Abstract
Cell membranes are comprised of multicomponent lipid and protein mixtures that exhibit a complex partitioning behavior. Regions of structural and compositional heterogeneity play a major role in the sorting and self-assembly of proteins, and their clustering into higher-order oligomers. Here, we use computer simulations and optical microscopy to study the sorting of transmembrane helices into the liquid-disordered domains of phase-separated model membranes, irrespective of peptide–lipid hydrophobic mismatch. Free energy calculations show that the enthalpic contribution due to the packing of the lipids drives the lateral sorting of the helices. Hydrophobic mismatch regulates the clustering into either small dynamic or large static aggregates. These results reveal important molecular driving forces for the lateral organization and self-assembly of transmembrane helices in heterogeneous model membranes, with implications for the formation of functional protein complexes in real cells.
Keywords: fluorescence microscopy, lipid rafts, membrane proteins, molecular dynamics, linactants
The heterogeneity of biological membranes plays an important role in cellular function (1, 2). Despite experimental progress in recent years (3), the characterization of lateral organization in biological membranes, however, remains challenging due to the lack of tools to study fluctuating nanoscale assemblies in living cells (4–6). Model membranes (7–10) as well as isolated plasma membranes (11, 12) are more frequently studied, because large-scale phase separation can be observed in these systems. In particular, at cholesterol concentrations reminiscent of biological membranes (10–30 mol % cholesterol), ternary mixtures of saturated and unsaturated lipids can segregate into coexisting lipid domains of different fluidity, a liquid-ordered (Lo) and a liquid-disordered (Ld) phase. Probing the structural and dynamical properties of these fluid domains has received a lot of attention, as it is presumably linked to the formation of lipid nanodomains (“rafts”) in real cells (13, 14).
The structure and function of membrane proteins is intimately connected with their lipid environment (15, 16). Because of the heterogeneity of the cell membrane, proteins partition between different lipid domains, are recruited to specific locations, and form functional complexes (17–19). This lateral organization is very important for various cellular processes, such as membrane fusion (20, 21), protein trafficking (22), and signal transduction (23–25). Although lipids and integral membrane proteins are well studied by themselves, the molecular properties that determine the specific interactions between them remain poorly understood. Interactions important for protein sorting and self-assembly are (specific) protein–protein and protein–lipid forces, and indirect lipid-mediated forces. The latter category includes, for instance, forces due to entropic confinement of lipid chains and forces arising from the mismatch between the hydrophobic parts of the protein and the bilayer. Recent computational studies highlight the importance of lipid-mediated interactions for membrane protein clustering (26–29).
Integral membrane proteins have a large structural variety, but all contain hydrophobic regions that span the bilayer; these regions are often single α-helices or α-helical bundles. Many properties of membrane proteins are thus determined by the interactions between these transmembrane (TM) helices and the surrounding lipids. Here, we investigate the molecular interactions between TM helices, specifically WALP peptides, and heterogeneous model membranes with coexisting Lo and Ld lipid domains. Synthetic WALP peptides were designed as probes to study the behavior of TM domains in model systems, such as their mode of insertion, aggregation, and partitioning (30, 31).
WALPs form α-helical TM domains comprised of alternating alanine and leucine residues, flanked by tryptophanes to anchor the peptide at the membrane interface. Varying the length of the peptide’s hydrophobic stretch allows for tailoring of desired properties, such as the hydrophobic mismatch. WALPs and other TM peptides were observed to be excluded from Lo domains in model membranes, irrespective of the hydrophobic mismatch, see, e.g., refs. 32 and 33. It was speculated that the TM peptides disturb the tight packing of lipids and cholesterol in Lo (14); however, the underlying thermodynamic driving forces are not well understood. Our study combines both large-scale molecular dynamics (MD) computer simulations of planar membranes and confocal fluorescence microscopy of giant unilamellar vesicles (GUVs) to investigate peptide sorting under different hydrophobic mismatch conditions. In addition, the simulations yield insight into the clustering of the peptides and the thermodynamic driving forces for lateral sorting.
Results
Simulations Show Partitioning of TM Helices into Ld Domain.
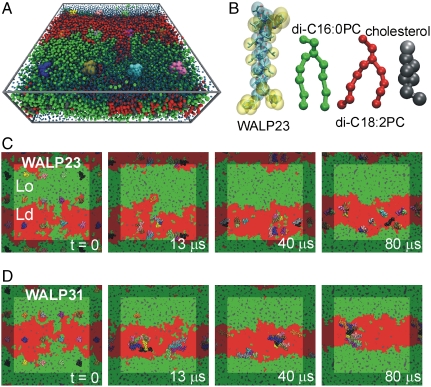
Fig. 1 A and B shows the simulation system, consisting of 12 WALP peptides inserted into a lipid bilayer patch and solvated by explicit water. All molecules are represented at a coarse-grained (CG) level, retaining near-atomic resolution. The bilayer consists of a ternary mixture of dipalmitoylphosphatidylcholine (di-C16∶0PC), dilinoleoylphosphatidylcholine (di-C18∶2PC), and cholesterol in a 0.42∶0.28∶0.30 molar ratio, about 2,000 lipids in total. The membrane is separated into two coexisting fluid domains that can spontaneously form from a random lipid distribution, as evidenced in previous simulations (34). Characterization of these domains revealed that the Lo membrane domain is mainly composed of the fully saturated di-C16∶0PC lipid and cholesterol, whereas the Ld domain is enriched in the doubly unsaturated di-C18∶2PC lipid and has a reduced cholesterol content. The Lo domain is thicker (PC headgroup–headgroup distance d0 ca. 4.6 nm) than the Ld domain (d0 ca. 3.8 nm). In the current work, we studied the partitioning of two different peptides, namely WALP23 [AW2L-(AL)8-W2A] and WALP31 [AW2L-(AL)12-W2A] in these domains. The shorter WALP23 (peptide length lp ca. 3.4 nm) is expected to fit well into the Ld domain, whereas the longer WALP31 (lp ca. 4.6 nm) could span the Lo domain and thus would display a rather large positive hydrophobic mismatch in the Ld domain. As shown in Fig. 1A, monomeric peptides were initially inserted into the membrane. CG-MD simulations using the MARTINI force field (35, 36) were then carried out for 80 μs for the WALP23 and WALP31 systems, respectively.
Fig. 1.
Simulation of the sorting and clustering of WALP peptides in a model bilayer with coexisting fluid domains. (A) WALP peptides (colored spheres) embedded in a ternary mixture of di-C16∶0PC (green), di-C18∶2PC (red), and cholesterol (gray), solvated by water (blue). (B) Coarse-grained representation of WALP23 (cyan and yellow), di-C16∶0PC (green), di-C18∶2PC (red), and cholesterol (gray), shown as spheres and sticks. The atomistic structure of the peptide is also shown (sticks). (C and D) Sorting and clustering of WALP23 (C) and WALP31 peptides (D) in the disordered lipid domain.
Fig. 1 C and D shows that the peptides adopt a distinct nonhomogeneous distribution during the simulation. Both the WALP23 and WALP31 peptides are expelled from the Lo domain and partition into the Ld domain, irrespective of their length. For the membrane studied here, in which the Ld domain spans about 10 nm, this sorting process is completed within 2 μs, after which most peptides are incorporated into the Ld phase. We never observed that peptides (transiently) returned to the Lo phase during the time span of our simulations, suggesting a rather large driving force for peptide sorting into the Ld phase. Attaching two palmitoyl anchors to the N terminus of WALP23 does not alter the partitioning, in agreement with experiments (32). The addition of eight palmitoyl anchors, however, leads to preferential binding to the domain boundary (see SI Appendix). The lateral segregation of the peptides leads to an increase of the effective peptide/lipid ratio to about 1∶50 in the disordered domain, where dynamic self-assembly of the peptides into oligomeric aggregates is observed. The size and stability of the aggregates depends on the mismatch, as is addressed below.
Confocal Microscopy Verifies That TM Helices Prefer the Ld Domain.
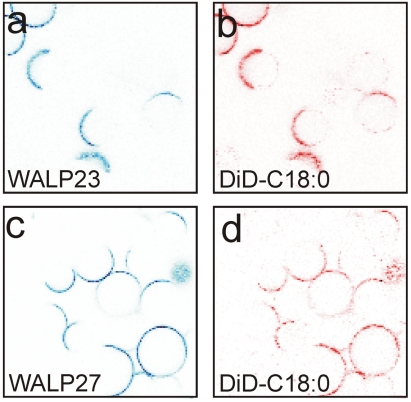
Next, we experimentally tested the predictions of our simulations by optical fluorescence microscopy. We incorporated Alexa Fluor 488-labeled WALP peptides into GUVs composed of cholesterol, di-C18∶1PC, and egg-sphingomyelin (1∶1∶1). The fluorescent lipid dye DiD-C18∶0 was added as a marker for the Ld phase (8, 11). First, we incorporated labeled WALP23 into the GUVs and recorded fluorescence images (Fig. 2A, blue channel). The confocal image in Fig. 2B shows the fluorescence of the Ld marker DiD-C18∶0 (red channel), confirming the phase separation. The colocalization of the WALP23 and DiD-C18∶0 fluorescence indicates that the peptides partition into the Ld domains (containing mainly di-C18∶1PC) and are expelled from the Lo domains (composed of mainly cholesterol and egg-sphingomyelin). In a next step, we repeated the experiments with a membrane in which di-C18∶1PC was replaced by di-C14∶1PC, thereby decreasing the hydrophobic thickness of the Ld phase by about 0.6 nm. To maximize the hydrophobic mismatch, we incorporated the WALP27 peptide, which is 0.6 nm longer than WALP23. Thus, taken together, the difference is 1.2 nm, identical to the length difference between the WALP23 and WALP31 peptides used in our MD simulations. Also here, the Lo domains match the hydrophobic length of the longer peptide much better than the thinner Ld domains. The confocal images shown in Fig. 2 C and D reveal that Alexa Fluor 488-labeled peptides and DiD-C18∶0 still colocalize, demonstrating that WALP peptides prefer the Ld phase over the Lo phase irrespective of the hydrophobic mismatch. These confocal images confirm our MD simulations and agree with previous detergent extraction experiments (32).
Fig. 2.
Confocal images of GUVs containing Alexa Fluor 488-labeled WALP peptides. (A and B) The vesicles were composed of di-C18∶1PC/cholesterol/egg-sphingomyelin (1∶1∶1) and WALP23 (blue fluorescence channel) and DiD-C18∶0 (red). (C and D) The vesicles contained di-C14∶1PC/cholesterol/egg-sphingomyelin (1∶1∶1) and WALP27 (blue) and DiD-C18∶0 (red).
Free Energy Calculations.
To more thoroughly understand the underlying molecular driving forces for the observed lateral sorting of the peptides, we calculated, by means of thermodynamic integration (TI), the partitioning free energies of a single WALP peptide between the Lo and Ld phase (for details, see SI Appendix). The results are summarized in Table 1. The free energies of transferring a peptide from the Lo to the Ld domain are ΔGLo/Ld = -63 ± 8 kJ/mol and -58 ± 8 kJ/mol for WALP23 and WALP31, respectively. The strong preference of both peptides for the Ld phase explains why during the free simulations, peptides once incorporated into the Ld domain never returned to the Lo domain. The transfer free energies ΔGLo/Ld result from large counteracting enthalpic and entropic contributions (Table 1). The enthalpy favors partitioning into the Ld phase, whereas the entropy would favor partitioning into Lo. This can be rationalized by the disorder introduced when a TM peptide is accommodated in the well-ordered Lo phase. However, lipid–lipid and lipid–cholesterol packing is optimal in the Lo phase, and any foreign inclusion comes with a large enthalpic penalty that is not outweighed by this increase in entropy. By contrast, the lipids in the Ld domain are already strongly disordered, and incorporation of a TM helix thus does not yield a significant increase in entropy in this case. At the same time, due to the less densely packed lipids in Ld, the inclusion of the TM helix is enthalpically more favorable in Ld than in Lo, again outplaying the entropic contribution.
Table 1.
Free energies, enthalpies, and entropies at 300 K of partitioning of single peptides between the Lo and Ld phases
All values are in kJ mol-1.
*ΔGLo/Ld was obtained from TI simulations.
†ΔHLo/Ld was estimated from the system’s potential energy, and -TΔSLo/Ld = ΔGLo/Ld - ΔHLo/Ld. Standard errors are given (see SI Appendix). Note, the decomposition of the free energy into enthalpy and entropy, and thus the absolute values of ΔHLo/Ld and ΔSLo/Ld, should be interpreted at a qualitative level (see SI Appendix).
The length of the peptide, and hence mismatch, does also play a role. The enthalpic gain of moving the shorter WALP23 peptide from the Lo to the Ld domain is significantly higher compared to that of the longer WALP31 peptide, which shows no hydrophobic mismatch with respect to the Lo phase (-594 kJ/mol and -276 kJ/mol for WALP23 and WALP31, respectively; see Table 1). These results are backed up by the calculation of lipid tail order parameters from CG and atomistic simulations; see SI Appendix. Taken together, for both peptides, the free energy gain upon moving from Lo to Ld results from the strong enthalpic contribution due to peptide-induced breaking up of the tight lipid packing in the Lo domain, which cannot be overcome by the entropic preference for the Lo phase.
Clustering of Peptides Depends on Mismatch.
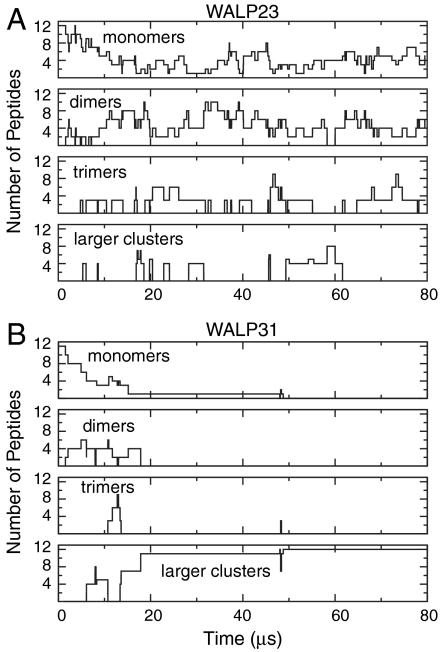
Fig. 3 A and B quantifies the cluster formation of the WALP23 and WALP31 peptides, respectively, revealing the distinct aggregation behavior of the two peptides. The shorter WALP23 peptides display a very dynamic clustering behavior. After an initial equilibration phase of ca. 15 μs, WALP23 peptides are mostly present as monomers, dimers, and trimers, and clusters continuously reorganize by exchanging peptides, with average cluster lifetimes of a few microseconds. Calculating the average number of monomers, dimers, and trimers over the last 60 μs of the trajectory yields nmon∶ndim∶ntrim = 2.8∶2.0∶1.0. Thus, the populations of monomer and dimer are similar at the effective peptide/lipid ratio of about 1∶50 in the Ld phase. For the dimers as the most abundant aggregate, and assuming that the law of mass action can be applied, we estimate the equilibrium association constant according to (37) K = (ndim/ntot)/(nmon/ntot)2, where ni is the average number of moles of species i in the Ld domain. Using ntot = 540 (number of lipids in the Ld domain), we obtain K ≈ 140 ± 60. The dimerization free energy is then ΔG = -RT ln K = -12 ± 2 kJ/mol, in agreement with experiments (38) and other simulations (39); see SI Appendix. Under experimental conditions, WALP peptides are found to be monomeric at low peptide concentrations (31). However, at peptide/lipid ratios of about 1∶25 to 1∶50, clustering (dimerization) was observed, a process that is additionally fostered by the presence of cholesterol (40). Thus, the presence of weakly bound WALP23 dimers as found in our simulations is in agreement with the available experimental data.
Fig. 3.
Clustering of WALP peptides during simulations. The number of WALP23 (A) or WALP31 (B) peptides that are present as a monomer, or part of a dimer, trimer, or a larger cluster are plotted over time. Two WALPs were considered bound if the distance between any two particles of the peptides was smaller than 0.7 nm.
Most of the WALP23 dimers observed in the simulation adopt a right-handed configuration, with a negative helix crossing angle θ (see SI Appendix). Although left-handed packing of helices is more abundant in known membrane protein structures (41, 42), right-handedness is also found, for example, in the glycophorin A (GpA) dimer (43). Similar to WALP, GpA is a single transmembrane α-helix, and its dimer has a rather small interface that does not involve hydrogen bonds or salt bridges. In the WALP23 trimers formed during the simulation, we mostly observed a linear arrangement of the peptides (i.e., the two outermost peptides within a trimer are not in direct contact with each other), again with a preference for a right-handed configuration. Left-handed helix–helix contacts are also occasionally present during the simulation; however, such aggregates are rather short-lived and rapidly dissociate again.
In contrast to WALP23, the longer WALP31 tends to form large clusters. In addition, the clustering behavior is much less dynamic as compared to WALP23, with fewer association/dissociation events observed. After 13 μs, a heptameric and a tetrameric cluster are formed, and a single WALP31 is not yet incorporated into the Ld domain (orange peptide, Fig. 1D); this peptide joins the heptameric cluster after 50 μs (Fig. 3B). Because the growing size of the clusters slows down their lateral diffusion, it takes until t = 70 μs for the encounter of the two clusters to occur, yielding a 12-mer that is stable for the rest of the simulation time. The large WALP31 cluster is rather unstructured; that is, it has no clear tertiary structure or long-range order. As shown in Fig. 1, the clusters occasionally approach the domain boundary, but get reincorporated into the core of the disordered domain, demonstrating that the clusters, like the monomers, also prefer the Ld over the Lo domain.
To check and validate our coarse-grained simulations, we transformed a snapshot (after t = 40 μs) from the CG-MD simulation of the WALP23 system to its underlying atomistic representation (44) and carried out a 100-ns atomistic MD simulation (see SI Appendix). During this simulation, the majority of the WALP23 clusters stay bound (a single dimer dissociates), consistent with the behavior shown in Fig. 3. The atomistic simulations thus support the clustering observed in the coarse-grained simulations.
Discussion
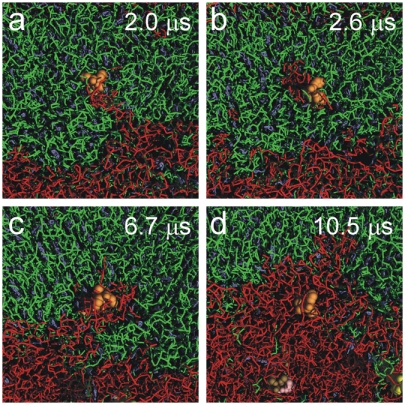
Our computational modeling data as well as the confocal microscopy results reveal that model transmembrane helices, such as the WALP peptides, do not like to be embedded in the liquid-ordered phase. Rather, they are observed to spontaneously partition into the liquid-disordered phase. The near-atomic resolution of our simulation approach allows us to study the sorting process in molecular detail. This is further illustrated in Fig. 4, showing a time series of the incorporation of a particular WALP23 peptide into the Ld domain. Although most peptides are rapidly expelled from the Lo domain, this particular peptide gets temporarily trapped and enters the Ld domain only after 10 μs. The incorporation of this peptide is facilitated by the transient formation of an island of unsaturated lipids pervading the ordered domain, emphasizing that it is the collective motion of both lipids and peptides that plays a role in the lateral sorting process.
Fig. 4.
Proteins moving lipids moving proteins. A WALP23 (orange spheres), initially located in the Lo phase (green), gets incorporated into the Ld phase (red) through concerted peptide and lipid motion. (A) A peninsula of di-C18∶2PC lipids bridges the peptide to the core of the Ld domain. (B) The peptide gathers an island of di-C18∶2PC lipids (red) in its environment. (C and D) The di-C18∶2PC island merges with the Ld domain, and the peptide subsequently diffuses away from the domain boundary.
Thus far, no TM peptides or proteins that partition into Lo domains in model membranes were reported (33, 45–49), irrespective of the amino acid sequence. However, the underlying molecular mechanisms and thermodynamic driving forces were poorly understood. Our simulations reveal that the driving force for this observed sorting is the enthalpic cost associated with the presence of a cylindrical object (the TM helix) inside the ordered lipid phase. Thus, although we used synthetic WALP peptides as generic models, our proposed mechanism is general and likely to be relevant for protein sorting, also in vivo. To test the generality, we additionally studied the partitioning of the α-helical TM domain of the syntaxin 1A protein (TM-Sx1A, residues 259–288) and of the linker for activation of T cells (TM-LAT, residues 2–32); see SI Appendix. LAT is frequently used as a raft marker, but differential partitioning behavior in model membranes and in plasma membranes was reported (48, 50). In our simulations, TM-Sx1A partitions into the Ld domain, in agreement with experimental findings (21, 45, 51–53). TM-LAT also prefers Ld, in agreement with experiments on model membranes (48). However, unlike TM-Sx1A, we find the larger TM-LAT clusters to be located close to the domain boundary interface.
In living cells, the differences between Lo and Ld domains are less pronounced than they can be in model membranes, due to the more complex lipid composition and high protein concentration in the plasma membrane, which may hamper the tight lipid packing in Lo (49, 50). In particular, in the model membrane used in our simulations, the almost complete depletion of unsaturated lipids from Lo yields a very ordered domain. Furthermore, the Ld domain is rather strongly disordered due to the doubly unsaturated di-C18∶2PC, explaining the observed large free energy differences (Table 1). McIntosh and coworkers (54) measured through detergent extraction experiments much smaller transfer free energies of TM peptides between lipid domains in bilayers comprised of dioleoylphosphatidylcholine, cholesterol, and sphingomyelin (1∶1∶1), which are closer to the critical point and might thus more closely resemble the situation in cell membranes. They report transfer free energies of only about 2–5 kJ/mol for the peptides P-23, P-29, and P-31 (α-helical peptides with a hydrophobic TM part and charged residues at the termini). However, such a quantitative analysis of detergent extraction experiments has to be taken with care (55), and larger free energy differences were estimated based on elastic theories (56).
For integral membrane proteins, a widely accepted assumption is that sorting is controlled by the length of the TM domains (18). This mechanism is supported by the finding that the TM parts of proteins in the Golgi membrane (enriched in unsaturated lipids) are on average spanned by 15 amino acids, whereas TM domains of plasma membrane proteins comprise on average 20 amino acids (22). We observed a preference for the Ld phase regardless of the length of the peptide and hence the degree of hydrophobic mismatch. One key difference between single TM helices, such as WALP peptides, and large integral membrane proteins is that the former can alleviate the energetic penalty due to positive hydrophobic mismatch through adopting a large tilt angle (according to our simulations, the average tilt angle of WALP31 in the Ld domain is indeed substantial, around 45°; see SI Appendix). The tilting of large integral membrane proteins as a whole is energetically unfavorable. Rather, they may induce stronger local bilayer deformations than single α-helices or adapt their hydrophobic thickness through structural rearrangements, such as changing the tilt of individual α-helices relative to each other. The associated free energy costs influence partitioning. To probe the effect of helix tilt, an additional simulation was carried out in which we restricted tilting of WALP31. The peptide preferentially adsorbs at the Lo/Ld domain interface (see SI Appendix). This result confirms that hydrophobic mismatch also plays an important role for the sorting of integral membrane proteins, and we predict that proteins that do not fit well into the Ld domain may display “linactant” (57, 58) behavior at the Lo/Ld interface. Also TM peptides with several palmitoyl anchors can accumulate at the domain interface (see SI Appendix). However, specific lipid factors, such as raft gangliosides, can also be important for sequestering into the Lo phase (12). Finally, protein–protein interactions can further alter the driving forces for Lo/Ld partitioning (59, 60) by modulating the number of lipid–protein contacts. Clustering of transmembrane proteins and peptides may occur as a response to hydrophobic mismatch, as it reduces the number of lipids bound to the nonmatching membrane inclusion. This was confirmed by experiments on various transmembrane proteins and peptides (31). The increased tendency to cluster observed for WALP31 supports this idea of mismatch driven self-aggregation.
By combining molecular dynamics simulations and confocal microscopy, we show why transmembrane helices partition into disordered lipid domains in heterogeneous model membranes, irrespective of helix–lipid mismatch. Our study demonstrates that computer simulations, in combination with experiments, can not only contribute to unravel the molecular forces driving complex phase behavior in biological systems, they are also poised to become a powerful tool for predicting the localization of proteins and lipids in heterogeneous cellular environments and foster the design of bioinspired molecules with tailored properties.
Materials and Methods
Molecular Dynamics Simulations.
All simulations were carried out with the Gromacs package, version 4.0.5 (61). In the coarse-grained simulations, the MARTINI force field (35, 36) was used. An integration time step of 20 fs was applied, together with the standard settings for the nonbonded interactions. Constant particle number, pressure, temperature ensembles were simulated within periodic boundary conditions, with peptides, lipids, and water coupled separately to a heat bath at 295 K (τT = 0.3 ps). The system was semiisotropically coupled to a pressure bath at 1 bar (τp = 3 ps). As the starting structure for the membrane platform, we took the phase-separated bilayer obtained after 20 μs of coarse-grained simulation from ref. 34. Subsequently, TM peptides were inserted, regularly distributed on a lateral grid. For the WALP simulations, 12 peptides were inserted (Fig. 1A), with neighboring peptides inserted in an alternating manner (i.e., 6 peptides were inserted “head up,” and 6 peptides “head down”). For TM-Sx1A and TM-LAT, 9 peptides were inserted in a parallel manner (see SI Appendix for details).
The final systems comprised of TM helices, 828 di-C16∶0PC, 540 di-C18∶2PC, and 576 cholesterol molecules, solvated by 12,600 CG water beads (one water bead represents four real water molecules). Prior to the free MD simulations, the systems were energy-minimized (1,000 steps steepest descent), followed by 1 ns of MD with positional restraints on the peptide beads (force constant 1,000 kJ mol-1 nm-2). For details about the resolution transformation and atomistic simulations, see SI Appendix. To convert simulation times to real times, a factor of 4 was used, obtained by comparing the lateral diffusion coefficient of a single WALP23 peptide in a bilayer to experimental data; see SI Appendix and ref. 62. All times reported in the text are real times.
Confocal Microscopy.
Cys-WALP23/27 [Acetyl-CGWW(LA)8/10LWWA-amide] were synthesized using Fmoc/tBu peptide solid-phase synthesis as described previously for related KALP peptides (63). The peptides were labeled with Alexa Fluor 488 C5-maleimide (Molecular Probes, Invitrogen Corp.) using procedures similar to the ones for another thiol-reactive fluorophore (64). A lipid mixture of cholesterol, sphingomyelin (extracted from egg-yolk), and 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine (di-C14∶1PC) or 1,2-dioleoyl-sn-glycero-3-phosphocholine (di-C18∶1PC) was prepared from lipid stock solution in chloroform with molar ratio 1∶1∶1 (lipids were purchased from Avanti Polar Lipids). The labeled peptides dissolved in trifluoroethanol and the fluorescent lipid marker DiD-C18∶0 (solid1,1′-di-octadecyl-3,3,3′,3′-tetramethyl-indodicarbocyanine, 4-chlorobenzene-sulfonate salt, Molecular Probes, Invitrogen Corp.), dissolved in methanol, were added to the lipid mixture at a ratio of 1∶100,000. The peptide/lipid mixture was applied to indium–tin oxide-coated microscope glasses (thickness 0.13–0.16 mm; 15–30 Ω, SPI Supplies), and solvents were evaporated. GUVs were produced using a homemade electroformation chamber assembled of a pair of indium–tin oxide-coated glasses filled with buffer (10 mM KPi, pH 7.0), to which an alternating current with 10 Hz and 2 V (corresponding to ca. 400 V/m) was applied for at least 2 h. For imaging, the electroformation chamber was directly mounted on the sample stage of a laser scanning confocal microscope equipped with two lasers. A blue argon ion laser (488 nm) and a red He-Ne laser (633 nm) were used to excite the Alexa 488 fluorophore and the lipid marker DiD-C18∶0, respectively.
Supplementary Material
Acknowledgments.
We thank M. Bulacu-Cioceanu, L. Monticelli, and H. J. Risselada for useful discussions, and Prof. T. Lang for carefully reading the manuscript prior to publication. We thank the Netherlands Organisation for Scientific Research for funding (Veni Grant 700.57.404 to L.V.S., Top Grant 700.57.303 to S.J.M., and Top Grant 700.56.302 to B.P.). Computer access was granted from the National Supercomputing Facilities (Grant SH-148) and the European Initiative for Supercomputing Applications (Grant XXLBIOMD). A.H. was supported through a Marie Curie Early Stage Research Training Fellowship (Biomem-MEST-CT 2004-007931).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009362108/-/DCSupplemental.
References
- 1.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 2.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 3.Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 4.Hancock JF. Lipid rafts: Contentious only from simplistic standpoints. Nat Rev Mol Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edidin M. The state of lipid rafts: From model membranes to cells. Annu Rev Biophys Biomol Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 6.Jacobson K, Mouritsen OG, Anderson RGW. Lipid rafts: At a crossroad between cell biology and physics. Nat Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 7.Baumgart T, Hess ST, Webb WW. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 8.Kahya N, Scherfeld D, Bacia K, Poolman B, Schwille P. Probing lipid mobility of raft-exhibiting model membranes by fluorescence correlation spectroscopy. J Biol Chem. 2003;278:28109–28115. doi: 10.1074/jbc.M302969200. [DOI] [PubMed] [Google Scholar]
- 9.Samsonov AV, Mihalyov I, Cohen FS. Characterization of cholesterol-sphingomyelin domains and their dynamics in bilayer membranes. Biophys J. 2001;81:1486–1500. doi: 10.1016/S0006-3495(01)75803-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veatch SL, Polozov IV, Gawrisch K, Keller SL. Liquid domains in vesicles investigated by NMR and fluorescence microscopy. Biophys J. 2004;86:2910–2922. doi: 10.1016/S0006-3495(04)74342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumgart T, et al. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci USA. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lingwood D, Ries J, Schwille P, Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc Natl Acad Sci USA. 2008;105:10005–10010. doi: 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown DA, London E. Structure and origin of ordered lipid domains in biological membranes. J Membr Biol. 1998;164:103–114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- 14.Simons K, Vaz WLC. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 15.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sachs JN, Engelman DM. Introduction to the membrane protein reviews: The interplay of structure, dynamics, and environment in membrane protein function. Annu Rev Biochem. 2006;75:707–712. doi: 10.1146/annurev.biochem.75.110105.142336. [DOI] [PubMed] [Google Scholar]
- 17.Hanzal-Bayer MF, Hancock JF. Lipid rafts and membrane traffic. FEBS Lett. 2007;581:2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 18.Sprong H, van der Sluijs P, van Meer G. How proteins move lipids and lipids move proteins. Nat Rev Mol Cell Biol. 2001;2:504–513. doi: 10.1038/35080071. [DOI] [PubMed] [Google Scholar]
- 19.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chamberlain LH, Burgoyne RD, Gould GW. SNARE proteins are highly enriched in lipid rafts in PC12 cells: Implications for the spatial control of exocytosis. Proc Natl Acad Sci USA. 2001;98:5619–5624. doi: 10.1073/pnas.091502398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang T, et al. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 2001;20:2202–2213. doi: 10.1093/emboj/20.9.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 23.Kawabuchi M, et al. Transmembrane phosphoprotein Cbp regulates the activities of Src-family tyrosine kinases. Nature. 2000;404:999–1003. doi: 10.1038/35010121. [DOI] [PubMed] [Google Scholar]
- 24.Moffett S, Brown DA, Linder ME. Lipid-dependent targeting of G proteins into rafts. J Biol Chem. 2000;275:2191–2198. doi: 10.1074/jbc.275.3.2191. [DOI] [PubMed] [Google Scholar]
- 25.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 26.Periole X, Huber T, Marrink SJ, Sakmar TP. G Protein-coupled receptors self-assemble in dynamics simulations of model bilayers. J Am Chem Soc. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt U, Guigas G, Weiss M. Cluster formation of transmembrane proteins due to hydrophobic mismatching. Phys Rev Lett. 2008;101:128104. doi: 10.1103/PhysRevLett.101.128104. [DOI] [PubMed] [Google Scholar]
- 28.Reynwar BJ, Deserno M. Membrane composition-mediated protein-protein interactions. Biointerphases. 2008;3:FA117–FA124. doi: 10.1116/1.2977492. [DOI] [PubMed] [Google Scholar]
- 29.de Meyer FJ-M, Venturoli M, Smit B. Molecular simulations of lipid-mediated protein-protein interactions. Biophys J. 2008;95:1851–1865. doi: 10.1529/biophysj.107.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Planque MRR, Killian JA. Protein-lipid interactions studied with designed transmembrane peptides: Role of hydrophobic matching and interfacial anchoring. Mol Membr Biol. 2003;20:271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- 31.Nyholm TKM, Özdirekcan S, Killian JA. How protein transmembrane segments sense the lipid environment. Biochemistry. 2007;46:1457–1465. doi: 10.1021/bi061941c. [DOI] [PubMed] [Google Scholar]
- 32.van Duyl BY, Rijkers DTS, de Kruijff B, Killian JA. Influence of hydrophobic mismatch and palmitoylation on the association of transmembrane alpha-helical peptides with detergent-resistant membranes. FEBS Lett. 2002;523:79–84. doi: 10.1016/s0014-5793(02)02939-3. [DOI] [PubMed] [Google Scholar]
- 33.Fastenberg ME, Shogomori H, Xu X, Brown DA, London E. Exclusion of a transmembrane-type peptide from ordered-lipid domains (rafts) detected by fluorescence quenching: extension of quenching analysis to account for the effects of domain size and domain boundaries. Biochemistry. 2003;42:12376–12390. doi: 10.1021/bi034718d. [DOI] [PubMed] [Google Scholar]
- 34.Risselada HJ, Marrink SJ. The molecular face of lipid rafts in model membranes. Proc Natl Acad Sci USA. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monticelli L, et al. The MARTINI coarse-grained force field: Extension to proteins. J Chem Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 36.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: Coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 37.Fleming KG. Standardizing the free energy change of transmembrane helix-helix interactions. J Mol Biol. 2002;323:563–571. doi: 10.1016/s0022-2836(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 38.Yano Y, Matsuzaki K. Measurement of thermodynamic parameters for hydrophobic mismatch 1: Self-association of a transmembrane helix. Biochemistry. 2006;45:3370–3378. doi: 10.1021/bi0522854. [DOI] [PubMed] [Google Scholar]
- 39.Ash WL. Calgary, AB, Canada: Univ of Calgary; 2009. Helix-helix interactions in membrane proteins probed with computer simulations. PhD Thesis. [Google Scholar]
- 40.Sparr E, et al. Self-association of transmembrane alpha-helices in model membranes: Importance of helix orientation and role of hydrophobic mismatch. J Biol Chem. 2005;280:39324–39331. doi: 10.1074/jbc.M502810200. [DOI] [PubMed] [Google Scholar]
- 41.Bowie JU. Helix packing in membrane proteins. J Mol Biol. 1997;272:780–789. doi: 10.1006/jmbi.1997.1279. [DOI] [PubMed] [Google Scholar]
- 42.Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 43.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: Structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 44.Rzepiela AJ, et al. Reconstruction of atomistic details from coarse-grained structures. J Comput Chem. 2010;31:1333–1343. doi: 10.1002/jcc.21415. [DOI] [PubMed] [Google Scholar]
- 45.Bacia K, Schuette CG, Kahya N, Jahn R, Schwille P. SNAREs prefer liquid-disordered over “raft” (liquid-ordered) domains when reconstituted into giant unilamellar vesicles. J Biol Chem. 2004;279:37951–37955. doi: 10.1074/jbc.M407020200. [DOI] [PubMed] [Google Scholar]
- 46.Hammond AT, et al. Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc Natl Acad Sci USA. 2005;102:6320–6325. doi: 10.1073/pnas.0405654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vidal A, McIntosh TJ. Transbilayer peptide sorting between raft and nonraft bilayers: Comparisons of detergent extraction and confocal microscopy. Biophys J. 2005;89:1102–1108. doi: 10.1529/biophysj.105.062380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shogomori H, et al. Palmitoylation and intracellular domain interactions both contribute to raft targeting of linker for activation of T cells. J Biol Chem. 2005;280:18931–18942. doi: 10.1074/jbc.M500247200. [DOI] [PubMed] [Google Scholar]
- 49.Nikolaus J, et al. Hemagglutinin of influenza virus partitions into the nonraft domain of model membranes. Biophys J. 2010;99:489–498. doi: 10.1016/j.bpj.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiser H-J, et al. Order of lipid phases in model and plasma membranes. Proc Natl Acad Sci U SA. 2009;106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laage R, Rohde J, Brosig B, Langosch D. A conserved membrane-spanning amino acid motif drives homomeric and supports heteromeric assembly of presynaptic SNARE proteins. J Biol Chem. 2000;275:17481–17487. doi: 10.1074/jbc.M910092199. [DOI] [PubMed] [Google Scholar]
- 52.Sieber JJ, Willig KI, Heintzmann R, Hell SW, Lang T. The SNARE motif is essential for the formation of syntaxin clusters in the plasma membrane. Biophys J. 2006;90:2843–2851. doi: 10.1529/biophysj.105.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sieber JJ, et al. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–1076. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- 54.McIntosh TJ, Vidal A, Simon SA. Sorting of lipids and transmembrane peptides between detergent-soluble bilayers and detergent-resistant rafts. Biophys J. 2003;85:1656–1666. doi: 10.1016/S0006-3495(03)74595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Munro S. Lipid rafts: Elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 56.Lundbaek JA, Andersen OS, Werge T, Nielsen C. Cholesterol-induced protein sorting: An analysis of energetic feasibility. Biophys J. 2003;84:2080–2089. doi: 10.1016/S0006-3495(03)75015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trabelsi S, Zhang S, Lee TR, Schwartz DK. Linactants: Surfactant analogues in two dimensions. Phys Rev Lett. 2008;100:037802. doi: 10.1103/PhysRevLett.100.037802. [DOI] [PubMed] [Google Scholar]
- 58.Schäfer LV, Marrink SJ. Partitioning of lipids at domain boundaries in model membranes. Biophys J. 2010;99:L91–L93. doi: 10.1016/j.bpj.2010.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong JH, Briggs MM, Mlaver D, Vidal A, McIntosh TJ. Sorting of lens aquaporins and connexins into raft and nonraft bilayers: Role of protein homo-oligomerization. Biophys J. 2009;97:2493–2502. doi: 10.1016/j.bpj.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buzhynskyy N, Girmens JF, Faigle W, Scheuring S. Human cataract lens membrane at subnanometer resolution. J Mol Biol. 2007;374:162–169. doi: 10.1016/j.jmb.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 61.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 62.Ramadurai S, et al. Influence of hydrophobic mismatch and amino acid composition on the lateral diffusion of transmembrane peptides. Biophys J. 2010;99:1447–1454. doi: 10.1016/j.bpj.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Planque MRR, et al. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane alpha-helical peptides. J Biol Chem. 1999;274:20839–20846. doi: 10.1074/jbc.274.30.20839. [DOI] [PubMed] [Google Scholar]
- 64.Holt A, et al. Tilt and rotation angles of a transmembrane model peptide as studied by fluorescence spectroscopy. Biophys J. 2009;97:2258–2266. doi: 10.1016/j.bpj.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.