKA strains exhibited modest increases (<8-fold) in MICs compared with the negative control (Table 3).
In general, there was a poor correlation between resistance and fitness in the absence of toxin. In some cases, fitter clones were also more resistant. For example, the YcgZ-expressing clone had the highest relative fitness of the 25 that we measured (W = 1.72), and it also showed a greater increase in aztreonam resistance (5.9-fold) than the other two clones that were isolated from the aztreonam-containing PM well (Table 3). However, there were also cases where the opposite was true. For example, overexpression of YdaC imparted a fourfold increase in erythromycin resistance and a small increase in fitness (W = 1.33); YdfW overexpression gave a smaller increase in resistance (threefold) but a larger increase in fitness (W = 1.51). Similarly, overexpressing NudB gave a higher MIC for sulfadiazine than LuxS overexpression (32 vs. 24 μg·mL−1), yet the NudB-expressing strain was considerably less fit (W = 0.68 vs. W = 1.11).
In a single case, expression of an ASKA ORF (an uncharacterized prophage gene, tfaX) decreased the MIC for the antibiotic (nitrofurantoin) in which it was selected. The TfaX-expressing strain showed a small but statistically significant fitness improvement in the absence of nitrofurantoin (W = 1.18). This suggests that increased fitness in the absence of an antibiotic can translate into a selective advantage when the drug is introduced; however, this appears to be uncommon. It is noteworthy that no single ASKA gene was isolated from more than seven PM wells (vide supra) (Table S1). It might be expected that one (or a handful) of general, growth-enhancing genes would have been isolated from a majority of the 237 toxin-containing environments, were such a gene present in the ASKA pool. Instead, our approach has revealed many latent resistance determinants in the E. coli genome.
Discussion
The aim of this study was to gauge the extent to which a simple adaptive response—overexpression of a preexisting protein—could impart new phenotypes on E. coli. A handful of analogous examples have been discovered adventitiously (2), but at the outset of this study the likelihood (and therefore relevance) of this route to evolutionary innovation was unclear. We used resistance to antibiotics and toxins as a readily selectable trait. Our survey was the most comprehensive to date, because we used the wells of 10 PM plates as our toxin-containing environments. We also used the ASKA ORF collection to directly identify proteins that could impart resistance. This process contrasts with previous studies where roles in resistance have been inferred indirectly, based on the increased sensitivity of mutants produced by transposon inactivation or gene knockouts (28, 30, 31).
In the first part of our experiment, we identified toxin-containing conditions where the pooled ASKA clones out-grew the negative control. In those PM wells, at least one member of the pool had gained an improved ability to survive and reproduce, through the overexpression of a single, wild-type ORF. Next, serial transfer was used to identify the fittest strains in the ASKA pool. Our approach was designed to focus on monogenic examples, and to exclude multicell phenomena (such as cross-feeding between members of the pool). Our experimental design also precluded the identification of ASKA clones with increased susceptibility to toxins, although these are likely to have been present in many (and perhaps all) of the PM wells. In total, we identified 61 ASKA ORFs that improved fitness in 86 of 237 toxin-containing environments. Eighteen of these ORFs were hits in multiple wells (Table S1); overall, we identified 115 examples of resistance. Hits were obtained from every class of toxic compound tested (Table 1). The hit rate (resistance to 36% of all toxins) was even higher than in our previous study (23), where only 20% of single-gene knockout strains were rescued by noncognate ASKA ORFs. As hypothesized previously (38), we have shown that a bacterium from a nonclinical environment (in this case, a laboratory strain of E. coli) can nevertheless possess a significant reservoir of latent resistance determinants. More generally, our study emphasizes just how likely evolutionary innovation can be. Our results imply that the overexpression of a randomly chosen E. coli protein will impart resistance to a novel toxin with a probability of (61/5,272) × (86/237) ≈ 0.4%.
The PM plates contained four concentrations of each toxin. Therefore, a hit could have been because of an increase in resistance (i.e., a strain undergoing division at a toxin concentration that was inhibitory to the other strains, including the negative control), or because of particularly rapid growth in a subinhibitory concentration of the toxin (i.e., dividing faster than the other strains in the pool). In this respect, our experiment mimics the dynamics of a clinical setting (37, 39). Almost exclusively, the proteins that we identified did facilitate increased resistance (Table 3). The majority of the measured MIC increases were modest (<8-fold); nevertheless, even small changes such as these can be responsible for increasing the severity of bacterial infections (40). In one case—cefuroxime—we showed that expression of a small stress response protein, YcgZ, increased the MIC from 6 to 16 μg·mL−1. The clinical breakpoint for resistance to this β-lactam is 8 μg·mL−1 (41); that is, our nonpathogenic host strain attained clinically relevant levels of resistance, simply through overexpression of a previously uncharacterized protein. Furthermore, most strains showed no fitness cost associated with expressing an ASKA ORF in the absence of toxin (and indeed, many out-competed the negative control) (Table 3). Strong selection pressure for cost-free resistance mutations has been observed in clinical isolates of Mycobacterium tuberculosis (36). Together, our MIC and fitness data suggest that up-regulating the expression of preexisting, latent resistance determinants may play an important role in the emergence of drug-resistant pathogens.
We discovered proteins acting through a variety of mechanisms to effect resistance (Table 2). Many of these mechanisms—such as toxin efflux/transport, stress responses and biofilm formation—were unsurprising, even if the identities of the individual resistance genes were less predictable. In 15 of 115 cases, overexpressed enzymes appeared to be acting promiscuously to impart resistance. In addition to these mechanisms, there was a prevalence of prophage genes and genes of unknown function in our list. Prophage genes are usually assumed to be cryptic or defective remnants of temperate bacteriophage genomes, although a recent study has shown that genes from the CP4-57 and DLP12 prophages can play roles in biofilm development (42). Here, we have reported 14 cases in which the overexpression of prophage genes (including two from DLP12) can improve growth in the presence of toxins. Twelve of these cases arose from the overexpression of only three prophage genes: ydaC, ydfW, and yfdO (Table S1). This result extends the previous finding (42), demonstrating that prophage genes can modulate broad, compound nonspecific cellular responses when they are activated from latency.
In contrast, we identified 20 uncharacterized, nonprophage genes that showed specificity in their actions; that is, they each imparted a growth advantage in the presence of a single toxin. These included genes for putative enzymes (wbbL_1, yeaD, ysgA), predicted transporters (yedA, yjeH), and a predicted transcriptional regulator (yidF), as well as 14 genes for which functional annotation was impossible due to insufficient homology with any gene of known function (Dataset S2). These results provide experimental evidence for the importance of cryptic genes as a reservoir of evolutionarily-accessible functions (43).
Our experiments have demonstrated the biochemical feasibility of the IAD model for the origins of new genes (11). Every plasmid-carrying ASKA clone approximated a nonplasmid-containing strain in which a single chromosomal locus had been amplified. Growth under selection began when the pooled ASKA clones were used to inoculate each PM well. To be scored positive, a PM well had to contain cells that out-grew the negative control, in which no ASKA ORF was being expressed. Therefore, we were specifically looking for cases in which proteins possessed activities that were valuable in the presence of a toxin and cases in which increases in their dosage were required to uncover the latent activities. We halted our selection experiments at this stage; that is, we applied selection, but we did not consider mutation beyond (artificial) gene amplification. The next step will be to determine feasible mutational routes for the divergence of amplified copies from their original function toward their new role in antibiotic resistance. These studies are on-going in our laboratory, and will allow related questions, such as the nature of the tradeoff between new and old enzymatic functions, to be addressed.
Concluding Remarks.
We have used two tools from functional genomics (PM plates and the ASKA collection) to conduct a comprehensive search for E. coli proteins that can impart improved growth in the presence of toxic compounds. The resulting catalog provides a unique picture of a bacterium's latent evolutionary potential and emphasizes the high frequency at which novel traits can evolve. By cataloguing sources of phenotypic innovation, we have revealed the diversity of adaptive mechanisms that can be underpinned by overexpression mutations such as gene amplification. Our results suggest that the IAD model (11) is a biochemically and evolutionarily feasible—and perhaps dominant—mechanism for the birth of new genes under selection.
Materials and Methods
Experimental methods are summarized below. Detailed protocols are provided in the SI Materials and Methods.
Library-on-Library Screen.
The library-on-library screening protocol is outlined in Fig. 1. Plates PM11 to PM20 were from Biolog Inc. After inoculation, the PM plates were incubated at 37 °C for 7 d and the growth in each well was scored daily. Two independent replicates of the screen were performed.
Serial Enrichment to Identify Resistance Genes.
For conditions in which the ASKA pool reproducibly out-grew the negative control, fresh aliquots of the pool were now used to inoculate the corresponding wells of new PM plates. At the first sign of growth (i.e., purple color development), the culture in that well was diluted 50-fold with fresh medium and transferred to the corresponding well of a new plate. After a second transfer, cells were spread on LB-chloramphenicol plates. PCR amplification and sequencing of the ASKA ORFs from at least eight of the resulting colonies revealed each ORF that was responsible for enhanced growth.
Relative Fitness Assays.
Twenty-five ASKA plasmids that conferred increased growth in the presence of antibiotics were purified from individual clones and used to transform fresh aliquots of E. coli DH5α-E. The fitness of each IPTG-induced strain, in the absence of antibiotics and in competition with the negative control harboring pCA24N-NoIns, was measured using Lenski's protocol (44). E. coli DH5α-E carries the lacZΔM15 mutation. To distinguish the two strains in each fitness assay, a mini-Tn7 system (45) was used to mark the negative control with a functional copy of lacZ. Control experiments showed that marking the strain had no effect on its fitness in the conditions used. The initial (t = 0 h) and final (t = 24 h) densities of each competitor could then be measured by spreading them on LB agar plates supplemented with X-gal. Each relative fitness value (W) in Table 3 is the mean of eight replicates.
Antibiotic Susceptibility Testing.
MICs were determined using either E-test strips (AB bioMérieux) or a broth microdilution method (46). Reported values are the mean of at least two independent experiments.
Supplementary Material
Acknowledgments
We thank Ilana Gerber, Susan Morton, and Laura Nigon for their technical assistance. We are also grateful to Monica Gerth for her advice on fitness assays and to John Roth for his comments on the manuscript. Financial support for this research was provided by the Maurice and Phyllis Paykel Trust, the Lottery Health Research Committee, the Auckland Medical Research Foundation, and the Marsden Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012108108/-/DCSupplemental.
See Commentary on page 1199.
References
- 1.Hegeman GD, Rosenberg SL. The evolution of bacterial enzyme systems. Annu Rev Microbiol. 1970;24:429–462. doi: 10.1146/annurev.mi.24.100170.002241. [DOI] [PubMed] [Google Scholar]
- 2.Mortlock RP. Microorganisms as Model Systems for Studying Evolution. New York, London: Plenum Press; 1984. [Google Scholar]
- 3.Lewis EB. Pseudoallelism and gene evolution. Cold Spring Harb Symp Quant Biol. 1951;16:159–174. doi: 10.1101/sqb.1951.016.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Ohno S. Evolution by Gene Duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- 5.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 6.Wagner A. Energy constraints on the evolution of gene expression. Mol Biol Evol. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- 7.Roth C, et al. Evolution after gene duplication: Models, mechanisms, sequences, systems, and organisms. J Exp Zoolog B Mol Dev Evol. 2007;308:58–73. doi: 10.1002/jez.b.21124. [DOI] [PubMed] [Google Scholar]
- 8.Conant GC, Wolfe KH. Turning a hobby into a job: How duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 9.Innan H, Kondrashov F. The evolution of gene duplications: Classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 10.Francino MP. An adaptive radiation model for the origin of new gene functions. Nat Genet. 2005;37:573–577. doi: 10.1038/ng1579. [DOI] [PubMed] [Google Scholar]
- 11.Bergthorsson U, Andersson DI, Roth JR. Ohno's dilemma: Evolution of new genes under continuous selection. Proc Natl Acad Sci USA. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khersonsky O, Tawfik DS. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 13.Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 14.O'Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 15.Anderson P, Roth J. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc Natl Acad Sci USA. 1981;78:3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reams AB, Kofoid E, Savageau M, Roth JR. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics. 2010;184:1077–1094. doi: 10.1534/genetics.109.111963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg CM, Wang MD, Vartak NB, Liu L. Acquisition of new metabolic capabilities: Multicopy suppression by cloned transaminase genes in Escherichia coli K-12. Gene. 1988;65:195–202. doi: 10.1016/0378-1119(88)90456-8. [DOI] [PubMed] [Google Scholar]
- 18.Christ D, Chin JW. Engineering Escherichia coli heat-resistance by synthetic gene amplification. Protein Eng Des Sel. 2008;21:121–125. doi: 10.1093/protein/gzm085. [DOI] [PubMed] [Google Scholar]
- 19.Morett E, et al. Sensitive genome-wide screen for low secondary enzymatic activities: The YjbQ family shows thiamin phosphate synthase activity. J Mol Biol. 2008;376:839–853. doi: 10.1016/j.jmb.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Miller BG, Raines RT. Identifying latent enzyme activities: Substrate ambiguity within modern bacterial sugar kinases. Biochemistry. 2004;43:6387–6392. doi: 10.1021/bi049424m. [DOI] [PubMed] [Google Scholar]
- 21.Patrick WM, Matsumura I. A study in molecular contingency: Glutamine phosphoribosylpyrophosphate amidotransferase is a promiscuous and evolvable phosphoribosylanthranilate isomerase. J Mol Biol. 2008;377:323–336. doi: 10.1016/j.jmb.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 23.Patrick WM, Quandt EM, Swartzlander DB, Matsumura I. Multicopy suppression underpins metabolic evolvability. Mol Biol Evol. 2007;24:2716–2722. doi: 10.1093/molbev/msm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bochner BR, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bochner BR. Global phenotypic characterization of bacteria. FEMS Microbiol Rev. 2009;33:191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keseler IM, et al. EcoCyc: A comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The UniProt Consortium. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breidenstein EBM, Khaira BK, Wiegand I, Overhage J, Hancock REW. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob Agents Chemother. 2008;52:4486–4491. doi: 10.1128/AAC.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duo M, Hou S, Ren D. Identifying Escherichia coli genes involved in intrinsic multidrug resistance. Appl Microbiol Biotechnol. 2008;81:731–741. doi: 10.1007/s00253-008-1709-6. [DOI] [PubMed] [Google Scholar]
- 31.Liu A, et al. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: Generating an antibiotic barcode. Antimicrob Agents Chemother. 2010;54:1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen SP, Hächler H, Levy SB. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz C, Levy SB. Many chromosomal genes modulate MarA-mediated multidrug resistance in Escherichia coli. Antimicrob Agents Chemother. 2010;54:2125–2134. doi: 10.1128/AAC.01420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gould SM, Tawfik DS. Directed evolution of the promiscuous esterase activity of carbonic anhydrase II. Biochemistry. 2005;44:5444–5452. doi: 10.1021/bi0475471. [DOI] [PubMed] [Google Scholar]
- 35.Andersson DI, Hughes D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 36.Sander P, et al. Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob Agents Chemother. 2002;46:1204–1211. doi: 10.1128/AAC.46.5.1204-1211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcusson LL, Frimodt-Møller N, Hughes D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 2009;5:e1000541. doi: 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martínez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 39.Martínez JL, Baquero F, Andersson DI. Predicting antibiotic resistance. Nat Rev Microbiol. 2007;5:958–965. doi: 10.1038/nrmicro1796. [DOI] [PubMed] [Google Scholar]
- 40.Drusano GL. Antimicrobial pharmacodynamics: critical interactions of 'bug and drug'. Nat Rev Microbiol. 2004;2:289–300. doi: 10.1038/nrmicro862. [DOI] [PubMed] [Google Scholar]
- 41.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). http://www.eucast.org/clinical_breakpoints.Accessed August 13, 2010. [DOI] [PubMed]
- 42.Wang X, Kim Y, Wood TK. Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J. 2009;3:1164–1179. doi: 10.1038/ismej.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall BG, Yokoyama S, Calhoun DH. Role of cryptic genes in microbial evolution. Mol Biol Evol. 1983;1:109–124. doi: 10.1093/oxfordjournals.molbev.a040300. [DOI] [PubMed] [Google Scholar]
- 44.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- 45.Choi KH, et al. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods. 2005;2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 46.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.