Abstract
The cytoplasmic polyhedrosis virus (CPV) from the family Reoviridae belongs to a subgroup of “turreted” reoviruses, in which the mRNA capping activity occurs in a pentameric turret. We report a full atomic model of CPV built from a 3D density map obtained using cryoelectron microscopy. The image data for the 3D reconstruction were acquired exclusively from a CCD camera. Our structure shows that the enzymatic domains of the pentameric turret of CPV are topologically conserved and that there are five unique channels connecting the guanylyltransferase and methyltransferase regions. This structural organization reveals how the channels guide nascent mRNA sequentially to guanylyltransferase, 7-N-methyltransferase, and 2′-O-methyltransferase in the turret, undergoing the highly coordinated mRNA capping activity. Furthermore, by fitting the deduced amino acid sequence of the protein VP5 to 120 large protrusion proteins on the CPV capsid shell, we confirmed that this protrusion protein is encoded by CPV RNA segment 7.
Viruses of the family Reoviridae have a segmented dsRNA genome enclosed by single, double, or triple capsid shells. They share a similar transcription mechanism in which the inner capsids (core) remain intact and serve as shelters protecting the transcriptional process from antiviral defense mechanisms inside the cytoplasm of the host cell during replication of their dsRNA genomes. Despite the absence of significant sequence homology among different genera of the Reoviridae, the innermost capsid shells, which are formed by two conformers of the one protein, of all members of the Reoviridae and the majority of dsRNA viruses share common functions (1).
The “turreted” reoviruses have been defined as a subgroup of the Reoviridae due to their similar protein composition and capsid architecture. For this subgroup of reoviruses, a pentameric turret formed by five copies of turret proteins on fivefold vertex of the innermost shell functions in the catalysis of mRNA 5′ cap synthesis (2–5). Cryo-EM or crystal structures are available for four turreted virus genera of the family Reoviridae: the Cypovirus (6), Orthoreovirus (2), Oryzavirus (7), and Aquareovirus (8, 9) genera. These viruses have a segmented genome consisting of 10–12 linear segments of dsRNA and their hosts include vertebrates (orthoreoviruses and aquareovriuses), invertebrates (cypoviruses), and plants (oryzaviruses).
The cytoplasmic polyhedrosis virus (CPV) belongs to the genus Cypovirus and has a dsRNA genome of 10 segments. It is unique among dsRNA viruses in having a single capsid layer (10), which corresponds to the core of orthoreoviruses and functions as a stable mRNA synthesis machine in the cytoplasm of host cells that transcribes mRNAs from the segmented double-stranded RNA templates (11). CPV virions are embedded in polyhedra capable of surviving dehydration, freezing, and chemical treatments that would denature most proteins (12). The polyhedra dissolve in the alkaline midgut of their hosts and release infectious virions during the infection process. CPV can infect silkworms causing significant losses to the silk industry, but also has potential as a biocontrol agent for some insect pests. SDS-PAGE has shown that CPV has three major capsid proteins: VP1, VP3, and VP5 (13). Previous studies have demonstrated that VP1 is the capsid shell protein encoded by RNA segment 1 and that VP3 is the turret protein encoded by RNA segment 4 (1). The identity of the RNA segment encoding VP5 is still unclear. Although a previous study (6) has presented a Cα model of VP1 and a partial Cα model of VP3, the incomplete traces of these Cα models and the lack of full atomic models still prevent us from understanding the mechanism behind the role of the unique spike-like protein complex on the pentameric turret of CPV in transcription.
In this study, we report a full atomic model of the CPV capsid built from a 3D structure obtained using cryo-EM and single-particle reconstruction. Based on the density map and the amino acid sequences of the capsid proteins, we built atomic models for all three major capsid proteins. The atomic models were built from a cryo-EM structure using data obtained exclusively from a CCD camera. It is noteworthy that all three major capsid proteins of CPV share no detectable sequence identity with any other reovirus for which atomic models have been resolved. Our structure reveals how the nascent mRNA sequentially passes by the mRNA guanylyltransferase (GTase), methyltransferase-1 (MTase-1), and methyltransferase-2 (MTase-2) domains of the pentameric turret in order to fulfill the highly coordinated mRNA capping activity. The structure also shows that the inner shell protein VP1 has a similar structural topology to its counterparts in other reoviruses except for a protrusion domain on the outer capsid surface. By fitting the deduced amino acid sequence of the protein VP5 to large protrusion protein, referred to as clamp protein, on the CPV capsid shell (14), we confirmed that this protein is encoded by CPV RNA segment 7.
Results and Discussion
Structure Determination.
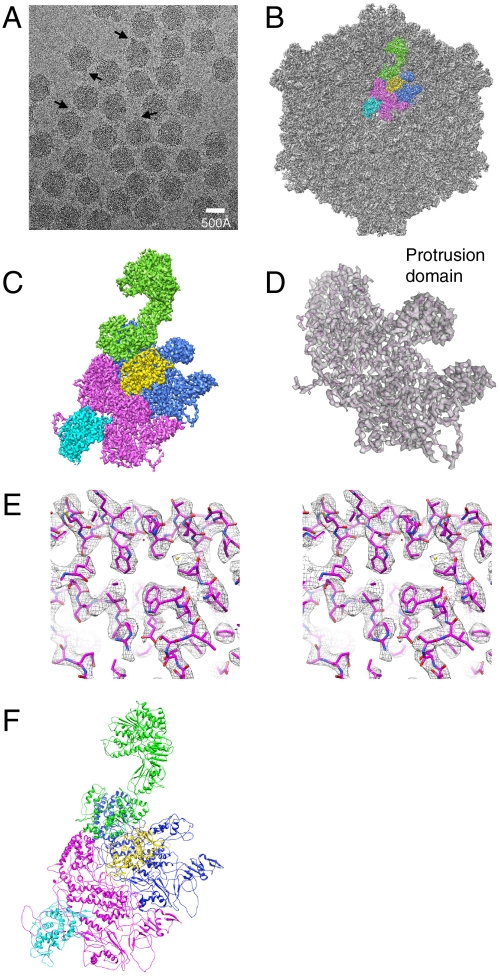
We have obtained the 3D structure of CPV by cryo-EM and single-particle reconstruction (Fig. 1 A–C). We reconstructed the structure of CPV using 29,000 virus particles selected from 1,600 micrographs collected by CCD with an FEI Titan Krios microscope. The estimated resolution of the reconstruction by Fourier shell correlation (FSC) criterion (15) is 4.3 Å at FSC = 0.5 and 3.9 Å at FSC = 0.143 (Fig. S1A). Based on the clarity of the resolved features, we estimated that the region at the inner radius, i.e., the capsid shell proteins, the clamp proteins, and the inner part of the turret proteins, is resolved to better than 3.9 Å (Fig. S1 B and C and Movies S1–S3). For the region at the outermost radius, a few side chains were not resolved as well as those in the inner region, but we can still follow the path of the main chain unambiguously (Fig. S1D). The density maps of the inner capsid proteins are of sufficient quality that we can follow nearly all the main chains of the capsid proteins. About 80% of the side chains of the amino acid residues were resolved, permitting us to build atomic models for all three major capsid proteins (Fig. 1 D–F and Movies S1–S3).
Fig. 1.
Overall structure of the CPV capsid. (A) A cryo-EM image of CPV. Arrowheads point to the heads of the spike-like complexes. (B) 3D reconstruction of the capsid of CPV. Capsid proteins VP3, VP1A, VP1B, VP5A, and VP5B in an asymmetric unit are segmented and colored with green, blue, magenta, yellow, and cyan, respectively. (C) Zoom-in view of an asymmetric unit. (D) Transparent view of the density map of VP1A with its atomic model (only the backbone is shown) superimposed. (E) Cross-eye stereo view of a part of VP1A density map (mesh) superimposed on the atomic model. (F) Atomic model (ribbon) of the asymmetric unit.
The CPV capsid exhibits a similar overall architecture to the core of orthoreoviruses (2) or aquareoviruses (9). The capsid shell of the CPV capsid is about 660 Å in diameter (excluding the fivefold turret) and there are 120 small protrusions and 120 large protrusions on the capsid shell. A total of 60 copies of turret proteins form 12 pentameric turrets, each of which has a size of 90 Å in height and 150 Å in width and sits on the top of one of the 12 fivefold vertices (16, 17). When the map is displayed at a slightly lower density threshold, a spike-like protein complex can be seen on each turret (13). The spike-like protein complexes can also be seen in the cryo-EM image in Fig. 1A.
Capsid Shell Protein VP1.
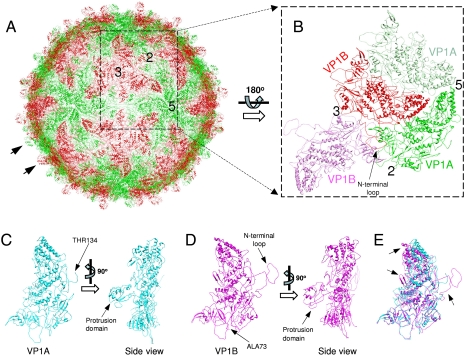
A total of 60 copies of VP1A and 60 copies of VP1B constitute the whole capsid shell of the CPV capsid (Fig. 2 A and B). By applying icosahedral symmetry to the models of VP1A and VP1B in an asymmetric unit, we obtained a model of the whole capsid shell (Fig. 2A). The organization of VP1A and VP1B in the capsid shell is identical to that of other reoviruses analyzed by X-ray crystallography or cryo-EM (2, 8, 9, 18–21). A previous structural study by Yu et al. has shown that the capsid shell protein VP1 has an overall thin-plate-like shape similar to that of orthoreovirus λ1 with the exception of a small protrusion domain (6). Our structure also shows that VP1 has a similar structural topology to the capsid shell proteins in other reoviruses in spite of a lack of detectable amino acid sequence identity. The distinct difference is that VP1 has an extra protrusion domain (residues 825–962) situated on the outer surface of the capsid shell (Figs. 1D and 2 A, C, and D). These protrusion domains are unique in single-layered cypoviruses and have not been observed in any reoviruses with double or triple layers. These outer layers serve to mediate the delivery of viral particles into the cytoplasm of host cells (22).
Fig. 2.
Whole capsid shell and two conformers of VP1. (A) The whole capsid shell formed by VP1A (green) and B (red). The arrows point to the protrusion domains of VP1A and B. (B) Higher magnification view of three copies of proteins in the box in A after 180° rotation shows the organization of two copies of VP1B and a copy of VP1A. The icosahedral 2-, 3-, and 5-fold axes are marked with 2, 3, and 5. (C and D) Ribbon views of atomic models of VP1A and B. (E) Superimposing VP1A (cyan) on VP1B (magenta). Their differences are indicated by arrowheads.
The overall structures of VP1A and VP1B are similar except for the N-terminal region and some peripheral regions that adapt asymmetric interactions (Fig. 2 C–E). Residues 1–133 of VP1A and residues 1–72 of VP1B were not resolved due to either flexibility or lack of fivefold symmetry. The inner capsid shell model shows that the resolved N-terminal segment (residues 73 to 145) of VP1B extends to the adjacent VP1A and VP1B (Fig. 2 B and D) in an “8”-like shape, and residues 80–82 turn back forming a β-sheet with residues 164–169 and residues 203–208 of the same copy of VP1B. The N-terminal segment of VP1B appears as a loop tethering the adjacent copy of VP1A and another copy of VP1B to itself (Fig. 2B). A similar type of organization to this kind of N-terminal extension can also be observed in capsid shell proteins of the other reoviruses: λ1 of orthoreoviruses (2), VP3 of aquareoviruses (9), and P3 of phytoreoviruses (23). These shell proteins are considered to enhance the stability of the capsid shell. However, the binding region of the VP1B N-terminal segment to the adjacent VP1A and VP1B appears broader than any of the other equivalent structures in these reoviruses. Indeed, it has been reported that VP1 is able to self-assemble into the inner capsid shell without the participation of other major capsid proteins (24), whereas the λ1 of orthoreoviruses has been shown to be unable to assemble in the absence of clamp protein σ2 (25). All of these observations suggest that reoviruses adopt a common assembly strategy to enhance the stability of their inner shell.
The N-terminal segments of the two conformers of the inner shell proteins that directly face the viral RNA genome have been shown to be of different lengths in all reoviruses with known structures. Previous studies have shown that the inner shell protein λ1 of orthoreovirus exhibits an affinity for both dsRNA and single-stranded RNA binding (26) and participates as a helicase during mRNA transcription (27). Sequence comparisons of our CPV with other cypoviruses and previous studies have shown that the N-terminal segment of the inner shell protein of the reovirus is the most divergent segment in the whole sequence (28, 29). Together, these results and the locations of the N-terminal segments imply that the N-terminal segment of reoviruses may play a role in mRNA transcription and capsid assembly.
The Turret and the Spike Form an Efficient mRNA Capping Machine.
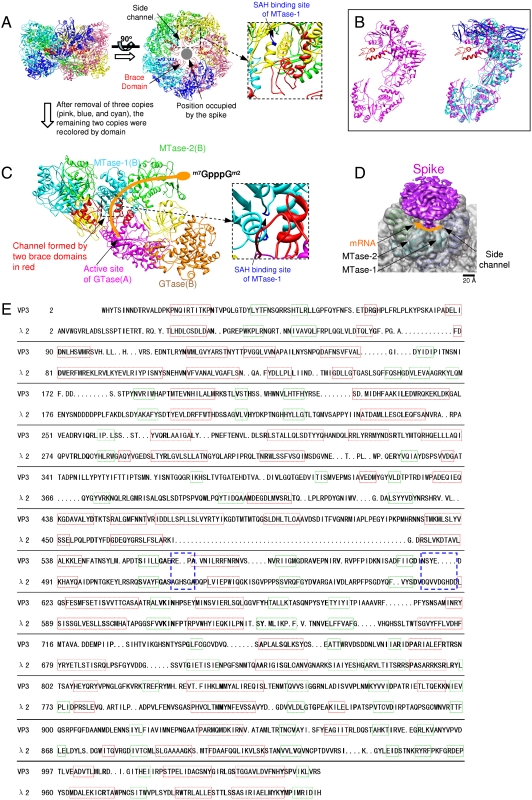
The turreted reoviruses cap their mRNA in the pentameric turrets that sit on top of the inner capsid shell before they release mRNA into the host cytoplasm, a process that is important for mRNA stability and efficient translation (2). The hollow pentameric turret of CPV on each icosahedral fivefold vertex consists of five copies of VP3 (Fig. 3 A–C). It is noteworthy that although VP3 shares no detectable amino acid sequence identity with its counterpart λ2 in orthoreoviruses, superimposing VP3 on λ2 reveals that they have a very similar structural topology except that VP3 does not have the flap-like structure present on the top of λ2 (Fig. 3B and Movie S4). Instead, there is a spike-like protein complex on the pentameric turret (13) (Fig. 3D). The density of the spike-like complex is visible only when the map is displayed at a slightly lower density threshold presumably due to a lack of icosahedral symmetry. Tertiary-structure-based amino acid sequence alignment (30) also shows that VP3 and λ2 have very similar distributions of α-helices and β-strands (Fig. 3E and Fig. S2A). The structural superimposition and the tertiary-structure-based amino sequence alignment between VP3 and λ2 reveals that VP3 also has four domains named according to the nomenclature of λ2 (2): the GTase domain (residues 1–366), the bridge domain (residues 367–418 and 726–832), the MTase-1 domain (residues 419–461 and 526–725), and the MTase-2 domain (residues 833–1058). The strikingly similar topology between VP3 and λ2 indicate that VP3 must serve the same functions as λ2 in catalyzing the three reactions in mRNA 5′ capping synthesis involving RNA GTase, 7-N-MTase, and 2′-O-MTase. However, VP3 has a small extra domain (residues 462–525) inserted in the MTase-1 domain that is not observed in λ2. We tentatively designate it the “brace domain” because five copies of the domain project to the center of the turret bracing the spike-like complex in the pentameric turret (Fig. 3 A–D). Five copies of the brace domain and the spike-like complex make the hollow turret look like a covered cavity where the RNA capping activity can take place.
Fig. 3.
Architecture of the pentameric turret and VP3. (A) Side view (Left) and top view (Middle) of the pentameric turret formed by the five copies of VP3 (in different colors). All five copies of the brace domains are colored in red. One of the five side channels formed by five copies of brace domain is pointed by an arrowhead. The position occupied by the spike protein is indicated by a gray plate. In the zoom-in view (Right), the related residues of the proposed SAH binding site are colored in blue. (B, Left) VP3 in purple except for the brace domain in red. (B, Right) VP3 is superimposed on orthoreovirus λ2 (in cyan except for the flap-like structure in blue). The brace domain of VP3 does not have a counterpart in λ2. (C) Two copies of VP3 [copy A (Left) and B (Right)] segmented from the left view in A. They have the same orientations as the two copies in green and yellow from the left view in A. The GTase domains in copy A and B are colored with magenta and orange for clarity. The Bridge, Brace, MTase-1, and MTase-2 domains of both copies of VP3 (A and B) are labeled and colored in yellow, red, cyan, and green, respectively. The pathway of mRNA is indicated by an orange strip. In the zoom-in view on the right, the related residues of the proposed SAH binding site are colored in blue. (D) Superimposing a 9-Å resolution density map of the turret (gray) on the top view of the turret in A after 45° rotation. The spike-like complex is in purple. The arrow in orange indicates the direction in which the mRNA passes by MTase-1 and MTase-2 successively after passing through the side channel formed by the brace domains. (E) Tertiary structure-based amino acid sequence alignment between CPV VP3 and orthoreovirus λ2. The α-helices are in red boxes and the β-strands are in green boxes. The related residues of the SAH binding site are in blue boxes. The residues after His1024 (flap-like structure) of λ2 are not shown.
Previous studies have identified the pathway taken by mRNA from the inner capsid chamber to the pentameric turret cavity and located the three mRNA capping enzymatic domains in the turret (2, 31). It has been reported that the channel for the nascent RNA of orthoreoviruses is not the pentameric channel at the fivefold axis on the inner shell but rather the channel located adjacent to it (31). The channel is formed by a copy of λ1B and two copies of λ1A. The outer end of the mRNA channel adjacent to the fivefold axis opens directly beneath the GTase domain in one copy of the turret protein λ2, and a groove on the underside of the λ2 leads into the turret cavity (31). This structural depiction for orthoreovirus is true for our CPV structure as well. A channel at the same position on the CPV capsid shell formed by a copy of VP1B and two copies of VP1A can be observed as well (Fig. S2B). In addition, in our CPV structure, the pentameric channel on the shell formed by five copies of VP1A connecting the inner capsid chamber and the turret cavity is blocked by a strong nodule-like structure that does not attach to any region around it (Fig. S2C). We attribute the structure to the bottom end of the spike-like complex. The absence of any attachment is probably due to the flexibility of the spike-like complex. Considering the same organization of the capsid shell proteins between CPV and orthoreovirus, and the highly conserved structural topology of the enzymatic domains between CPV VP3 and orthoreovirus λ2, we suggest that CPV has similar mRNA capping mechanisms.
How does the structure of CPV ensure that mRNA reaches the three enzymes in proper sequence? Our structure of the pentameric turret shows that five side channels with a diameter of 15–20 Å are formed by the five copies of the brace domain and that each side channel is right above a copy of the GTase domain and below the MTase-1 domain (Fig. 3 A, C, and D and Movies S5 and S6). Considering that the central channel formed by the five copies of the brace domain is blocked by the spike-like complex (Fig. 3A), these side channels are the only pathways connecting the pentameric GTase region and the pentameric MTase-1 region (Fig. 3C, Fig. S2D, and Movie S5). In this organization of the CPV turret, after the GTase reaction occurs in the GTase domain (VP3 A in Fig. 3C), the side channel guides the mRNA to the MTase-1 domain of another copy of VP3 first (VP3 B in Fig. 3C) and then to the MTase-2 domain of the same copy of VP3 (VP3 B in Fig. 3C). This structural organization of the CPV turret indicates that the MTase-1 is the 7-N-MTase and the MTase-2 is the 2′-O-MTase as N7 methylation precedes 2′O methylation (32). This assignment of MTase-1 and MTase-2 to 7-N-MTase and 2′-O-MTase in CPV is in agreement with that suggested by Reinisch et al. for orthoreovirus (2) although CPV has a different turret organization than that of orthoreovirus. Furthermore, the tertiary-structure-based amino acid sequence alignment (Fig. 3E) showed that the corresponding S-adenosyl-L-homocysteine (SAH) binding site of MTase-1 in the CPV VP3 is located just inside the exit of the proposed mRNA side channel (Fig. 3 A and C and Movie S6). The CPV cryo-EM image in Fig. 1A shows that the heads of the spike-like complexes occupy the top of the turrets. The locations of the exits of the side channels and the positions of the spike-like complexes indicate that the spike can affect the path that the mRNA takes to pass by the MTase-1 and MTase-2 domains when mRNA transcripts exit the side channels (Fig. 3D)
In contrast to the flap-like structure on the orthoreovirus λ2 or the aquareovirus VP1, the spike-like complex of CPV sits on the turret substituting for the flap-like structure of orthoreoviruses or aquareoviruses. The turret of orthoreoviruses and aquareoviruses can adopt both closed and open states (8, 20, 33–35). The biological mechanism behind the open/closed state of the turret is still elusive.
The Clamp Protein Is a Cleavage Product.
The 120 larger protrusion proteins (Fig. 1 B and C) are located on the outer surface of the CPV capsid shell. These kind of protrusion proteins exist on the inner capsid shells of all turreted reoviruses (orthoreovirus has 150 copies including 30 extra copies on its twofold axes) and serve as clamp proteins that enhance the stability of the capsid shell (16, 25). Conformers A and B of the clamp protein in an asymmetric unit differ only in the regions interacting with the capsid shell protein VP1 (Fig. S3). It is interesting that the clamp protein of CPV is smaller than its counterpart protein σ2 in orthoreoviruses and presents a totally different structural topology although they occupy similar positions on the capsid shell. Presumably, the divergence of clamp protein is because the clamp proteins of the multilayered turreted reoviruses have an additional function of binding the outer layer capsid proteins.
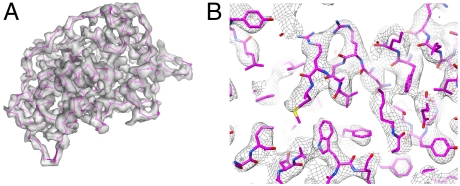
Which RNA segment encodes the clamp protein? Based on SDS-PAGE analysis (13), we postulated that a ∼31 kD VP5 band, whose molecular weight is a little smaller than that of orthoreovirus σ2 (47.1 kD), is the best candidate for the clamp protein of CPV. In addition, it has been suggested that the ∼31 kD VP5 of CPV is a cleavage product of protein P50 encoded by RNA segment 7 of CPV (36). Our model building for the clamp protein of CPV indicates that the size of the side chains in the density map of the clamp protein fits well with residues 1–291 of the P50 (Fig. 4, Fig. S1C, and Movie S3), confirming that the clamp protein is produced by the cleavage of P50. Summing the masses of residues 1–291 of P50 gives a protein with a molecular weight of 32.3 kD, consistent with the estimation of ∼31 kD determined by SDS-PAGE (13). The cleavage site might be at or close to Asn291-Ala292.
Fig. 4.
Structure of VP5A. (A) Transparent view of the density map of VP5A with its atomic model (only the backbone is shown) superimposed. (B) Zoom-in view of a part of VP5A (mesh) superimposed with its atomic model.
Conclusion
From the above results, we draw three conclusions. First, despite the major capsid proteins of CPV lacking significant amino acid sequence identity with other turreted reoviruses, CPV is structurally conserved in its enzymatic protein VP3 and capsid shell protein VP1, indicating that the turreted reoviruses adopt a common mechanism for mRNA transcription and capsid assembly. Second, the distinct structural organization on the top of the CPV pentameric turret indicates that CPV has adopted an effective strategy for guiding mRNA efficiently through the three mRNA capping reactions. Third, the clamp protein VP5 of CPV is produced by posttranslational cleavage of the protein P50 encoded by RNA segment 7.
Materials and Methods
Virus Purification.
Fourth-instar larvae of Bombyx mori were fed with mulberry leaves smeared with Bombyx mori CPV polyhedra for 6 h, then fed with the normal mulberry leaves. Silkworms were dissected before maturation. Midgut tissues were ground in a pestle and mortar and then filtered. The solution was centrifuged at 5,000 × g with 1% sodium dodecyl sulpate and sucrose at different density gradients (40%–50%–60%). The resulting white precipitate (purified polyhedra) was collected and diluted in distilled water. Virus particles were purified from the polyhedra as described (16).
Cryo-EM and 3D Reconstruction.
An aliquot of 3.5 μL purified CPV sample was applied to a 200 mesh holey grid (constructed by us and named GiG) and blotted for 4 s in a chamber at 100% humidity using an FEI Vitrobot Mark IV. Viruses were imaged with an FEI 300-kV Titan Krios cryoelectron microscope equipped with a Gatan UltraScan4000 (model 895) 16-megapixel CCD. The viruses were imaged at 300 kV with an absolute magnification of 125,390× corresponding to a pixel size of 1.19 Å, and the dose for each micrograph was about 20 e-/Å2. The contrast transfer function for each micrograph was determined by the ctfit program from the EMAN package (37) based on incoherently averaged Fourier transforms of each image. The orientations and centers of the virus particles were determined using the IMIRS package (38) based on the common-line strategy (39); then all selected particles were combined and reconstructed using a reconstruction program based on the icosahedral symmetry-adapted functions (ISAF) (40).
Atomic Model Building and Structural Analysis.
We built the atomic models of all proteins using Coot (41). The real-space electron density maps were converted to reciprocal space to generate simulated crystallographic diffraction data. The models were then refined in a pseudocrystallographic manner using Crystallography and NMR System (42), with a process that included simulated annealing refinement, crystallographic conjugate gradient minimization, and group B-factor refinement, using maximum likelihood refinement targets with amplitude and phase probability distribution. The geometry of the models was validated by PROCHECK (43) and MOLPROBITY (44) (see SI Text). Refinement statistics are given in Table S1. The individual proteins were segmented from the density map of the virus capsid using Chimera, and the density maps of the individual proteins and the PDB maps were superimposed and visualized using Chimera (45).
Supplementary Material
Acknowledgments.
We thank Professor Qibin Yang and Dr. Hongrong Liu (Xiangtan University, Xiangtan, Hunan, China) for providing the ISAF program, Zhongjun Hu for assistance in cryoelectron microscopy, and Dr. Joy Fleming for help with manuscript editing. We thank Kenneth Taylor and Kenneth Roux for critical reading of this manuscript. This research was supported by the National Basic Research Program of China (2010CB912403, 2006CB911002 and 2009CB825503), the National Natural Science Foundation of China (31070663, 30871828, U0832604, 90919041, 30872345, and 31000333), and the Chinese Academy of Sciences (KSCX2-YW-R-142).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The electron density maps and atomic models have been deposited in the EBI, http://www.ebi.ac.uk/pdbe, and Protein Data Bank, www.pdb.org (accession code EMD-5233 and PDB ID code 3IZ3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014995108/-/DCSupplemental.
References
- 1.Mertens P. The dsRNA viruses. Virus Res. 2004;101:3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Reinisch KM, Nibert M, Harrison SC. Structure of the reovirus core at 3.6 Å resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 3.Fausnaugh J, Shatkin AJ. Active site localization in a viral mRNA capping enzyme. J Biol Chem. 1990;265:7669–7672. [PubMed] [Google Scholar]
- 4.Luongo CL, Reinisch KM, Harrison SC, Nibert ML. Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein lambda 2. J Biol Chem. 2000;275:2804–2810. doi: 10.1074/jbc.275.4.2804. [DOI] [PubMed] [Google Scholar]
- 5.Luongo CL, Contreras CM, Farsetta DL, Nibert ML. Binding site for S-adenosyl-L-methionine in a central region of mammalian reovirus lambda 2 protein—Evidence for activities in mRNA CAP methylation. J Biol Chem. 1998;273:23773–23780. doi: 10.1074/jbc.273.37.23773. [DOI] [PubMed] [Google Scholar]
- 6.Yu XK, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazaki N, et al. Structural evolution of Reoviridae revealed by Oryzavirus in acquiring the second capsid shell. J Virol. 2008;82:11344–11353. doi: 10.1128/JVI.02375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 Å Cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng LP, et al. Backbone model of an Aquareovirus virion by Cryo-electron microscopy and bioinformatics. J Mol Biol. 2010;397:852–863. doi: 10.1016/j.jmb.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill CL, et al. The structure of a cypovirus and the functional organization of dsRNA viruses. Nat Struct Biol. 1999;6:565–568. doi: 10.1038/9347. [DOI] [PubMed] [Google Scholar]
- 11.Nibert ML, Baker TS. CPV, a stable and symmetrical machine for mRNA synthesis. Structure. 2003;11:605–607. doi: 10.1016/s0969-2126(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 12.Coulibaly F, et al. The molecular organization of cypovirus polyhedra. Nature. 2007;446:97–101. doi: 10.1038/nature05628. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, et al. Visualization of protein-RNA interactions in cytoplasmic polyhedrosis virus. J Virol. 1999;73:1624–1629. doi: 10.1128/jvi.73.2.1624-1629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou ZH, Zhang H, Jakana J, Lu XY, Zhang JQ. Cypovirus structure at 8 Å by electron cryomicroscopy—Structural basis of capsid stability and multifunctional RNA processing apparatus. Biophys J. 2003;84:281a. doi: 10.1016/s0969-2126(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 15.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Zhou ZH, Zhang H, Jakana J, Lu XY, Zhang JQ. Cytoplasmic polyhedrosis virus structure at 8 Å by electron cryomicroscopy: Structural basis of capsid stability and mRNA processing regulation. Structure. 2003;11:651–663. doi: 10.1016/s0969-2126(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 17.Xia Q, Jakana J, Zhang JQ, Zhou ZH. Structural comparisons of empty and full cytoplasmic polyhedrosis virus—Protein-RNA interactions and implications for endogenous RNA transcription mechanism. J Biol Chem. 2003;278:1094–1100. doi: 10.1074/jbc.M205964200. [DOI] [PubMed] [Google Scholar]
- 18.Grimes JM, et al. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 19.McClain B, Settembre E, Temple BRS, Bellamy AR, Harrison SC. X-ray crystal structure of the Rotavirus inner capsid particle at 3.8 Å resolution. J Mol Biol. 2010;397:587–599. doi: 10.1016/j.jmb.2010.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng LP, Fang Q, Shah S, Atanasov IC, Zhou ZH. Subnanometer-resolution structures of the grass carp reovirus core and virion. J Mol Biol. 2008;382:213–222. doi: 10.1016/j.jmb.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakagawa A, et al. The atomic structure of Rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure. 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC. Structure of the reovirus membrane-penetration protein, mu 1, in a complex with its protector protein, sigma 3. Cell. 2002;108:283–295. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagiwara K, et al. The amino-terminal region of major capsid protein P3 is essential for self-assembly of single-shelled core-like particles of Rice dwarf virus. J Virol. 2004;78:3145–3148. doi: 10.1128/JVI.78.6.3145-3148.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagiwara K, Naitow H. Assembly into single-shelled virus-like particles by major capsid protein VP1 encoded by genome segment S1 of Bombyx mori cypovirus 1. J Gen Virol. 2003;84:2439–2441. doi: 10.1099/vir.0.19216-0. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, et al. The hydrophilic amino-terminal arm of reovirus core shell protein lambda 1 is dispensable for particle assembly. J Virol. 2002;76:12211–12222. doi: 10.1128/JVI.76.23.12211-12222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bisaillon M, Lemay G. Molecular dissection of the reovirus lambda 1 protein nucleic acids binding site. Virus Res. 1997;51:231–237. doi: 10.1016/s0168-1702(97)00092-0. [DOI] [PubMed] [Google Scholar]
- 27.Bisaillon M, Bergeron J, Lemay G. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus lambda 1 protein. J Biol Chem. 1997;272:18298–18303. doi: 10.1074/jbc.272.29.18298. [DOI] [PubMed] [Google Scholar]
- 28.Shen PC, et al. Genetic variation of the lambda A and lambda C protein encoding genes of avian reoviruses. Res Vet Sci. 2007;83:394–402. doi: 10.1016/j.rvsc.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Harrison SJ, et al. Mammalian reovirus L3 gene sequences and evidence for a distinct amino-terminal region of the lambda 1 protein. Virology. 1999;258:54–64. doi: 10.1006/viro.1999.9707. [DOI] [PubMed] [Google Scholar]
- 30.Gille C, Frommel C. STRAP: Editor for STRuctural Alignments of Proteins. Bioinformatics. 2001;17:377–378. doi: 10.1093/bioinformatics/17.4.377. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat Struct Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furuichi Y, Muthukrishnan S, Tomasz J, Shatkin AJ. Mechanism of formation of Reovirus messenger-RNA 5'-terminal blocked and methylated sequence, M7gpppgmpc. J Biol Chem. 1976;251:5043–5053. [PubMed] [Google Scholar]
- 33.Chandran K, et al. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. J Virol. 2001;75:5335–5342. doi: 10.1128/JVI.75.11.5335-5342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dryden KA, et al. Early steps in Reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation—analysis of virions and subviral particles by cryoelectron microscopy and image-reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nason EL, Samal SK, Prasad BVV. Trypsin-induced structural transformation in aquareovirus. J Virol. 2000;74:6546–6555. doi: 10.1128/jvi.74.14.6546-6555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagiwara K, Matsumoto T. Nucleotide sequences of genome segments 6 and 7 of Bombyx mori cypovirus 1, encoding the viral structural proteins V4 and V5, respectively. J Gen Virol. 2000;81:1143–1147. doi: 10.1099/0022-1317-81-4-1143. [DOI] [PubMed] [Google Scholar]
- 37.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 38.Liang YY, Ke EY, Zhou ZH. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J Struct Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 39.Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles—The uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 40.Liu HR, et al. Symmetry-adapted spherical harmonics method for high-resolution 3D single-particle reconstructions. J Struct Biol. 2008;161:64–73. doi: 10.1016/j.jsb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 44.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen EF, et al. UCSF chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.