Abstract
The importance of the EGF receptor (EGFR) signaling pathway in the development and progression of nonsmall cell lung carcinomas (NSCLC) is widely recognized. Gene sequencing studies revealed that a majority of tumors responding to EGFR kinase inhibitors harbor activating mutations in the EGFR kinase domain. This underscores the need for novel biomarkers and diagnostic imaging approaches to identify patients who may benefit from particular therapeutic agents and approaches with improved efficacy and safety profiles. To this goal, we developed 4-[(3-iodophenyl)amino]-7-{2-[2-{2-(2-[2-{2-([18F]fluoroethoxy)-ethoxy}-ethoxy]-ethoxy)-ethoxy}-ethoxy]-quinazoline-6-yl-acrylamide ([18F]F-PEG6-IPQA), a radiotracer with increased selectivity and irreversible binding to the active mutant L858R EGFR kinase. We show that PET with [18F]F-PEG6-IPQA in tumor-bearing mice discriminates H3255 NSCLC xenografts expressing L858R mutant EGFR from H441 and PC14 xenografts expressing EGFR or H1975 xenografts with L858R/T790M dual mutation in EGFR kinase domain, which confers resistance to EGFR inhibitors (i.e., gefitinib). The T790M mutation precludes the [18F]F-PEG6-IPQA from irreversible binding to EGFR. These results suggest that PET with [18F]F-PEG6-IPQA could be used for the selection of NSCLC patients for individualized therapy with small molecular inhibitors of EGFR kinase that are currently used in the clinic and have a similar structure (i.e., iressa, gefitinib, and erlotinib).
Keywords: positron emission tomography, epidermal growth factor receptor, activation mutation
Lung cancer is the most common cancer in both men and women, and it is responsible for 1.3 million deaths worldwide annually (1). In 2009, 219,440 Americans learned they had lung cancer. Nonsmall cell lung carcinoma (NSCLC) represents the majority of lung cancers (2). NSCLC treatments involving nonspecific, nonselective cytotoxic chemotherapy result in only a modest increase in survival and cause significant toxicity to the patient (3). Therapy with EGF receptor (EGFR) kinase inhibitors results in dramatic antitumor activity in ∼10% of Caucasian patients (young women and nonsmokers) with NSCLC and 30% of patients of East Asian ethnicities (4–11). Sequencing of the EGFR gene revealed that most tumors that respond to EGFR kinase inhibitors harbor activating mutations in the EGFR kinase domain (6, 12, 13). In patients harboring such mutations, the overall response rate is as high as 80% compared with <10% in patients with WT EGFR (6, 7–12,30). Also, the presence of these EGFR kinase mutations is associated with improved progression-free survival and overall survival after therapy with EGFR kinase inhibitors compared with patients with WT EGFR (16, 18, 20, 23, 24, 27).
The most frequently detected alterations were small deletions in exon 19 (35–45%) that eliminate amino acids 747–750 (Leu-Arg-Glu-Ala), located around the active site of the kinase, and point mutations in exon 21 that result in the amino acid substitution L858R, a residue located in the activation loop (6). A secondary T790M mutation is present in ∼50% of cases of NSCLC with acquired resistance to EGFR kinase inhibitors (31–34). A recent study showed that the EGFR T790M mutation was associated with significantly shorter progression-free survival compared with patients without detectable T790M before treatment (35). Thus, activating mutations in EGFR kinase domain and T790M secondary mutation are useful biomarkers for identifying patients who will benefit from or became resistant to therapy with EGFR kinase inhibitors (i.e., gefitinib and erlotinib).
The complexity and limitations of invasive methods of analysis of EGFR mutations prompted the development of novel radiolabeled agents for noninvasive PET imaging, which may help in identification of NSCLC patients who may benefit from EGFR kinase inhibitors. The majority of the previously reported radiolabeled agents for PET imaging of EGFR at the kinase level have been derived from 4-(anilino) quinazoline pharmacophore (36, 37), which includes the ML series of 18F-labeled five 4-(anilino)quinazoline derivatives (18F-ML01) (38), N-{4-[(4,5-dichloro-2-fluorophenyl)-amino]quinazolin-6-yl}-acrylamide labeled with 11C(11C-ML03) (39, 40), 4-dimethylamino-but-2-enoic acid [4-(phenylamino)-quinazoline-6-yl]-amides (ML04) labeled with 11C (11C-ML04) (40), 18F (18F-ML04) (41), and fluoro-4×PEG-ML04 (41, 42, as well as [124I]IPQA (43), [18F]gefinitib (44), [11C]PD153035 (45), and [11C]erlotinib (46). Recent PET imaging studies in mice using several PEG-ylated anilinoquinazoline derivatives labeled with 124I, 18F, and 11C failed to show the accumulation of these radiotracers in s.c. tumor xenografts overexpressing WT EGFR (47). Clinical PET imaging studies with [11C]PD153035 have shown some promise for imaging EGFR expression in NSCLC patients (48). However, none of the imaging agents reported to date had exhibited selectivity for mutant EGFR kinases that confer sensitivity or resistance to inhibitors that are currently used in the clinic (49).
Here, we report that the 4-[(3-iodophenyl)amino]-7-{2-[2-{2-(2-[2-{2-([18F]fluoroethoxy)-ethoxy}-ethoxy]-ethoxy)-ethoxy}-ethoxy]-quinazoline-6-yl-acrylamide ([18F]F-PEG6-IPQA) has increased selectivity and irreversible binding to the L858R active mutant EGFR kinase compared with WT and T790M mutant EGFR kinases. PET/computed tomography (CT) imaging with [18F]F-PEG6-IPQA allowed for noninvasive detection of NSCLC xenografts harboring L858R activating EGFR mutations and their discrimination from NSCLC xenografts expressing the WT or L858R/T790M dual-mutant EGFR kinases.
Results
Differential Sensitivity of NSCLC Cell Lines to EGFR Kinase Inhibition.
In vitro cell growth of different NSCLC cell lines was differentially inhibited by iressa in a dose-dependent manner. The most sensitive to iressa were the H3255 cells expressing L858R mutant EGFR (IC50 1.8 μm), whereas the H1975 cells expressing both the L858R and T790M EGFR mutations were significantly more resistant (IC50 24.4 μm). The H441 and PC14 cells expressing WT EGFR exhibited significant resistance to iressa (IC50 46.0 and 46.3 μm, respectively).
Differential Expression of EGFR in NSCLC Cell Lines.
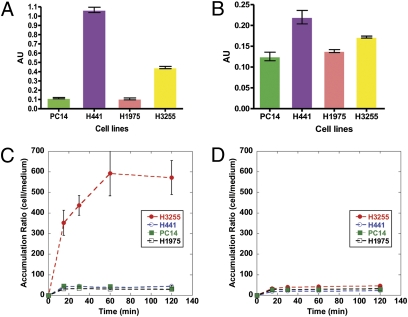
The levels of total EGFR expression, as measured by ELISA, were similar in H3255 and H441 cells and higher than those in PC14 and H1975 cells (Fig. 1A). However, the level of phospho-EGFR expression varied less than twofold among the tested cell lines (Fig. 1B).
Fig. 1.
Characterization of PC14, H441, H3255, and H1975 tumor cells lines in vitro. The level of (A) total EGFR and (B) phospho-EGFR. Time-dependent accumulation of [18F]F-PEG6-IPQA (C) at baseline and (D) in the presence of 100 μM/L iressa.
Preferential Accumulation of [18F]F-PEG6-IPQA in NSCLC Cells with L858R EGFR Mutation.
All four cell lines showed a rapid uptake of [18F]F-PEG6-IPQA during the initial phase (first 20 min). Thereafter, the accumulation of [18F]F-PEG6-IPQA reached a plateau in H441, H1975, and PC14 cells at about 30–40 cells/medium concentration ratio. In contrast, in H3255 cells, the accumulation of [18F]F-PEG6-IPQA continued to increase up for to 1 h and thereafter, reached a plateau at a cells per medium concentration ratio of 400–600 (Fig. 1C). The magnitude of [18F]F-PEG6-IPQA accumulation in H3255 cells was more than 10-fold higher than in H441 cells, despite the twofold lower levels of total EGFR and phospho-EGFR expression in H3255 cells than in H441 cells (Fig. 1 A and B). The radiotracer washout studies showed significant retention of [18F]F-PEG6-IPQA in H3255 cells (cells per medium concentration ratio of about 400), which accounted for 65% of the total accumulated radioactivity between 60 and 120 min of incubation (Fig. S1). In contrast, almost 50% of [18F]F-PEG6-IPQA radioactivity could be washed out from H441 cells, more than 60% could be washed out from PC14 cells, and more than 70% could be washed out from H1975 cells. After washout in PC14 and H1975 cells, the magnitude of [18F]F-PEG6-IPQA retention was less than 10 cells/medium concentration ratio. The magnitude of [18F]F-PEG6-IPQA accumulation in all tested cell lines was significantly decreased in the presence of iressa (100 μM/L) in the culture medium (Fig. 1D).
Preferential and Irreversible Binding of [18F]F-PEG6-IPQA to the L858R Mutant EGFR Kinase.
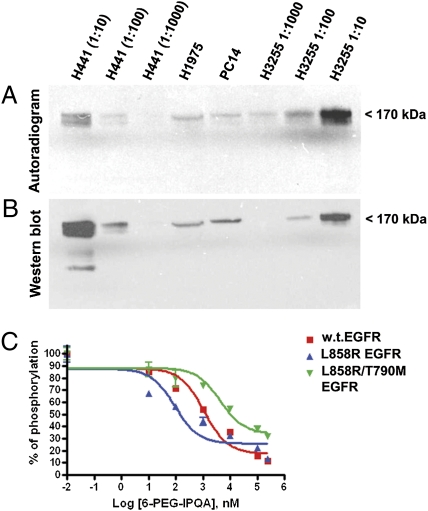
The autoradiograhic and Western blot analysis of electrophorograms of protein extracts from different NSCLC cells incubated with [18F]F-PEG6-IPQA showed preferential covalent binding of [18F]F-PEG6-IPQA to the L858R mutant EGFR kinase domain, which corresponds to the 72-kDa protein band (Fig. 2). In H441 cells, the level of irreversible binding to the WT EGFR kinase was significantly lower than in H3255 cells. This observation became more evident when comparing the intensity of radioactive bands (Fig. 2A) and corresponding Western blot bands (Fig. 2B) of different dilutions of cellular protein extracts. In particular, the 1:10 dilution of H441 extract contained significantly more EGFR protein than the 1:10 diluted extract from H3255 cells; however, the intensity of the radioactive band corresponding to the 1:10 dilution of H3255 cells was significantly greater (at least 1 log order) than that in the 1:10 diluted extract of H441 cells. Autoradiographic detection was more sensitive than immunoblotting and chemoluminescent detection, which was evidenced by the presence of a faint radioactive band in 1:1,000 diluted extracts of H3255 cells, whereas the corresponding Western blot band was not detectable. The covalent binding of [18F]F-PEG6-IPQA to the EGFR kinase domain was barely detectable in undiluted extracts of PC14 cells expressing low levels of WT EGFR and in undiluted extracts of H1975 cells expressing L858R/T790M dual-mutant EGFR.
Fig. 2.
Assessment of irreversible binding of [18F]F-PEG6-IPQA to WT and mutant EGFR kinases. (A) The autoradiogram of a Western blot membrane and (B) chemiluminescent image of the same membrane stained with anti-EGFR kinase antibody. Individual lanes are labeled with corresponding cell lines and dilutions of protein extracts (the H1975 and PC14 extracts were undiluted). (C) Inhibition of substrate phosphorylation by recombinant WT (red squares), L858R active mutant (blue triangles), and L585R/T790M dual-mutant (green triangles) EGFR kinases in the presence of different concentrations of F-PEG6-IPQA; the IC50 concentrations are indicated next to the enzyme labels.
F-PEG6-IPQA Preferentially Inhibits L858R EGFR Mutant Kinase Activity.
In vitro assays with recombinant WT and mutant EGFR kinases showed (Fig. 2C) preferential irreversible inhibitory activity of cold (nonradiolabeled) F-PEG6-IPQA against the L858R mutant EGFR (IC50 = 92 nM) compared with the WT EGFR (IC50 = 1.1 μM) and L858R/T790M mutant EGFR (IC50 = 4.4 mM).
In Vivo PET/CT Imaging with [18F]F-PEG6-IPQA Enables Discrimination of NSCLC Expressing L858R Mutant Vs. WT and L858R/T790M Dual-Mutant EGFR.
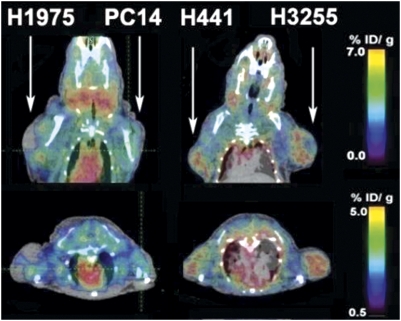
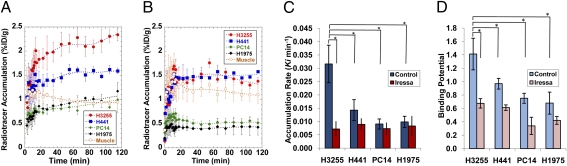
In vivo dynamic PET/CT imaging was performed in 12 mice (6 per each tumor pair) before and after therapy with iressa. PET/CT imaging showed rapid accumulation of [18F]F-PEG6-IPQA in H3255 and to a lesser degree, in H441 s.c. tumor xenografts (Fig. S2). The [18F]F-PEG6-IPQA accumulation at 120 min post-i.v. injection reached 2.34 ± 0.13% injected dose (ID)/g in H3255 tumors and 1.59 ± 0.44% ID/g in H441 tumors (Fig. 3). Tumor to muscle ratios of [18F]F-PEG6-IPQA accumulation at 120 min post-i.v. injection reached the level of 2.08 ± 0.19 in H3255 tumors and 1.47 ± 0.08 in H441 tumors (Fig. S3A). In contrast, the level of [18F]F-PEG6-IPQA accumulation was insignificant in PC14 and H1975 s.c. tumors, reaching 0.90 ± 0.11% ID/g and 1.05 ± 0.09% ID/g, respectively (Figs. 3 and 4A), which was similar to the 0.94 ± 0.22% ID/g observed in the muscle (reference tissue). Thus, the magnitude of [18F]F-PEG6-IPQA accumulation in H3255 was 1.4, 6.3, and 7.6 times higher (P < 0.001) than in H441, PC14, and H1975, respectively. Tumor to muscle ratios of [18F]F-PEG6-IPQA accumulation in PC14 and H1975 tumor xenografts reached equilibrium of about 1 starting at 20 min post-i.v. injection (Fig. S3A). The unidirectional rate of [18F]F-PEG6-IPQA accumulation (Ki), calculated using the Patlak graphical analysis, was significantly higher in H3255 tumors than in H441, PC14, and H1975 tumors (P < 0.05) (Fig. 4C and Fig. S4A). Logan's graphical analysis showed a significantly increased binding potential (BP) of [18F]F-PEG6-IPQA in H3255 tumors compared with other tumor xenografts (P < 0.05) (Fig. 4D and Fig. S5A). A summary of quantitative measures of [18F]F-PEG6-IPQA accumulation in different tumor xenografts at 120 min post-i.v. injection is provided in Table 1.
Fig. 3.
Representative coronal and axial PET/CT images obtained 120 min postinjection of [18F]F-PEG6-IPQA in two different mice bearing tumor xenografts in the opposite shoulders (arrows). Color coding of the images is set to maximize the visualization of tumors in each projection.
Fig. 4.
Time-activity curves of [18F]F-PEG6-IPQA–derived radioactivity (percent ID per gram) in different tumors (A) at baseline and (B) after pretreatment of animals with iressa. Quantification of [18F]F-PEG6-IPQA accumulation in different tumor xenografts at baseline (blue bars) and after pretreatment with iressa (red bars). (C) The rate of unidirectional accumulation (Ki) of [18F]F-PEG6-IPQA determined using Patlak graphical analysis and (D) binding potential (BP) of [18F]F-PEG6-IPQA determined by Logan model-independent graphical analysis. Bars are SEs; statistically significant differences (P < 0.05) are indicated by an asterisk.
Table 1.
Accumulation of [18F]-PEG6-IPQA in different tumor xenografts in mice
| Before treatment |
After treatment |
|||||||
| PET/CT |
QAR |
PET/CT |
QAR |
|||||
| Tumor type | % ID/g | T/M | % ID/g | T/M | % ID/g | T/M | % ID/g | T/M |
| H441 | 1.59 ± 0.06 | 1.47 ± 0.09 | 2.53 ± 0.15 | 2.84 ± 0.17 | 1.58 ± 0.04 | 1.77 ± 0.52 | 0.78 ± 0.06 | 0.94 ± 0.06 |
| H3255 | 2.34 ± 0.13 | 2.08 ± 0.19 | 3.01 ± 0.84 | 3.64 ± 1.01 | 1.38 ± 0.43 | 1.55 ± 0.37 | 0.92 ± 0.13 | 1.11 ± 0.16 |
| H1975 | 1.17 ± 0.18 | 1.05 ± 0.10 | 1.27 ± 0.12 | 1.68 ± 0.16 | 0.43 ± 0.17 | 0.46 ± 0.11 | 1.13 ± 0.10 | 1.19 ± 0.16 |
| PC14 | 0.99 ± 0.18 | 0.9 ± 0.11 | 1.41 ± 0.13 | 1.54 ± 0.14 | 0.52 ± 0.19 | 0.33 ± 0.08 | 1.17 ± 0.10 | 1.05 ± 0.01 |
| Muscle | 0.95 ± 0.22 | — | 0.83 ± 0.07 | — | 1.10 ± 0.18 | — | 0.96 ± 0.10 | — |
Treatment with Iressa Significantly Inhibits [18F]F-PEG6-IPQA Accumulation in NSCLCs Expressing L858R EGFR Mutant.
In vivo dynamic PET/CT imaging showed a significant reduction in [18F]F-PEG6-IPQA accumulation in H3255 tumor xenografts after treatment with iressa (100 mg/kg 1 h before administration of [18F]F-PEG6-IPQA) to the level of 1.38 ± 0.43% ID/g (Fig. 4B) and a tumor to muscle ratio of 1.56 ± 0.36 at 120 min post-i.v. injection (Fig. S3B). In contrast, the level of [18F]F-PEG6-IPQA accumulation in H441 tumor xenografts was 1.58 ± 0.01% ID/g (Fig. 4B), and the tumor to muscle ratio was 1.77 ± 0.53 at 120 min post-i.v. injection (Fig. S3B), which was similar to that before treatment (Fig. 4A and Fig. S3A). In PC14 and H1975 tumor xenografts, the magnitude and tumor to muscle ratios of [18F]F-PEG6-IPQA accumulation further decreased (Fig. 4B and Fig. S3B). Both Patlak and Logan graphical analyses (Figs. S4 and S5) showed a statistically significant reduction in Ki and BP of [18F]F-PEG6-IPQA in H3255 tumor xenografts (P < 0.05) after treatment with iressa, whereas the reduction of [18F]F-PEG6-IPQA accumulation in other tumor xenografts was not statistically significant (Fig. 4 C and D).
Pharmacokinetics of [18F]F-PEG6-IPQA in Tumor-Bearing Mice.
After i.v. injection, [18F]F-PEG6-IPQA exhibited biexponential plasma clearance kinetics with half-lives for the rapid and slow phases of 1.15 and 21.32 min, respectively (Fig. S3). At 30 min postinjection (n = 3), 16.2 ± 8.6% radioactivity in blood plasma was in the form of the parent [18F]F-PEG6-IPQA, and the remaining fraction was [18F]F-PEG6. At 60 min postinjection (n = 3), 100% of plasma radioactivity was caused by [18F]F-PEG6 metabolite. The initial clearance of [18F]F-PEG6-IPQA–derived radioactivity from the circulation was through the hepato-billiary route; the clearance was monoexponential, with a half-life of 31.88 min (Figs. S2 and S3 C and D). In the urine, at 30 min postinjection, only 14.6 ± 4.5% of radioactivity was in the form of [18F]F-PEG6-IPQA, and the rest was [18F]F-PEG6; at 60 min, almost 96% of radioactivity was in the form of [18F]PEG6, and the rest was [18F]F-PEG6-IPQA and traces of [18F]-fluoride. Therefore, increasing bladder radioactivity during the first 30 min predominantly represents the rate of renal clearance of [18F]F-PEG6-IPQA (29.92% ID/mL per minute), whereas after some lag phase between 30 and 40 min postinjection, an increase in bladder radioactivity at the rate of 24.12% ID/mL per minute is predominantly because of the renal clearance of [18F]F-PEG6 metabolite.
Quantitative Autoradiography Confirms the Results of in Vivo PET/CT Imaging with [18F]F-PEG6-IPQA.
In a separate group of animals (n = 4 per tumor pair and n = 4 per treatment condition), quantitative autoradiography (QAR) showed preferential accumulation of [18F]F-PEG6-IPQA in viable portions of H3255 tumor xenografts (Fig. S6). Also, QAR revealed intratumoral heterogeneity of [18F]F-PEG6-IPQA. High levels of [18F]F-PEG6-IPQA–derived radioactivity accumulated in the viable portions of H3255 tumor xenografts (3.01 ± 0.84% ID/g). In contrast, no specific [18F]F-PEG6-IPQA accumulation was observed inside the necrotic tumor areas. The accumulation of [18F]F-PEG6-IPQA in H441 tumors (2.53 ± 0.15% ID/g) was lower than that in H3255 xenografts, whereas in H1975 (1.27 ± 0.12% ID/g) and PC14 (1.54 ± 0.14% ID/g) tumors, it was significantly lower. Pretreatment with iressa resulted in a fourfold decrease in [18F]F-PEG6-IPQA accumulation in H3255 tumors (to 0.92 ± 0.13% ID/g; P < 0.05) and a threefold decrease in H441 tumors (to 0.78 ± 0.06% ID/g; P < 0.05). There were no statistically significant differences in [18F]F-PEG6-IPQA accumulation in PC14 and H1975 tumor xenografts before and after treatment with iressa (1.17 ± 0.10 and 1.05 ± 0.01% ID/g, respectively).
Discussion
Noninvasive PET/CT imaging of EGFR expression activity and mutation status in NSCLC could aid in the selection of patients for individualized therapy with EGFR kinase inhibitors by complimenting the existing gene sequencing (24), immunohistochemical (50), and FISH analyses (51). More importantly, because repetitive biopsies of multiple tumor lesions in patients with advanced NSCLC are prohibitively traumatic, whole-body noninvasive PET/CT imaging could provide measures of EGFR expression activity in the individual tumor lesions and estimate treatment-responsive vs. -resistant tumor burden before the initiation of therapy with EGFR inhibitors (i.e., gefitinib and erlotinib); potentially, it could predict the overall outcome.
This study showed significantly increased accumulation of [18F]F-PEG6-IPQA in H3255 cells compared with H441, PC14, and H1975 cells in vitro. Because levels of expression of L858R EGFR in H3255 cells and WT EGFR in H441 cells were similar, the preferential accumulation of [18F]F-PEG6-IPQA in H3255 tumors can be explained, at least in part, by the presence of the L585R activating mutation in the EGFR kinase domain (6, 52, 53). The L858R mutation dramatically enhances the magnitude of EGFR signaling activity in H3255 cells compared with H441 cells that overexpress WT EGFR (6, 53) and PC14 cells that express WT EGFR at a low level.
Our structure–activity relationship modeling studies (SI Materials and Methods and Fig. S7) indicate that the activation status of EGFR kinase seems to be very important for the binding mode of [18F]F-PEG6-IPQA and the formation of a covalent bond with Cys773. Other studies with structurally similar 4-anilinoquinazoline–based reversible inhibitors (gefitinib and erlotinib) have also shown that these compounds had a more than a 20-fold increase in affinity and inhibitory activity for the L858R mutant compared with WT EGFR kinase (54). A shift in equilibrium to the active state in L858R mutant EGFR kinase and the much stronger alternative binding of [18F]F-PEG6-IPQA to the L858R mutant are thought to be the reasons for the more rapid covalent bond formation that leads to faster irreversible entrapment of [18F]F-PEG6-IPQA in cells expressing the L858R mutant EGFR. Similar explanations apply to the interpretation of in vivo PET/CT imaging studies in mice, which showed that [18F]F-PEG6-IPQA preferentially accumulates in s.c. tumor xenografts established from H3255 cells compared with moderate accumulation of [18F]F-PEG6-IPQA in H441 tumor xenografts, low accumulation in PC14 tumors, and very low accumulation in H1975 tumors.
Although H1975 cells carry the L858R EGFR mutation, the second T790M mutation causes resistance to reversible EGFR inhibitors, such as gefitinib and erlotinib, by interfering with the binding of these inhibitors to the ATP binding site of EGFR kinase (32, 55). The T790 residue is located at the back of the ATP binding site and is often called the gatekeeper residue (56). This mechanism is analogous to that resulting from the T315L mutation in Bcr-Abl, which leads to imatinib resistance (57). In the EGFR–erlotinib structure, the side chain of T790 makes a water-mediated hydrogen bond to N3 of the quinazoline. In the EGFR–lapatinib structure, the threonine side chain is rotated relative to its position in the erlotinib structure and makes a number of direct and water-mediated hydrogen bonds to the protein. Mutation of this residue to methionine is predicted to sterically block inhibitor binding, because the methionine side chain would occupy part of the same space as the quinazoline ring for both inhibitors (58). This also explains the lack of [18F]F-PEG6-IPQA accumulation in H1975 cells in vitro and in H1975 tumors in vivo.
Autoradiographic and Western blot analyses of electrophorograms of proteins extracted from different NSCLC cells incubated in vitro with [18F]F-PEG6-IPQA also showed preferential irreversible binding of [18F]F-PEG6-IPQA to the L858R mutant EGFR kinase domain compared with WT EGFR kinase and lack of irreversible binding to L858R/T790M dual-mutant EGFR kinase. Similarly, enzyme assays using recombinant WT and mutant EGFR kinases showed that nonradiolabeled cold F-PEG6-IPQA has 20-fold more potent inhibitory activity against the L858R mutant EGFR kinase compared with the WT EGFR kinase, and more than 1 log order lower inhibitory activity against the L858R/T790M dual-mutant EGFR. These data also explain the lack of [18F]F-PEG6-IPQA accumulation in H1975 cells in vitro and in H1975 tumors in vivo. Although H1975 cells are known to be sensitive to other irreversible inhibitors, such as EKB569 and CI1033 (55, 59), these cells exhibited significant resistance to F-PEG6-IPQA in vitro. This also suggests a different and unique mode of binding of F-PEG6-IPQA to the ATP binding site of the H1975 mutant EGFR kinase compared with EKB569 and CI1033.
Preincubation of H3255 cells in vitro with the EGFR inhibitor iressa in biologically effective concentrations effectively blocked the accumulation of [18F]F-PEG6-IPQA in these cells because of the competition of iressa for binding to the same site in the L858R EGFR mutant kinase domain. Similarly, pretreatment of tumor-bearing mice with pharmacologically effective doses of iressa blocked the accumulation of [18F]F-PEG6-IPQA in H3255 tumors expressing L858R EGFR mutant and to a lesser degree, in H441 tumors overexpressing WT EGFR. Therefore, it should be feasible to use PET imaging with [18F]F-PEG6-IPQA to assess pharmacodynamic characteristics and determine biologically effective doses of small molecular EGFR kinase inhibitors in individual tumors.
Compared with other small molecular agents for imaging of EGFR at the kinase level reported by us previously, such as morpholino-[124I]-IPQA (43), [18F]F-PEG6-IPQA exhibits improved radiotracer pharmacokinetic characteristics. Notably, the biexponential clearance half-time of [18F]F-PEG6-IPQA from the blood is shorter than that of morpholino-[124I]-IPQA (rapid and slow phases = 1.15 and 21.32 min vs. 4.8 and 552 min, respectively). This is partially because of the better water solubility of [18F]F-PEG6-IPQA (LogD = 1.44) compared with morpholino-[124I]-IPQA (LogD = 3.8). These results support the predictions made by us and other investigators (42) that improved water solubility of the 7-oligoPEGylated aminophenylquinazoline analogs will improve the potential of this class of radiolabeled compounds to be used as radiotracers for molecular imaging of EGFR overexpressing tumors.
Another reason for its superior radiotracer characteristics is that [18F]F-PEG6-IPQA is radiolabeled on the 7-(PEG6) side chain, which, on metabolic cleavage from the quinazoline core, is rapidly eliminated through renal clearance, as evidenced by the early appearance of radioactivity in the bladder. On metabolic degradation of [18F]F-PEG6-IPQA and similar 7-(PEG6) analogs, the 6-acrylamide aminophenyl-quinazoline pharmacophore is known to bind to both inactive and activated EGFR kinase and does not discriminate between the WT and active mutant EGFR kinases. Therefore, the aminophenyl moiety of this class of compounds is not suitable for radiolabeling, because it would render compounds that, on metabolic cleavage of 7-(PEG6) moiety, would have much higher lipophilicity and longer half-time circulation and would not adequately discriminate between the WT and L858R active mutant EGFR kinase. In contrast, [18F]-labeling of the 7-(PEG6) side chain ensures that radioactivity accumulation in the tumor tissue is caused only by the parent [18F]F-PEG6-IPQA compound, which irreversibly and covalently binds to the active mutant EGFR kinase domain, as was confirmed by our current in vitro studies. This is because the cleaved [18F]F-PEG6 side chain does not cross the cell membrane and is rapidly washed out from the extracellular space and eliminated from circulation predominantly by the renal clearance. The latter explains the low background levels in tissues that are not involved in the clearance of [18F]F-PEG6-IPQA and [18F]F-PEG6 from the body. Lower background levels of [18F]F-PEG6-IPQA compared with morpholino-[124I]-IPQA (43) and other previously reported agents (36–46, 48) can also be explained in part by the lower affinity of [18F]F-PEG6-IPQA to the WT and inactive EGFR, expressed by normal tissues, such as the lungs, intestine, and skin.
In conclusion, PET/CT with [18F]F-PEG6-IPQA could potentially be used to detect tumors expressing L858R active mutant EGFR and to select patients for individualized therapy with small molecular inhibitors of EGFR kinase that have a pharmacophore structure similar to [18F]F-PEG6-IPQA (i.e., gefitinib and erlotinib). Identification of tumors with high uptake and retention levels of [18F]F-PEG6-IPQA should predict favorable responses to therapy with EGFR kinase inhibitors, whereas the lack of [18F]F-PEG6-IPQA accumulation should predict resistance. Because mutations in EGFR and Kirsten rat sarcoma viral oncogene homolog (KRAS) genes are almost mutually exclusive (60, 61), tumors that accumulate [18F]F-PEG6-IPQA are less likely to be driven by KRAS mutations and therefore, will be more likely to respond to therapy with EGFR kinase inhibitors. Also, after the initial course of treatment with EGFR inhibitors and before an increase in tumor growth is observed by computed tomography, repeat imaging with [18F]F-PEG6-IPQA could potentially be used to identify tumor lesions with acquired T790M mutations in the EGFR kinase domain, which should exhibit significantly lower accumulation of [18F]F-PEG6-IPQA, and to justify triage to therapy with novel irreversible EGFR kinase inhibitors that are effective against T790M mutants (62). Additional studies in NSCLC expressing other activating mutations of EGFR kinase (i.e., exon 19 del) should be conducted to fully assess the diagnostic potential of PET/CT imaging with [18F]F-PEG6-IPQA. Imaging with [18F]F-PEG6-IPQA PET/CT may add to the toolbox of molecular imaging approaches to the differential diagnosis of molecular–genetic subtypes of NSCLC and other EGFR-driven tumor types and for the selection and monitoring of individualized therapies with EGFR inhibitors.
Materials and Methods
The [18F]F-PEG6-IPQA was prepared in high specific activity and high purity following the method described elsewhere (63). Four NSCLC cell lines expressing different levels of WT and active mutant EGFR and different sensitivity iressa were used: (i) H441 (WT EGFR; resistant) (6, 53), (ii) PC14 (WT EGFR; resistant) (64–66), (iii) H3255 (mutant L858R EGFR; sensitive) (6, 52, 53), and (iv) H1975 (L858R, T790M double-mutant EGFR; resistant) (32, 55). In vitro radiotracer uptake and washout studies were performed in monolayer cultures of these different tumor cell lines as previously described (43, 67). SI Materials and Methods contains the details for methods used to assess the sensitivity of different tumor cell lines to iressa, ELISA for measurements of EGFR expression in different tumor cell lines, irreversible covalent binding of [18F]F-PEG6-IPQA to the WT, L858R, and T790M double-mutant EGFR kinases, in silico docking of [18F]F-PEG6-IPQA with WT and different EGFR kinase mutants, inhibition of EGFR kinase activity in vitro, development of different s.c. tumor xenografts in mice, and PET imaging protocol in mice and data analysis. Statistical analyses included ANOVA to assess differences in the magnitude of [18F]F-PEG6-IPQA accumulation (percent ID per gram) in different tumors established from different NSCLC cell lines and to assess differences in [18F]F-PEG6-IPQA accumulation before and after treatment with iressa. Mean ± SD values were calculated for each tumor type and compared using the t test for group averages. Paired Student t test was used to assess differences in [18F]F-PEG6-IPQA accumulation in pairs of tumors grown in the same animal (H441 vs. H3255 and H1975 vs. PC14); P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Karen Yoas for excellent coordination of studies. This work was supported by Grants W81XWH-05-2-0027 from the Department of Defence (DoD) (to J.G.G.), 5U24CA126577 (to J.G.G.), and Cancer Center Support Grant NIH-NCI CA-16672 from the National Institutes of Health (NIH).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010744108/-/DCSupplemental.
References
- 1.World Health Organization Cancer. 2009. Fact sheet No.297. Available at http://www.who.int/mediacentre/factsheets/fs297/en/index.html. Accessed December 30, 2010.
- 2.ACS Cancer Reference Information. 2009. Available at http://www.cancer.org/Cancer/LungCancer-Non-SmallCell/DetailedGuide/non-small-cell-lung-cancer-key-statistics=. Accessed December 30, 2010.
- 3.Pirker R, Filipits M. Targeted therapies in lung cancer. Curr Pharm Des. 2009;15:188–206. doi: 10.2174/138161209787002915. [DOI] [PubMed] [Google Scholar]
- 4.Fukuoka M, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 5.Kris MG, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: A randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Huang CM, et al. Epidermal growth factor receptor (EGFR) gene Bsr I polymorphism is associated with systemic lupus erythematosus. Lupus. 2004;13:773–776. doi: 10.1191/0961203304lu1081oa. [DOI] [PubMed] [Google Scholar]
- 8.Thatcher N, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: Results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 9.Shepherd FA, Tsao MS. Unraveling the mystery of prognostic and predictive factors in epidermal growth factor receptor therapy. J Clin Oncol. 2006;24:1219–1220. doi: 10.1200/JCO.2005.04.4420. [DOI] [PubMed] [Google Scholar]
- 10.Riely GJ, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 11.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 12.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 13.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mu XL, Li LY, Zhang XT, Wang SL, Wang MZ. Evaluation of safety and efficacy of gefitinib (‘iressa’, zd1839) as monotherapy in a series of Chinese patients with advanced non-small-cell lung cancer: Experience from a compassionate-use programme. BMC Cancer. 2004;4:51–59. doi: 10.1186/1471-2407-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell DW, et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: Molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23:8081–8092. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- 16.Cappuzzo F, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 17.Chou TY, et al. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11:3750–3757. doi: 10.1158/1078-0432.CCR-04-1981. [DOI] [PubMed] [Google Scholar]
- 18.Cortes-Funes H, et al. Epidermal growth factor receptor activating mutations in Spanish gefitinib-treated non-small-cell lung cancer patients. Ann Oncol. 2005;16:1081–1086. doi: 10.1093/annonc/mdi221. [DOI] [PubMed] [Google Scholar]
- 19.Eberhard DA, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 20.Han SW, et al. Epidermal growth factor receptor (EGFR) downstream molecules as response predictive markers for gefitinib (Iressa, ZD1839) in chemotherapy-resistant non-small cell lung cancer. Int J Cancer. 2005;113:109–115. doi: 10.1002/ijc.20550. [DOI] [PubMed] [Google Scholar]
- 21.Kim JC, et al. Correlation of HER1/EGFR expression and degree of radiosensitizing effect of the HER1/EGFR-tyrosine kinase inhibitor erlotinib. Indian J Biochem Biophys. 2005;42:358–365. [PubMed] [Google Scholar]
- 22.Kondo M, et al. Mutations of epidermal growth factor receptor of non-small cell lung cancer were associated with sensitivity to gefitinib in recurrence after surgery. Lung Cancer. 2005;50:385–391. doi: 10.1016/j.lungcan.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Mitsudomi T, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 24.Takano T, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 25.Taron M, et al. Activating mutations in the tyrosine kinase domain of the epidermal growth factor receptor are associated with improved survival in gefitinib-treated chemorefractory lung adenocarcinomas. Clin Cancer Res. 2005;11:5878–5885. doi: 10.1158/1078-0432.CCR-04-2618. [DOI] [PubMed] [Google Scholar]
- 26.Tomizawa Y, et al. Clinicopathologic significance of the mutations of the epidermal growth factor receptor gene in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11:6816–6822. doi: 10.1158/1078-0432.CCR-05-0441. [DOI] [PubMed] [Google Scholar]
- 27.Tsao MS, et al. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 28.Zhang XT, et al. The EGFR mutation and its correlation with response of gefitinib in previously treated Chinese patients with advanced non-small-cell lung cancer. Ann Oncol. 2005;16:1334–1342. doi: 10.1093/annonc/mdi340. [DOI] [PubMed] [Google Scholar]
- 29.Shih HC, Hsiao YP, Wu MF, Yang JH. Gefitinib-induced acute generalized exanthematous pustulosis in two patients with advanced non-small-cell lung cancer. Br J Dermatol. 2006;155:1101–1102. doi: 10.1111/j.1365-2133.2006.07511.x. [DOI] [PubMed] [Google Scholar]
- 30.Uramoto H, et al. Epidermal growth factor receptor mutations are associated with gefitinib sensitivity in non-small cell lung cancer in Japanese. Lung Cancer. 2006;51:71–77. doi: 10.1016/j.lungcan.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 32.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosaka T, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 34.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S24–S31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishani E, Abourbeh G. Cancer molecular imaging: Radionuclide-based biomarkers of the epidermal growth factor receptor (EGFR) Curr Top Med Chem. 2007;7:1755–1772. doi: 10.2174/156802607782507457. [DOI] [PubMed] [Google Scholar]
- 37.Mishani E, Hagooly A. Strategies for molecular imaging of epidermal growth factor receptor tyrosine kinase in cancer. J Nucl Med. 2009;50:1199–1202. doi: 10.2967/jnumed.109.062117. [DOI] [PubMed] [Google Scholar]
- 38.Bonasera TA, et al. Potential (18)F-labeled biomarkers for epidermal growth factor receptor tyrosine kinase. Nucl Med Biol. 2001;28:359–374. doi: 10.1016/s0969-8051(01)00200-1. [DOI] [PubMed] [Google Scholar]
- 39.Ortu G, et al. Labeled EGFr-TK irreversible inhibitor (ML03): In vitro and in vivo properties, potential as PET biomarker for cancer and feasibility as anticancer drug. Int J Cancer. 2002;101:360–370. doi: 10.1002/ijc.10619. [DOI] [PubMed] [Google Scholar]
- 40.Mishani E, et al. Novel carbon-11 labeled 4-dimethylamino-but-2-enoic acid [4-(phenylamino)-quinazoline-6-yl]-amides: Potential PET bioprobes for molecular imaging of EGFR-positive tumors. Nucl Med Biol. 2004;31:469–476. doi: 10.1016/j.nucmedbio.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 41.Abourbeh G, et al. Evaluation of radiolabeled ML04, a putative irreversible inhibitor of epidermal growth factor receptor, as a bioprobe for PET imaging of EGFR-overexpressing tumors. Nucl Med Biol. 2007;34:55–70. doi: 10.1016/j.nucmedbio.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Dissoki S, et al. The effect of the [18F]-PEG group on tracer qualification of [4-(phenylamino)-quinazoline-6-YL]-amide moiety—an EGFR putative irreversible inhibitor. Appl Radiat Isot. 2007;65:1140–1151. doi: 10.1016/j.apradiso.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 43.Pal A, et al. Molecular imaging of EGFR kinase activity in tumors with 124I-labeled small molecular tracer and positron emission tomography. Mol Imaging Biol. 2006;8:262–277. doi: 10.1007/s11307-006-0049-0. [DOI] [PubMed] [Google Scholar]
- 44.Su H, et al. Evaluation of [(18)F]gefitinib as a molecular imaging probe for the assessment of the epidermal growth factor receptor status in malignant tumors. Eur J Nucl Med Mol Imaging. 2008;35:1089–1099. doi: 10.1007/s00259-007-0636-6. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, et al. Further characterization of the epidermal growth factor receptor ligand 11C-PD153035. Chin Med J (Engl) 2007;120:960–964. [PubMed] [Google Scholar]
- 46.Memon AA, et al. Positron emission tomography (PET) imaging with [11C]-labeled erlotinib: A micro-PET study on mice with lung tumor xenografts. Cancer Res. 2009;69:873–878. doi: 10.1158/0008-5472.CAN-08-3118. [DOI] [PubMed] [Google Scholar]
- 47.Pantaleo MA, et al. Evaluation of modified PEG-anilinoquinazoline derivatives as potential agents for EGFR imaging in cancer by small animal PET. Mol Imaging Biol. 2010;12:616–625. doi: 10.1007/s11307-010-0315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu N, et al. PET-based biodistribution and radiation dosimetry of epidermal growth factor receptor-selective tracer 11C-PD153035 in humans. J Nucl Med. 2009;50:303–308. doi: 10.2967/jnumed.108.056556. [DOI] [PubMed] [Google Scholar]
- 49.Gelovani JG. Molecular imaging of epidermal growth factor receptor expression-activity at the kinase level in tumors with positron emission tomography. Cancer Metastasis Rev. 2008;27:645–653. doi: 10.1007/s10555-008-9156-5. [DOI] [PubMed] [Google Scholar]
- 50.Cappuzzo F, et al. Gefitinib in pretreated non-small-cell lung cancer (NSCLC): Analysis of efficacy and correlation with HER2 and epidermal growth factor receptor expression in locally advanced or metastatic NSCLC. J Clin Oncol. 2003;21:2658–2663. doi: 10.1200/JCO.2003.01.039. [DOI] [PubMed] [Google Scholar]
- 51.Hirsch FR, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: A Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 52.Tracy S, et al. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- 53.Mukohara T, et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J Natl Cancer Inst. 2005;97:1185–1194. doi: 10.1093/jnci/dji238. [DOI] [PubMed] [Google Scholar]
- 54.Yun CH, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo A, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci USA. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Shah K, Yang F, Witucki L, Shokat KM. Engineering Src family protein kinases with unnatural nucleotide specificity. Chem Biol. 1998;5:91–101. doi: 10.1016/s1074-5521(98)90143-0. [DOI] [PubMed] [Google Scholar]
- 57.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 58.Blencke S, Ullrich A, Daub H. Mutation of threonine 766 in the epidermal growth factor receptor reveals a hotspot for resistance formation against selective tyrosine kinase inhibitors. J Biol Chem. 2003;278:15435–15440. doi: 10.1074/jbc.M211158200. [DOI] [PubMed] [Google Scholar]
- 59.Kwak EL, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McDermott U, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci USA. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou W, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pal ABJ, et al. Radiosynthesis and initial in vitro evaluation of [(18)F]F-PEG (6)-IPQA-A novel PET radiotracer for imaging EGFR expression-activity in lung carcinomas. Mol Imaging Biol. 2010 doi: 10.1007/s11307-010-0408-8. September 22 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arao T, et al. Small in-frame deletion in the epidermal growth factor receptor as a target for ZD6474. Cancer Res. 2004;64:9101–9104. doi: 10.1158/0008-5472.CAN-04-2360. [DOI] [PubMed] [Google Scholar]
- 65.Noro R, et al. Gefitinib (IRESSA) sensitive lung cancer cell lines show phosphorylation of Akt without ligand stimulation. BMC Cancer. 2006;6:277–289. doi: 10.1186/1471-2407-6-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hosaka T, et al. Mutant epidermal growth factor receptor undergoes less protein degradation due to diminished binding to c-Cbl. Anticancer Res. 2007;27:2253–2263. [PubMed] [Google Scholar]
- 67.Najjar AM, et al. Molecular-genetic PET imaging using an HSV1-tk mutant reporter gene with enhanced specificity to acycloguanosine nucleoside analogs. J Nucl Med. 2009;50:409–416. doi: 10.2967/jnumed.108.058735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.