Abstract
Modulation of DNA repair proteins by small molecules has attracted great interest. An in vitro helicase activity screen was used to identify molecules that modulate DNA unwinding by Werner syndrome helicase (WRN), mutated in the premature aging disorder Werner syndrome. A small molecule from the National Cancer Institute Diversity Set designated NSC 19630 [1-(propoxymethyl)-maleimide] was identified that inhibited WRN helicase activity but did not affect other DNA helicases [Bloom syndrome (BLM), Fanconi anemia group J (FANCJ), RECQ1, RecQ, UvrD, or DnaB). Exposure of human cells to NSC 19630 dramatically impaired growth and proliferation, induced apoptosis in a WRN-dependent manner, and resulted in elevated γ-H2AX and proliferating cell nuclear antigen (PCNA) foci. NSC 19630 exposure led to delayed S-phase progression, consistent with the accumulation of stalled replication forks, and to DNA damage in a WRN-dependent manner. Exposure to NSC 19630 sensitized cancer cells to the G-quadruplex–binding compound telomestatin or a poly(ADP ribose) polymerase (PARP) inhibitor. Sublethal dosage of NSC 19630 and the chemotherapy drug topotecan acted synergistically to inhibit cell proliferation and induce DNA damage. The use of this WRN helicase inhibitor molecule may provide insight into the importance of WRN-mediated pathway(s) important for DNA repair and the replicational stress response.
Keywords: genomic instability, human disease
Inhibition of DNA repair has been proposed as a strategy for combating cancer (1). Synthetic lethality is an approach that exploits preexisting DNA repair deficiencies in certain tumors to develop inhibitors of DNA repair pathways that compensate for the tumor-associated repair deficiency. Because helicases play critical roles in the DNA damage response and DNA repair, particularly in actively dividing and replicating cells, characterization of synthetic lethal relationships of DNA helicases may be of value in developing improved anticancer treatment strategies (2); moreover, small molecules that specifically target a given DNA helicase may be useful for understanding the role of the helicase in cellular nucleic acid metabolism.
We sought to identify and characterize a small-molecule inhibitor of a human RecQ helicase that is important for the cellular response to replicational stress. The RecQ family of DNA helicases has been implicated in the maintenance of genomic stability and human disease. Mutations in the Werner syndrome (WRN), Bloom’s syndrome (BLM), and RecQ protein-like 4 (RECQL4) genes are responsible for the human disorders Werner syndrome, Bloom’s syndrome, and Rothmund–Thomson syndrome, respectively (3). RecQ helicases are required for normal cellular DNA replication, repair, and recombination. Although this interesting class of enzymes has been studied extensively, the precise functions of these proteins in vivo still are not well understood.
We chose to focus on the WRN helicase as a target for small-molecule inhibition based on the plethora of evidence that WRN plays an important role in cell proliferation and DNA repair (4–6). Werner syndrome is a premature aging disorder that displays many clinical symptoms of aging at an accelerated rate (7). The WRN gene product that is defective in the chromosomal instability disorder has DNA helicase and exonuclease activities and interacts with a number of nuclear proteins to maintain genomic stability (8). We investigated the hypothesis that a potent and specific WRN helicase inhibitor could be identified and used to inhibit WRN-dependent functions in vivo. Our findings provide evidence that a small molecule can modulate in vivo the function of a human helicase in the DNA damage response.
Results
In Vitro WRN Helicase Activity Screen of National Cancer Institute Diversity Set Compounds.
The National Cancer Institute (NCI) Diversity Set library was screened for inhibitors of WRN helicase activity using an in vitro radiometric assay with a 19-bp forked duplex DNA substrate. Initially, a single 50-μM concentration of the compound was tested (Fig. S1A). Approximately 70% of the DNA substrate was unwound by WRN incubated with 5% DMSO in the control reaction (Fig. S1A). Compound 5 strongly inhibited WRN helicase activity. WRN helicase data from a larger group of 500 compounds from the NCI Diversity Set are shown in Fig. S1B. Seven of the small molecules identified from the initial screen were chosen for further analysis based on their ability to inhibit WRN helicase activity by 75% or more (Table S1). IC50 values were determined from compound titrations as shown by NSC 19630 (Fig. S1 C and D), giving a range of 2–20 μM for the small molecules tested (Fig. S1E). To examine the specificity of WRN helicase inhibition, we tested the selected compounds on DNA unwinding catalyzed by two related human RecQ helicases (RECQ1 and BLM), Fanconi anemia group J (FANCJ) helicase mutated in Fanconi anemia, and three Escherichia coli helicases (RecQ, UvrD, and DnaB) (Table S2). Based on results from DNA unwinding assays with WRN and other helicases, two compounds (NSC 19630 and NSC 2805) inhibited WRN helicase activity but not the other six DNA helicases assayed.
Cell Proliferation Assays to Screen WRN Helicase Inhibitors.
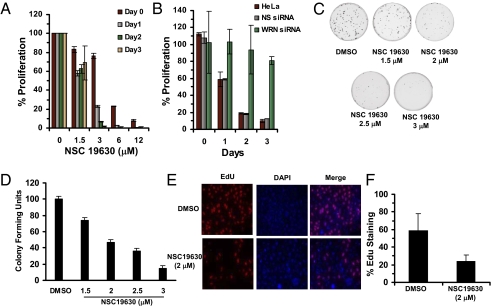
To determine if the small molecules identified by the in vitro WRN helicase activity screen were biologically active, we examined their effect on the proliferation of the human cervical cancer cell line HeLa 1.2 11 (hereafter abbreviated “HeLa”). HeLa cells were exposed to DMSO (as a control) or to increasing concentrations of selected small molecules for 0–3 d. Proliferation of compound-treated cells was compared with the DMSO-treated cells. Of the compounds tested, NSC 19630 showed the greatest inhibition of cell proliferation at the lower concentrations (Fig. 1A). NSC 19630 (3 μM) inhibited proliferation by 95% after day 2 (Fig. 1A). Higher concentrations of NSC 19630 (6 μM and 12 μM) inhibited proliferation by 99% after day 1. Because NSC 19630 inhibited proliferation of p53-inactivated HeLa cells at the lowest concentration among all tested compounds, we examined its effect on proliferation of U2OS cells that have a wild-type p53 gene. As shown in Fig. S2, 80% and 90% inhibition of U2OS cell proliferation was observed after exposure to NSC 19630 for 2 or 3 d, respectively.
Fig. 1.
NSC 19630 inhibits cell proliferation in a WRN-specific manner and impairs cell growth and DNA synthesis. HeLa cells (A) or siRNA-transfected HeLa cells (B) were treated with DMSO (control) or the indicated concentration of specified compound for 0–3 d. Percent cell proliferation determined by WST-1 was calculated as the ratio of OD450 values obtained for HeLa cells grown in the presence of a small-molecule inhibitor compared with the presence of DMSO. Day 0 represents effect of treatment on cell proliferation after 4 h. (C) Effect of NSC 19630 on cell growth. HeLa cells were treated with DMSO or the indicated concentration of NSC 19630 for 3 d. Cells were washed and allowed to grow in fresh medium lacking DMSO or NSC 19630 for 7 d and stained with methylene blue. (D) Percent colony-forming units was calculated as the ratio of colonies formed from cultures grown in the presence of NSC 19630 to cultures grown in the presence of DMSO, which was considered 100%. (E) Effect of NSC 19630 on DNA synthesis. HeLa cells were treated with DMSO or 2 μM NSC 19630 for 3 d. Cells were processed for EdU staining as described in SI Materials and Methods. Merged picture shows cells stained with EdU (red) or DAPI (blue). (F) Percent EdU-staining cells was calculated as the ratio of cells showing EdU staining compared with total number of cells with DAPI-stained nuclei.
Specificity of NSC 19630-Mediated Inhibition of Cell Proliferation.
To determine if the antiproliferative effect of NSC 19630 was mediated through inhibition of WRN cellular function, we compared its effect on WRN-depleted cells and on control cells. First, we established that the WRN protein level depleted by siRNA interference remained low throughout the 4-d time course of the experiment. Western blot analysis demonstrated that WRN was reduced by ≥90% throughout the 4 d after siRNA-WRN treatment (Fig. S3). WRN-depleted HeLa cells were treated with DMSO or 3 μM NSC 19630 for 0–3 d (Fig. 1B). WRN-depleted HeLa cells grown in the presence of 3 μM NSC 19630 were resistant to its antiproliferative effects, whereas control siRNA HeLa cells were highly sensitive to NSC 19630 (Fig. 1B). The other compounds tested (NSC 83224, NSC 42352, and NSC 2805) showed less significant inhibition of cell proliferation, and the antiproliferative effect was not dependent on WRN status, because WRN-depleted cells’ sensitivity to the compound tested was similar to that of control cells (Fig. S3). To assess if recovery of WRN expression after siRNA-mediated suppression reestablished NSC 19630 sensitivity, the WRN-depleted HeLa cells were allowed to continue to grow until WRN expression (through siRNA dilution by cell division) returned to a normal level (Fig. S4A). At this stage, cells were treated with 3 μM NSC 19630 and were found to be sensitive to the compound as measured by cell proliferation (Fig. S4B).
Because NSC 19630 exerted a WRN-dependent effect on cell proliferation, we evaluated whether BLM status affected cellular sensitivity to the compound. The results demonstrate that BLM-null and BLM-corrected cells display similar sensitivity to NSC 19630 (Fig. S4C), indicating that BLM does not play a role in the antiproliferative effects of NSC 19630.
Effect of NSC 19630 on the Growth of NCI 60 Cancer Cell Lines.
To evaluate further the biological effect of NSC 19630, existing data from the NCI Developmental Therapeutics Program were mined. This database contains information on the effects of small molecules from the NCI Diversity Set on growth of 60 human tumor cell lines from different cancer types. The most notable effect of NSC 19630 on cell growth was observed for leukemia cell lines, in which all six lines tested were inhibited by NSC 19630 (Fig. S5). Growth also was suppressed by exposure to NSC 19630 in the majority of renal cancer cell lines and in certain human cell lines from other forms of cancer (colon, ovarian, and breast). Western blot analysis of WRN in cell lysates from the NCI collection suggests that, although there are differences in WRN protein levels, these differences do not correlate with the sensitivity of the cell lines to NSC 19630 (Fig. S5).
Because some of the cancer cell lines were sensitive to NSC 19630, we examined the effect of the compound on noncancerous breast epithelial cells (MCF10A) and telomerase-immortalized normal fibroblasts (NHF1-hTERT), both of which were found to be resistant to the antiproliferative effects of NSC 19630 as measured by the WST-1 proliferation assay (Fig. S6). T47D breast cancer cells were sensitive to NSC 19630, but MCF7 cells were not (Fig. S6), in agreement with the NCI60 data.
DNA Binding by NSC 19630 and Its Effect on WRN ATPase and Exonuclease Activities.
To determine if NSC 19630 mediated its inhibitory effect on WRN helicase activity by binding the DNA substrate, we performed Thiazole Orange displacement assays using the forked duplex helicase substrate and either NSC 19630 or Hoechst 33258, a compound that intercalates and inserts a moiety in the minor groove of B-form duplex DNA. NSC 19630 did not displace Thiazole Orange from the forked duplex DNA molecules, whereas displacement of Thiazole Orange by 10 μM Hoechst 33258 was observed (Fig. S7A). At a concentration of NSC 19630 in which significant WRN helicase inhibition was observed, no binding to the forked duplex DNA molecules by the compound was detected.
Because helicase activity is coupled to ATP hydrolysis, we examined the effect of NSC 19630 on WRN ATPase activity. NSC 19630 concentrations up to 50 μM had only a modest effect (19% reduction) on the kcat for ATP hydrolysis by WRN (Fig. S7B). In contrast, WRN helicase activity was reduced by 75% at an NSC 19630 concentration of 50 μM. These results suggest that WRN helicase activity (IC50 ∼20 μM) was more sensitive than WRN ATPase activity to inhibition by NSC 19630. We also observed that WRN exonuclease activity was reduced only mildly in the presence of 25 μM NSC 19630 (Fig. S7C).
NSC 19630 Inhibits Cell Survival, Induces Apoptosis, and Causes Double-Strand Breaks.
To evaluate the ability of the WRN helicase inhibitor NSC 19630 to affect cell growth, colony-formation assays were performed. HeLa cells were incubated with the small molecule or with DMSO for 3 d, subsequently allowed to grow in medium free of DMSO or NSC 19630 for 7 d, and then stained with methylene blue. A significant reduction in both the size and number of colonies was observed in cells exposed to NSC 19630 compared with control DMSO-treated cells (Fig. 1 C and D). Quantitative analysis demonstrated a 55% reduction in colony number as a consequence of exposure to 2 μM NSC 19630 (Fig. 1 C and D). At 3 μM NSC 19630, an 85% reduction in colonies was observed (Fig. 1 C and D), indicating that the effect on cell growth was dependent on NSC 19630 concentration.
To test the effect of NSC 19630 on cellular mitogenic efficiency, we evaluated 5-ethynyl-2′-deoxyuridine (EdU) incorporation as a measure of DNA synthesis. HeLa cells exposed to NSC 19630 were impaired in their ability to synthesize DNA compared with control (DMSO) cells (Fig. 1 E and F).
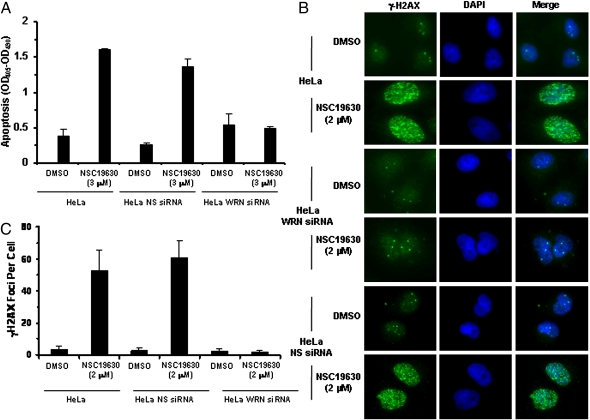
The negative effect of NSC 19630 exposure on cellular proliferation suggested that the compound also might affect apoptotic potential. HeLa cells exposed to 3 μM NSC 19630 displayed a fivefold increase in apoptosis compared with DMSO-treated cells (Fig. 2A). There was no difference in the level of apoptosis in WRN siRNA HeLa cells treated with NSC 19630 and those treated with DMSO (Fig. 2A), suggesting that NSC 19630 induction of apoptosis is WRN dependent.
Fig. 2.
NSC 19630 induces apoptosis and γ-H2AX foci. (A) Effect of NSC 19630 on apoptosis. Untransfected or WRN siRNA-transfected HeLa cells were treated with DMSO or 3 μM NSC 19630 for 3 d. Cells were assayed for histone-associated DNA fragments indicative of apoptosis as described in SI Materials and Methods. (B and C) NSC 19630 induces γ-H2AX foci in a WRN-dependent manner. Untransfected or WRN siRNA-transfected HeLa cells were treated with DMSO or 2 μM NSC 19630 for 3 d. Cells were stained for γ-H2AX as described in SI Materials and Methods. (B) Merged picture shows cells stained with anti–γ-H2AX antibody (green) and DAPI (blue). (C) Number of γ-H2AX foci per cell in cells treated with DMSO or 2 μM NSC 19630.
RecQ helicases are proposed to maintain and stabilize replication forks in response to endogenous stress or exogenous DNA damage (3, 9). Failure to stabilize forks can lead to fork collapse and double-strand breaks (DSB). Therefore, we analyzed formation of γ-H2AX foci in NSC 19630-treated or control cells. Exposure of HeLa cells to 2 μM NSC 19630 elevated γ-H2AX foci ∼17-fold compared with DMSO-treated cells (Fig. 2 B and C). WRN siRNA-treated HeLa cells showed intact nuclei and a similar number of γ-H2AX foci in NSC 19630-treated cells and in control (DMSO-treated) cells. These results suggest that inhibition of WRN activity in vivo by NSC 19630 leads to the accumulation of DSB. Consistent with this idea, exposure of HeLa cells to 2 μM NSC 19630 led to the activation of ataxia telangiectasia mutated (ATM) as detected by accumulation of autophosphorylated ATM at Ser-1981 (Fig. S8A).
NSC 19630 Induces S-Phase Delay in a WRN-Dependent Manner.
In the absence of DNA damage, WRN-depleted cells progress through S phase with kinetics similar to that in mock-depleted cells (6). However, WRN is required for normal replication fork progression after DNA damage or replication fork arrest (6). Because NSC 19630 exposure resulted in DSB accumulation, inhibition of DNA synthesis, and elevated apoptosis, we reasoned that cells treated with NSC 19630 might be delayed in their progression through S phase. To determine the effect of NSC 19630 on cell-cycle progression, WRN siRNA or control siRNA HeLa cells were exposed to 2 μM compound for 72 h and analyzed by flow cytometry. HeLa cells exposed to NSC 19630 displayed a significant increase in S-phase population (42% compared with 24% in DMSO-treated control cells) (Fig. S8B). Mock-depleted cells exposed to NSC 19630 also displayed a significant increase in S-phase population similar to that observed with HeLa. However, no significant difference in S-phase population was seen in NSC 19630- versus DMSO-treated WRN-depleted cells, suggesting that NSC 19630 induces S-phase delay in a WRN-dependent manner. WRN depletion in HeLa cells did not affect the length of the S phase (Fig. S8C), suggesting that inhibition of WRN helicase activity by NSC 19630 exerts a dominant negative effect on S-phase progression.
Proliferating Cell Nuclear Antigen Staining Revealed the Accumulation of Blocked Replication Forks in NSC 19630-Exposed Cells.
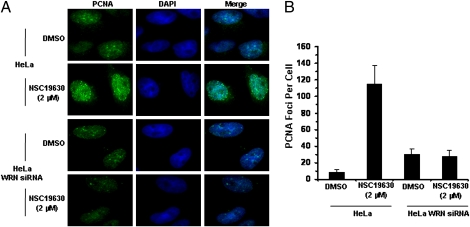
Mammalian cells exposed to replication inhibitors or DNA damage that blocks replication fork progression display an enrichment of proliferating cell nuclear antigen (PCNA) foci that correspond to stalled replication forks and/or assembly sites of DNA repair proteins (10, 11). Because cells exposed to the WRN helicase inhibitor NSC 19630 display a delayed S phase and accumulation of γ-H2AX foci, we wanted to determine whether PCNA foci staining would be increased and, if so, whether this increase occurred in a WRN-dependent manner. HeLa cells exposed to 2 μM NSC 19630 showed a significantly increased (∼20-fold) number of PCNA-staining foci compared with DMSO-treated cells (Fig. 3 A and B). In contrast, WRN-depleted cells showed similar levels of PCNA staining for the NSC 19630- and DMSO-treated cultures. These results suggest that NSC 19630-induced accumulation of PCNA foci is WRN dependent.
Fig. 3.
Elevated PCNA staining of NSC 19630-treated cells is WRN dependent. (A) Untransfected or WRN siRNA-transfected HeLa cells were treated with DMSO or 2 μM NSC 19630 for 3 d. Cells were stained for PCNA as described in SI Materials and Methods. Merged picture shows cells stained with PCNA antibody (green) and DAPI (blue). (B) Number of PCNA foci per cell in cells treated with DMSO or 2 μM NSC 19630.
Synergistic Effect of NSC 19630 and Telomestatin on Cell Proliferation.
Exposure of human cells to small molecules such as telomestatin (TMS) that specifically bind G4 structures in vitro induce the dissociation of shelterin proteins [e.g., protection of telomeres 1 (POT1) and TATA box binding protein-related factor 2 (TRF2)] or telomere-associated proteins [e.g., topoisomerase III (TOP3)] from their telomeric sites (12–14). TMS is proposed to compete with such proteins for binding to G4 DNA or stabilizing a G4 structure that is not favorably bound by the telomere-interacting protein, leading to telomere uncapping in cells with alternative lengthening of telomere (12, 14). TMS also can reduce proliferation of telomerase-positive tumor cells effectively by inhibiting telomerase (13, 15).
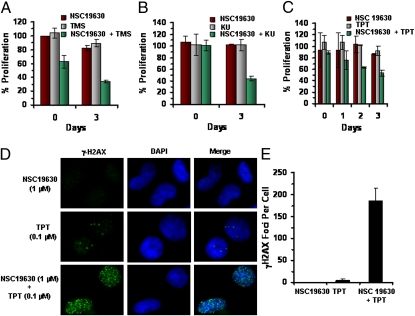
Because WRN can unwind G4 DNA substrates in vitro (16, 17), and evidence suggests that WRN helicase activity has a specialized role in unwinding alternate DNA structures that form in the G-rich strand of the telomere end (18), we sought to determine if NSC 19630 and TMS might act synergistically to inhibit cell proliferation. Limited concentrations of TMS (0.6 μM) and of NSC 19630 (1 μM) exerted only mild effects on cell proliferation (Fig. 4A). However, cotreatment of U2OS cells with both NSC 19630 and TMS inhibited cell proliferation by 70%, demonstrating an apparent synergistic effect.
Fig. 4.
NSC19630 exposure enhances the cells’ sensitivity to agents that induce replicational stress or DNA damage. (A) U2OS cells were treated with NSC 19630 (1 μM), TMS (0.6 μM), or both compounds for 3 d. (B) HeLa cells were treated with NSC 19630 (1 μM), PARP inhibitor KU0058948 (1 nM), or both compounds for 3 d. (C) HeLa cells were treated with NSC 19630 (1 μM), TPT (0.1 μM), or both compounds for 3 d. Cell proliferation was determined with WST-1 reagent as described in Materials and Methods. Percent proliferation was calculated as the ratio of OD450 values obtained for HeLa cells grown in the presence of the indicated compounds compared with values for cells grown in the presence of DMSO. Day 0 represents the effect of treatment on cell proliferation after 4 h. (D) Cells were stained for γ-H2AX. Merged picture shows cells stained with anti–γ-H2AX antibody (green) or DAPI (blue). (E) Number of foci per cell in cells treated with NSC 19630 (1 μM), TPT (0.1 μM), or both NSC 19630 (1 μM) and TPT (0.1 μM).
NSC 19630 Increases Cellular Sensitivity to Poly(ADP Ribose) Polymerase Inhibitor.
It was reported previously that inhibitors of the single-stranded break DNA repair protein poly(ADP ribose) polymerase 1 (PARP1) are synthetic lethal in homologous recombination (HR)-deficient cells with mutations in the breast cancer susceptibility proteins, breast cancer 1 (BRCA1) and breast cancer 2 (BRCA2) (19, 20). It is believed that when PARP1 is inhibited, single-stranded breaks persist, leading to the collapse of replication forks in dividing cells that ultimately result in potentially toxic DSB. In BRCA-deficient cells, the absence of HR, which normally provides an error-free pathway to deal with replication fork-associated lesions, results in cell killing by concentrations of PARP inhibitor that are not toxic to HR-proficient cells.
Given the prominence of the HR pathway of DSB repair in dealing with replication-associated lesions that accumulate in rapidly proliferating cells, we hypothesized that WRN helicase, which is known to have a role in HR, represented a target to enhance the cellular sensitivity to a known PARP inhibitor, KU0058948. HeLa cells were exposed to sublethal concentrations of KU0058948 (1 nM), NSC 19630 (1 μM), or a combination of the two compounds and were evaluated for cell proliferation. Cotreatment of HeLa cells with both NSC 19630 and the PARP inhibitor resulted in an ≈60% reduction in cell proliferation, whereas neither compound alone had any detectable effect (Fig. 4B), suggesting that the PARP inhibitor and NSC 19630 act synergistically to inhibit cell proliferation.
NSC 19630 Enhances the Effect of Topotecan on Cell Proliferation and DNA Damage Induction.
To assess the effect of NSC 19630 on the ability of cells to cope with replication stress induced by the chemotherapeutic drug topotecan (TPT), HeLa cells were cotreated with the topoisomerase inhibitor and NSC 19630. Results from cell proliferation assays and analyses of γ-H2AX foci formation were compared in cells exposed to TPT or NSC 19630 alone. Treatment with either NSC 19630 (1 μM) or TPT (0.1 μM) exerted only a mild effect on cell proliferation (Fig. 4C). However, treatment of cells with both NSC 19630 (1 μM) and TPT (0.1 μM) resulted in an ≈50% reduction in cell proliferation (Fig. 4C).
Cellular exposure to 1 μM NSC 19630 did not induce γ-H2AX foci formation, whereas exposure to 0.1 μM TPT resulted in, on average, four to five γ-H2AX foci per cell (Fig. 4 D and E). However, in the presence of both TPT and NSC 19630, a significant (40-fold) increase in the number of γ-H2AX foci per cell was observed (Fig. 4 D and E). These results demonstrate an apparent synergistic effect of NSC 19630 and TPT on DSB accumulation.
Discussion
Because of the importance of WRN in the replicational stress response, we sought to identify and characterize a small molecule that would modulate its biological function. To accomplish this goal, we screened the NCI Diversity Set using an in vitro helicase assay. A compound designated NSC 19630 (IC50 = 20 μM) appeared to be specific based on its reduced ability to affect DNA unwinding by two other human RecQ helicases (BLM and RECQ1) and four additional helicases (FANCJ, RecQ, UvrD, and DnaB). Consistent with the data from the in vitro helicase activity screen, inhibition of WRN helicase activity by NSC 19630 is not likely to be mediated by a NSC 19630–DNA interaction. Moreover, WRN ATPase and exonuclease activities were affected only mildly by NSC 19630 compared with WRN helicase inhibition.
NSC 19630 inhibited proliferation of human tumor cells grown in culture. The antiproliferative effect of NSC 19630 is likely to be mediated by a direct effect on WRN, because WRN-depleted cells were resistant to the antiproliferative effect of NSC 19630. NSC 19630 inhibited cell growth and induced apoptosis in a WRN-dependent manner. Spontaneous γ-H2AX foci, a marker of DSB, and PCNA foci, which form at sites of blocked replication forks or DNA repair, were elevated dramatically in cells by sublethal dosage of NSC 19630. Cotreatment of cells with NSC 19630 and the topoisomerase inhibitor TPT resulted in a synergistic inhibition of cell proliferation and induction of DSB. This report of a human DNA helicase inhibitor demonstrates the efficacy of screening small-molecule compound libraries for modulators of helicase function in cell-based assays. NSC 19630 and related compounds may be useful tools to interrogate further WRN function in cellular DNA metabolism and to target WRN to enhance existing or developing DNA-damaging anticancer therapies.
Mining the NCI60 cancer cell line database showed that NSC 19630 inhibited growth of cell lines from various cancers with a particularly strong representation from leukemia (five of six cell lines) and renal cancer (five of eight cell lines). However, additional cell lines from other cancer types (e.g., colon and breast) also were sensitive to the antiproliferative effect of NSC 19630. The NCI Diversity Set has been distributed widely and tested in many high-throughput screens. In a study published in 2005, Bykov et al. (21) reported that NSC 19630 and related maleimide analogs induced apoptosis in mutant p53 cell lines by restoring wild-type conformation and function to mutant p53. In the current study, NSC 19630 induced apoptosis in both wild-type and mutant p53 backgrounds, suggesting that the effect of the molecule on WRN helicase activity may have contributed to the effects observed in the p53-mutant cell lines reported previously. Because the NCI Diversity Set has been incorporated into chemical libraries used by the National Institutes of Health Roadmap initiative, a substantial screening database exists for member compounds, including NSC 19630, and is accessible online via PubChem (http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=227681&loc=ec_rcs). Although some activity was noted in particular screens, it is not apparent that such activity bears any relationship to the activity reported here.
Because NSC 19630 exerts its antiproliferative effects in the absence of exogenous DNA damage, the compound may interfere with the action of WRN on cellular DNA replication or repair intermediates that arise from endogenous DNA damage that accumulates in rapidly dividing cells. The difference in NSC 19630 concentration required for activity in cell culture versus biochemical assay may reflect conditions in the cell that render WRN more accessible to the compound compared with conditions used for in vitro biochemical reactions. Conceivably, NSC 19630 inhibits S-phase progression by forming a helicase-inactive WRN complex with other proteins or key DNA replication intermediates. Accumulation of DNA damage and PCNA foci, as well as ATM activation, are consistent with a model in which WRN helicase inhibition derails normal cellular DNA replication. WRN is unique among the RecQ helicases because the protein has dual helicase and exonuclease activities; however, the relative importance of WRN helicase and exonuclease activities in vivo is not well understood. Cell-based assays suggest a role for the WRN exonuclease activity in aspects of DSB end joining (22), but the enzyme is proposed to have pleiotropic functions. The WRN helicase inhibitor may provide a tool to investigate the relative contributions of WRN helicase and exonuclease activities in DNA repair or replication. Further exploration of the synergistic effects of NSC 19630 and related compounds with DNA-damaging therapies (e.g., chemotherapy drugs and radiation) are warranted to define the WRN-mediated pathway(s) important for proper DNA repair or replicational stress response. Indeed, the abilities of the WRN helicase inhibitor NSC 19630 to sensitize cells to a PARP inhibitor or to a potential telomere-directed cancer therapy represent advances in this area. For the PARP inhibitor, we hypothesize that accumulation of strand breaks leads to genomic instability and inhibition of cell proliferation because the compromised HR repair pathway is compromised when WRN helicase is inhibited. Synergistic interaction between NSC 19630 and TMS in telomerase-negative cells probably reflects the importance of WRN helicase activity in resolving G4 and other alternate structures such as the T-loop associated with telomeres or other G-rich sequence elements.
In cancer therapeutics there is heightened interest in strategies amplifying tumor-specific replicative lesions by inhibiting specific DNA repair pathways (23). Our laboratory has taken a particular interest in directed cancer therapy through helicase-targeted synthetic lethality (24). Tumor cells with DNA repair deficiencies, either BRCA or mismatch repair, might be particularly vulnerable to therapeutic strategies targeting WRN. Certainly there is clinical precedent for this approach in the case of PARP inhibitors (25). The cancer- or tumor-specific delivery of a small-molecule WRN helicase inhibitor remains an ongoing area of investigation. Future studies probably will address the use of helicase-specific inhibitors to target tumors with an existing DNA repair deficiency that rely more heavily on a WRN-dependent pathway for coping with endogenous or exogenously induced DNA damage.
Materials and Methods
Helicase Assays.
Compounds were obtained as 10-mM stocks dissolved in DMSO and were stored at −20 °C. Initially, a single 50-μM concentration of compound was tested for effect on WRN helicase activity. Compounds were diluted in DMSO to a concentration of 1,000 μM. Two microliters of diluted stock were added to 20 μL of reaction mixture so that final compound concentration was 50 μM. WRN helicase reactions using a forked duplex (19 bp) DNA substrate (0.5 nM) were initiated by the addition of 1.2 nM WRN and incubation at 37 °C for 15 min as described (26). RECQ1 (27) and FANCJ (28) helicase reactions were performed as described. Reaction conditions for BLM, DnaB, and UvrD were the same as for FANCJ. Helicase reactions were quenched, and reaction products were resolved on native 12% polyacrylamide gels as described (26).
Cell Proliferation Assays.
The effect of compounds on HeLa cell proliferation was determined using the WST-1 assay (Roche). Compounds were diluted in DMSO so that 10 μL of diluted stock in a 1-mL aliquot of HeLa cells (50,000 cells/mL) yielded a desired concentration of compound at 1% DMSO. HeLa cell cultures containing the specified compound were plated onto a 96-well microtiter plate at 5,000 cells per well in duplicate. As a control, 5,000 cells per well were seeded in medium containing 1% DMSO in duplicate. For background subtraction, plates lacking cells but containing medium and compound were used. Plates were incubated at 37 °C for 0–3 d, followed by addition of WST-1 reagent for 2 h. OD450 was measured at indicated time intervals using a microplate reader (Bio-Rad). Similar assays were performed to determine the effect of selected compounds on proliferation of NHF1-hTERT, U2OS, BLM-null (PSNG13) or BLM-corrected (PSNF5), MCF10A, T47D, MCF7, and HeLa WRN siRNA and control-siRNA cells.
Supplementary Material
Acknowledgments
We thank Dr. Bob Wersto and Jade Scheers of the National Institute on Aging Flow Cytometry Unit for assistance, Dr. Ian Hickson for providing purified recombinant BLM protein and BLM−/− (PSNG13) and BLM+/+ (PSNF5) cells, Dr. Daniel Kaplan for purified recombinant DnaB helicase, and Dr. Kazuo Shin-ya and Dr. Bernardo Reina San-Martin for TMS and for the PARP inhibitor KU0058948, respectively. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging, and National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006423108/-/DCSupplemental.
References
- 1.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 2.Gupta R, Brosh RM., Jr. Helicases as prospective targets for anti-cancer therapy. Anticancer Agents Med Chem. 2008;8:390–401. doi: 10.2174/187152008784220339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma S, Doherty KM, Brosh RM., Jr. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu WK, Hickson ID. RecQ helicases: Multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 5.Kudlow BA, Kennedy BK, Monnat RJ., Jr. Werner and Hutchinson-Gilford progeria syndromes: Mechanistic basis of human progeroid diseases. Nat Rev Mol Cell Biol. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- 6.Sidorova JM, Li N, Folch A, Monnat RJ., Jr. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin GM. Genetic syndromes in man with potential relevance to the pathobiology of aging. Birth Defects Orig Artic Ser. 1978;14:5–39. [PubMed] [Google Scholar]
- 8.Brosh RM, Jr., Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickson ID. RecQ helicases: Caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 10.Hübscher U. DNA replication fork proteins. Methods Mol Biol. 2009;521:19–33. doi: 10.1007/978-1-60327-815-7_2. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S, et al. WRN helicase and FEN-1 form a complex upon replication arrest and together process branch migrating DNA structures associated with the replication fork. Mol Biol Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez D, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 13.Tahara H, et al. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 14.Temime-Smaali N, et al. The G-quadruplex ligand telomestatin impairs binding of topoisomerase IIIalpha to G-quadruplex-forming oligonucleotides and uncaps telomeres in ALT cells. PLoS ONE. 2009;4:e6919. doi: 10.1371/journal.pone.0006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tauchi T, et al. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: In vitro and in vivo studies in acute leukemia. Oncogene. 2006;25:5719–5725. doi: 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- 16.Fry M, Loeb LA. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 17.Mohaghegh P, Karow JK, Brosh RM, Jr., Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 19.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 20.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 21.Bykov VJ, et al. Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J Biol Chem. 2005;280:30384–30391. doi: 10.1074/jbc.M501664200. [DOI] [PubMed] [Google Scholar]
- 22.Perry JJ, et al. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- 23.Helleday T. Amplifying tumour-specific replication lesions by DNA repair inhibitors - a new era in targeted cancer therapy. Eur J Cancer. 2008;44:921–927. doi: 10.1016/j.ejca.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 24.Aggarwal M, Brosh RM., Jr. Hitting the bull's eye: Novel directed cancer therapy through helicase-targeted synthetic lethality. J Cell Biochem. 2009;106:758–763. doi: 10.1002/jcb.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 26.Brosh RM, Jr., Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J Biol Chem. 2002;277:23236–23245. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- 27.Sharma S, et al. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 28.Gupta R, et al. Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J Biol Chem. 2005;280:25450–25460. doi: 10.1074/jbc.M501995200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.