Abstract
WNK [with no lysine (K)] protein kinases are found in all sequenced multicellular and many unicellular organisms. WNKs influence ion balance. Two WNK family members are associated with a single gene form of hypertension. RNA interference screens have implicated WNKs in survival and growth, and WNK1 is essential for viability of mice. We found that the majority of WNK1 is localized on cytoplasmic puncta in resting cells. During cell division, WNK1 localizes to mitotic spindles. Therefore, we analyzed mitotic phenotypes in WNK1 knockdown cells. A large percentage of WNK1 knockdown cells fail to complete cell division, displaying defects in mitotic spindles and also in abscission and cell survival. One of the best-characterized WNK1 targets is the protein kinase OSR1 (oxidative stress responsive 1). OSR1 regulates ion cotransporters, is activated in response to osmotic stress by WNK family members, and is largely associated with WNK1. In resting cells, the majority of OSR1, like WNK1, is on cytoplasmic puncta. OSR1 is also in nuclei. In contrast to WNK1, however, OSR1 does not concentrate around spindles during mitosis and does not show a WNK1-like localization pattern in mitotic cells. Knockdown of OSR1 has only a modest effect on cell survival and does not lead to spindle defects. We conclude that decreased cell survival associated with loss of WNK1 is attributable to defects in chromosome segregation and abscission and is independent of the effector kinase OSR1.
Keywords: cytokinesis, microtubules, serine-, proline-, alanine-rich kinase
The serine/threonine protein kinase WNK1 [with no lysine (K)] is important for blood pressure regulation (1–4). Positional cloning identified two of the four WNK family members as the genes mutated in a form of hypertension known as pseudohypoaldosteronism type II (5). Disease-causing mutations in WNK1 are large deletions of the first intron, which are thought to lead to the increased expression of the protein. Consistent with this idea, WNK1 heterozygous mice have low blood pressure (6). Little is understood about the regulation of WNKs. The only known WNK activators are increased and decreased ionic strength (7, 8), consistent with their roles in regulating ion transport.
WNKs differ from other members of the protein kinase superfamily because of the unusual location of the catalytic lysine (7, 9). WNKs are large, varying in size from nearly 1,200 to almost 2,400 residues (10). In addition to a protein kinase domain, WNKs contain an autoinhibitory domain that suppresses protein kinase activity, at least two coiled-coil regions, and many protein-interaction motifs but no other currently known folded domains. Outside of the kinase domain, strong sequence similarity among the WNKs is limited to conserved interaction motifs and a C-terminal coiled coil. WNK1 is found ubiquitously in tissues but has several splice variants that are differentially distributed, suggesting both common and tissue-specific functions (7, 10–12).
Transport functions attributed to WNKs include regulation of several ion cotransporters and ion channels. Among these, few mechanisms have been well defined, but the best understood is the regulation of sodium, potassium, two-chloride cotransporters mediated by the WNK substrates oxidative stress responsive 1 (OSR1) and its homolog, the serine-, proline-, alanine-rich kinase (SPAK or PASK) (13–17). One or the other of these related kinases is expressed in most if not all tissues. In addition to their kinase and regulatory domains, OSR1 and SPAK have a C-terminal PF2 (for PASK and Fray, the fly ortholog) protein-protein interaction domain. Through their PF2 domains, these kinases bind stably to WNK proteins and are activated by WNK-mediated phosphorylation in response to osmotic stress (14–19).
WNK family members have also been identified in screens of proteins involved in development and cancer (20–22). In addition, WNKs have been implicated in cell-based screens for survival and proliferation (23–25). Loss of WNK3 has been found to accelerate apoptosis by enhancing caspase-3 activation (26). Mechanisms that might account for the identification of WNK1 in cell-based screens are unknown.
In this study, we investigated the localization of endogenous WNK1 and its target OSR1. We found the unexpected concentration of WNK1 on mitotic spindles. By examining phenotypes of cells in which these proteins were individually knocked down, we identified a unique property of WNK1 that has an impact on cell survival.
Results
WNK1 and OSR1 Display a Punctate Localization Pattern.
The localization of endogenous WNK1 in HeLa cells by immunofluorescence demonstrated a decidedly punctate cytoplasmic pattern (Fig. 1A and Fig. S1B). This punctate staining pattern was also observed in other cell types, including MCF7 breast and HT29 colon cancer lines (Fig. S1 D and E). The specificity of the WNK1 antibody was verified by blocking the signal with peptide antigen and by siRNA knockdown of WNK1. Both peptide block and knockdown almost completely eliminated the antibody signal, indicating that the immunofluorescent staining pattern was attributable to WNK1 (Fig. 1 B and C). Previously, WNK1 localization was examined using heterologously expressed WNK1 fusion proteins and was described as diffuse cytoplasmic (7, 27). We examined GFP-tagged WNK1, which also appeared in a punctate pattern as long as it was expressed at low levels (Fig. S1 A and C). Expression of larger amounts resulted in diffuse staining that did not represent the endogenous pattern.
Fig. 1.
WNK1 and OSR1 display a punctate localization pattern. Immunostaining of HeLa cells: WNK1 or OSR1 (red) and DAPI (blue). (A) Projected Z-slices of HeLa cells stained for WNK1. The specificity of the WNK1 antibody was verified by peptide antigen block (B) and siRNA knockdown (C). (Scale bar: 5 μm.) (D and E) Projected Z-slices of HeLa cells stained for OSR1. (E and F) Specificity of the OSR1 antibody was verified by siRNA knockdown. (Scale bar: 10 μm.)
OSR1 is one of the best-characterized WNK1 substrates. Biochemical studies have shown that WNK1 and OSR1 are tightly bound to each other in cells (16, 17). Therefore, we examined the localization of OSR1 in HeLa cells by immunofluorescence and found that OSR1, like WNK1, displayed a punctate cytoplasmic staining pattern; unlike WNK1, OSR1 was also prominent in the nucleus (Fig. 1D). The specificity of the OSR1 immunofluorescent signal was confirmed by OSR1 knockdown (Fig. 1 E and F).
WNK1 Localizes to Mitotic Spindles During Mitosis.
In WNK1 knockdown experiments, we occasionally observed some cell division defects. Therefore, we characterized the localization of endogenous WNK1 at different stages of mitosis. Unexpectedly, we found that WNK1 was concentrated on mitotic spindles during mitosis. In early prophase, WNK1 was located mainly in the cytoplasm; however, there was a population of WNK1 in proximity to astral microtubules (Fig. 2 A–C and Fig. S1 F–I, compare merged signal with computer-generated colocalization in the latter). In metaphase, WNK1 was evident on spindle pole microtubules. The signal density was higher near the spindle poles than at the kinetochore ends (Fig. 2 D–F). For clarity, Fig. 2 G–I shows data from Fig. 2 D–F as a 3D volume rendering that weights the transparency of each voxel according to its depth in the volume. In anaphase, WNK1 signals clearly remained on spindle microtubules as the sister chromatids were separated, was evident on polar microtubules, and was detectable at the midzone (Fig. 2 J and L). In telophase, WNK1 was present around astral microtubules and prominent at the spindle midzone between the two daughter cells (Fig. 2 M–O).
Fig. 2.
WNK1 localizes on mitotic spindles during mitosis. Immunostaining of HeLa cells: WNK1 (red) and tubulin (green). The colocalized channel (yellow) was generated by Imaris software. (A–C) Early prophase. (D–F) Metaphase. (G–I) Images same as in D–F, shown in volume-rendered projection. (J–L) Anaphase. (M–O) Telophase. (Scale bar: 5 μm.)
Depletion of WNK1 Causes Aberrant Mitotic Spindles and Defective Abscission.
Localization on mitotic spindles strongly suggests that WNK1 may have a function in mitosis. Indeed, knockdown of WNK1 resulted in aberrant mitotic spindles and chromosome segregation. Cells transfected with scrambled oligonucleotides showed well-aligned spindles and chromosomes (Fig. 3 A–C). In contrast, cells with WNK1 knocked down displayed a range of defects, including longer and misaligned spindles and chromosomes (Fig. 3 D–F); tripolar spindles and L-shaped chromosomes (Fig. 3 G–I); symmetrical tetrapolar spindles and cross-shaped chromosomes (Fig. 3 J–L); and multipolar spindles with misaligned chromosomes (Fig. 3 M–O). Multipolar spindles often result in multinucleated cells (28). Consistent with this idea, cells lacking WNK1 were frequently multinucleate (Fig. S2).
Fig. 3.
Depletion of WNK1 causes aberrant mitotic spindles. Immunostaining of HeLa cells 48 h after transfection with scrambled (A–C) or WNK1 siRNA (D–O) oligonucleotides. WNK1 (red), tubulin (green), and DAPI (blue channel). (Scale bar: 5 μm.)
In late telophase, mitosis is completed once separation of the two daughter cells occurs by abscission of the midbody (29). In control cells before abscission, the length of the bridge between two daughter cells was 14.1 ± 0.4 μm, whereas the average length of the bridge in WNK1-depleted cells was more than twice as long at 31.8 ± 0.9 μm (Fig. 4 A–C). Time-lapse microscopy supported the conclusion that abscission failure might account for the longer bridge in WNK1-depleted daughter cells. Control cells successfully severed the bridge and formed two independent cells within 2 h (Fig. 4D, yellow broken lines). Knockdown of WNK1 caused the daughter cells to remain connected even after 17.5 h (Fig. 4E, yellow arrowheads). As cells attempted to move away from each other, the bridge lengthened (Movies S1–S3).
Fig. 4.
RNAi of WNK1 resulted in a defect in abscission. Immunostaining of HeLa cells 72 h after transfection with scrambled (A) or WNK1 siRNA (B) oligonucleotides. WNK1 (red), tubulin (green), and DAPI (blue channel). In A and B, the bridge between two daughter cells is overlaid with a bar indicating length. (Scale bar: 15 μm.) (C) Average length of bridges in scrambled control and WNK1 knockdown cells is plotted. Sample images were taken from four independent experiments (scrambled control, n = 80; WNK1 knockdown, n = 90). Statistical analysis was performed by an unpaired t test. Individual frames from time-lapse microscopy of scrambled control (D) and WNK1 knockdown (E) cells. Regions between separating cells are indicated by yellow arrowheads. Perimeters of separated daughter cells in the scrambled control are indicated by yellow lines in D.
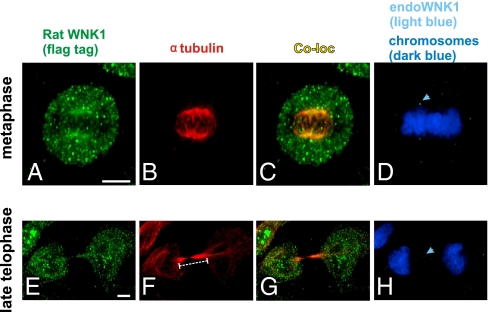
Rat WNK1 Rescues Mitotic Phenotypes Caused by Knockdown of WNK1.
To provide additional support that phenotypes caused by siRNA are not attributable to off-target effects, rat WNK1 was expressed in HeLa cells and tested for its ability to rescue mitotic spindle and abscission phenotypes. Cells expressing rat WNK1 (Fig. 5 A and E) displayed normal mitotic spindles and well-aligned chromosomes, in spite of little endogenous WNK1 (Fig. 5 B–D). Likewise, the bridge between daughter cells was of normal length (Fig. 5 F–H). Rat WNK1 was localized, like endogenous HeLa WNK1, on mitotic spindles (Fig. 5 C and G). These observations support the conclusion that depletion of WNK1 results in several defective mitotic phenotypes.
Fig. 5.
Rescue of mitotic phenotypes by rat WNK1. Rat Flag-WNK1 was expressed in cells in which the endogenous protein was knocked down. Rat Flag-tagged WNK1 (green), endogenous WNK1 (light blue), tubulin (red), and DAPI (dark blue). (A–D) Metaphase. (E–H) Late telophase. (Scale bar: 5 μm.) The bridge between two daughter cells is overlaid with a bar indicating length. Arrowheads in D and H point to staining of endogenous WNK1.
Knockdown of OSR1 Does Not Show Mitotic Defects and Has Little Effect on Cell Viability.
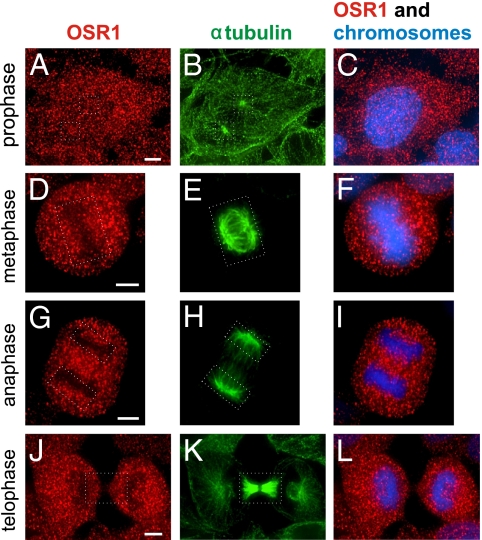
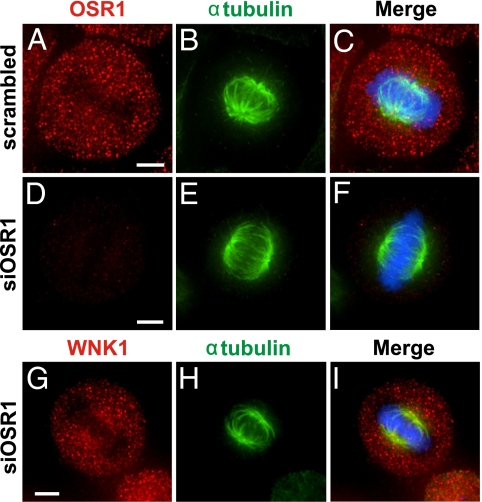
Because OSR1 is associated with WNK1 in cells and is a WNK1 effector (14–19), we wanted to determine if it mediates the effects of WNK1 on mitosis. Unlike WNK1, OSR1 was distributed relatively evenly in all stages of mitosis (Fig. 6) and was not concentrated on mitotic spindles (Fig. 6 A, B, D, E, G, H, J, and K, areas marked by broken white lines). Furthermore, OSR1 knockdown did not cause the aberrant mitotic phenotypes observed in WNK1 knockdown cells; spindles and chromosomes were well aligned despite little OSR1 (Fig. 7 D–F). Finally, the localization of WNK1 on mitotic spindles was unchanged by OSR1 depletion (Fig. 7 G–I). Based on our previous knockdown experiments examining sodium, potassium, two-chloride cotransporter activity, there is insufficient SPAK to replace OSR1 function under these conditions (17).
Fig. 6.
OSR1 displays a localization distinct from WNK1 during mitosis. Immunostaining of HeLa cells: OSR1 (red), tubulin (green), and DAPI (blue). (A–C) Early prophase. (D–F) Metaphase. (G–I) Anaphase. (J–L) Telophase. The broken white lines generally mark mitotic spindles (A, B, D, E, G, H, J, and K). (Scale bar: 5 μm.)
Fig. 7.
Depletion of endogenous OSR1 does not cause defects in mitotic spindles. Immunostaining of HeLa cells 48 h after transfection with scrambled (A–C) or OSR1 siRNA (D–I) oligonucleotides. OSR1 (A and D; red) or WNK1 (G; red), tubulin (B, E, and H; green), and DAPI (C, F, and I; blue). (Scale bar: 5 μm.)
We have consistently noted decreased survival of most cell types following WNK1 knockdown. To quantify cell survival, the relative number of viable cells following knockdown of WNK1 or OSR1 was measured. We found that nearly 80% of OSR1-depleted HeLa cells were viable (Fig. 8 A and B). Conversely, the survival of WNK1-depleted cells was below 35%.
Fig. 8.
Effect of depletion of WNK1 or OSR1 on cell survival. (A) Percentage of viable cells remaining following transfection with scrambled, WNK1 siRNA, or OSR1 siRNA oligonucleotides is shown as the mean ± SEM. Three independent experiments were performed. Statistical analysis was by one-way ANOVA with a Bonferroni test. *P < 0.05; ***P < 0.001. (B) Immunoblotting of HeLa cells transfected with scrambled, WNK1 siRNA, or OSR1 siRNA oligonucleotides.
Discussion
Here, we report a unique property of WNK1, a requirement for localization to spindles during mitosis, not anticipated by any previous biochemical or physiological studies. Depletion of WNK1 showed multipolar spindles, disorganized chromosomes, failed abscission, multinucleated cells, and reduced cell survival. WNK1 binds a number of other proteins, including protein kinases, through its noncatalytic domains (30–32). Thus, WNK1 may exert its mitotic function through its enzymatic activity, protein-protein associations, or likely both.
WNK1 is essential for viability in mice (6). Later analysis of embryos has suggested that failure to complete development is attributable to a defect in formation of the cardiovascular system, which can be corrected by expression of WNK1 specifically in endothelial cells (33). These results suggest that the role of WNK1 in mitosis does not primarily account for the lethality of its deletion. In reconstitution assays, multiple WNK family members can regulate several common effectors not limited to OSR1 and SPAK (1–4). In this regard, we have been unable to detect other WNKs in HeLa cells by immunoblotting. Thus, it seems possible that the lack of obvious mitotic phenotypes in the WNK1 null embryos may be attributable to the capacity of other WNK isoforms to compensate for loss of this WNK1 function.
Examination of the localizations of endogenous WNK1 and OSR1 reveals punctate and largely cytoplasmic distributions. Although the puncta are too small to be identified unequivocally as vesicles by light microscopy, the data suggest that WNK1 and OSR1 may be on intracellular vesicles in resting cells and support the strong biochemical data showing WNK1-OSR1 association. Conversely, when GFP-WNK1 was overexpressed, not only did we observe a diffuse rather than punctate staining pattern but several intense GFP spots, which suggested aggregation. Therefore, it seems unlikely that the diffuse GFP-WNK1 signal is an accurate reflection of WNK1 localization. From live cell imaging, we also found that expression of WNK1 had to be carefully titrated. Overexpression caused cell death within hours posttransfection.
Because OSR1 is a major WNK1 effector and is bound to WNK1 in resting cells (14–19), it is surprising that it was not colocalized on mitotic spindles. Consistent with the distinct mitotic localization of OSR1, knockdown showed that it was apparently not involved in WNK1-dependent mitotic events. The OSR1 homolog SPAK is present at nearly undetectable levels in HeLa cells (Fig. S3), making the possibility that we missed relevant phenotypes as a result of functional redundancy unlikely. The difference in survival between WNK1-depleted and OSR1-depleted cells may simply mean that OSR1 is not involved in WNK1-mediated cell survival functions. Conversely, it is possible that WNK1 exerts its function on cell survival through multiple downstream targets, of which OSR1 is just one, and the assays used here did not reveal its contribution.
Our findings suggest that WNK1 is a potential mitotic kinase regulating both the formation of mitotic spindles and the completion of abscission. WNK1 was found in a shRNA knockdown screen for multipolar spindle phenotypes among human kinases (25), further supporting our discovery of a mitotic function of WNK1. During the initial stages of mitosis, proper nucleation of microtubules at centrosomes forms bipolar spindles. Centrosome maturation and centriole duplication, key steps leading to microtubule nucleation, are controlled by several mitotic kinases (34, 35). Aurora and Polo-like kinases (PLKs) differentially regulate recruitment of multiple microtubule-associated proteins and motor proteins required for centrosome maturation (36–38). These kinases also regulate steps in mitotic exit and contractile ring formation, the first step of cytokinesis (34, 35). Although the localization of WNK1 resembles that of Aurora A and PLK1, especially on spindle poles, WNK1 localizes differently from Aurora B during mitosis. Knocking down Aurora B results in multipolar spindles, depletion of PLK1 causes monopolar spindles, but overexpression of PLK1 or Aurora A results in multipolar spindles (34, 39–41). Given the phenotypic distinctions, perhaps WNK1 negatively regulates PLK1 or Aurora kinases during formation of bipolar spindles. Alternatively, WNK1 may act independent of PLK1 or Aurora kinases. Abscission finally separates the two daughter cells and involves diverse processes, including membrane delivery, microtubule remodeling, and membrane fusion (42). How WNK1 is involved in this final step requires further investigation. Clues to the underlying molecular mechanisms may be obtained from identification of potential WNK1 interactors during mitosis. The present study sets the stage to explore this unique mitotic function of WNK1.
Materials and Methods
Constructs.
Rat Flag-WNK1 cDNA was created by PCR using the following forward and reverse primers: GAATTCATGTCTGACGGCACCGCAGAGAAGC and GGATCCCTAGGTGGTCCGTAGGTTGGAACT with Myc WNK1 as a cDNA template. PCR products were digested with EcoRI and BamHI and ligated into p3XFlagCMV7.1.
Antibodies and Reagents.
Antibodies to the following proteins were as indicated: WNK1 immunofluorescence (no. 4979; Cell Signaling) and immunoblotting [Q256 (7)], OSR1 immunofluorescence (no. 3729; Cell Signaling), SPAK (no. 2281; Cell Signaling), α-tubulin (clone DM1A; Sigma Aldrich or clone YL1/2; Abcam), and Flag epitope (clone M2; Sigma Aldrich). Secondary antibodies for immunofluorescence were goat anti-mouse Alex 488 or 546, goat anti-rabbit Alex 488 or 546, and goat anti-rat Alex 647 (all from Invitrogen). DAPI, dextran conjugated with Alex 488, and 10% (vol/vol) normal goat serum were also purchased from Invitrogen. Antibody U5438 raised against His6-OSR1 residues 345–527 recognizes both OSR1 and SPAK as described (43).
Cell Culture Immunofluorescence and Immunoblotting.
Cells were cultured in DMEM with 10% FBS (vol/vol) and 1% l-glutamine at 37 °C under 5% (vol/vol) CO2. For imaging, cells were plated on 35-mm or six-well glass-bottom plates (MatTeK Corp.). For immunofluorescence, cells were washed with PBS at 37 °C; fixed with 4% paraformaldehyde (vol/vol) in 60 mM Pipes, 25 mM Hepes (pH 6.9), 10 mM EGTA, and 2 mM MgCl2 for 10 min; and washed twice for 5 min each time with PBS. Cells were permeabilized with 0.1% Triton X-100 in PBS at 4 °C for 5 min, washed as above, incubated with 10% normal goat serum (vol/vol) at room temperature for 30 min, and then incubated with primary antibody at 4 °C overnight. Cells were then washed with PBS, incubated with Alexa fluor-conjugated secondary antibody at room temperature for 1 h, washed twice with PBS, and imaged by fluorescent microscopy.
For immunoblotting, confluent cells were harvested in lysis buffer containing 1% Triton X-100, 50 mM Hepes (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol (vol/vol), 100 mM NaF, 0.2 mM Na3VO4, 50 mM β-glycerophosphate, 1 mM DTT, 1 mM benzamidine hydrochloride, 10 mg/L leupeptin, 0.5 mg/L pepstatin A, and 1.5 mg/L aprotinin. The cells were vortexed for 30 s and incubated on ice for 10 min. The lysates were clarified by centrifugation at 21,300 × g in a microfuge for 15 min.
RNAi.
HeLa cells were transfected with dsRNA oligonucleotides using Lipofectamine RNAiMAX according to the manufacturer's instructions (Invitrogen). After 48 h, protein localization was examined. The following oligonucleotides were used: WNK1: sense, GGAUCAAGUGCGAGAAAUUTT, and antisense, AAUUUCUCGCACUUGAUCCTT; OSR1: sense, GGAACAGGUCCGUGGUUAUTT, and antisense, AUAACCACGGACCUGUUCCTT; and scrambled control, Silencer Negative control no. 1 siRNA (Ambion).
Time-Lapse Microscopy.
Cells were transfected as above with small interfering WNK1 or scrambled control oligonucleotides. After 24 h, cells were imaged with a Deltavision RT deconvolution microscope (Applied Precision) with an environmental control chamber (Solent). Images were acquired using a 20× phase contrast objective lens every 30 min for 72 h. Data were processed with ImageJ (National Institutes of Health).
Image Analysis.
Fluorescent Z-stacks were acquired and deconvolved using the Deltavision RT deconvolution microscope. Colocalization and 3D models were analyzed using Imaris software (v. 6.4; Bitplane). All data shown are displayed as projections through the Z-stack. The colocalization channels depicting voxels showing statistically significant colocalization were generated using the Coloc module of Imaris, which is based on the method of Costes et al. (44).
Cell Survival Assay.
Cells were transfected twice, 24 h apart, with WNK1, OSR1, or scrambled control oligonucleotides. After 72 h, cells excluding Trypan blue were counted. Cell survival was normalized to that for the scrambled control. Three independent experiments were performed. Statistical analysis was by one-way ANOVA with a Bonferroni test.
Supplementary Material
Acknowledgments
We thank Richard Anderson (University of Texas Southwestern) for comments in the early stages of this work. We thank A.-Young Lee, Elma Zaganjor, and other members of the Cobb laboratory for critical review of this manuscript and Dionne Ware for administrative assistance. S.T. performed this work in partial fulfillment of the requirements for the PhD degree. This work was supported by Grant GM53032 from the National Institutes of Health and Grant I1243 from the Welch Foundation (to M.H.C.). Fluorescence imaging was carried out in the Live Cell Imaging Facility at the University of Texas Southwestern Medical Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018567108/-/DCSupplemental.
References
- 1.Kahle KT, et al. WNK kinases: Molecular regulators of integrated epithelial ion transport. Curr Opin Nephrol Hypertens. 2004;13:557–562. doi: 10.1097/00041552-200409000-00012. [DOI] [PubMed] [Google Scholar]
- 2.Xie J, Craig L, Cobb MH, Huang C-L. Role of with-no-lysine [K] kinases in the pathogenesis of Gordon's syndrome. Pediatr Nephrol. 2006;21:1231–1236. doi: 10.1007/s00467-006-0106-6. [DOI] [PubMed] [Google Scholar]
- 3.Deaton SL, Sengupta S, Cobb MH. WNK kinases and blood pressure control. Curr Hypertens Rep. 2009;11:421–426. doi: 10.1007/s11906-009-0072-z. [DOI] [PubMed] [Google Scholar]
- 4.Welling PA, Chang YP, Delpire E, Wade JB. Multigene kinase network, kidney transport, and salt in essential hypertension. Kidney Int. 2010;77:1063–1069. doi: 10.1038/ki.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson FH, et al. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- 6.Zambrowicz BP, et al. Wnk1 kinase deficiency lowers blood pressure in mice: A gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci USA. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu B, et al. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 8.Lenertz LY, et al. Properties of WNK1 and implications for other family members. J Biol Chem. 2005;280:26653–26658. doi: 10.1074/jbc.M502598200. [DOI] [PubMed] [Google Scholar]
- 9.Min X, Lee BH, Cobb MH, Goldsmith EJ. Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure. 2004;12:1303–1311. doi: 10.1016/j.str.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 10.Veríssimo F, Jordan P. WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene. 2001;20:5562–5569. doi: 10.1038/sj.onc.1204726. [DOI] [PubMed] [Google Scholar]
- 11.Delaloy C, et al. Multiple promoters in the WNK1 gene: One controls expression of a kidney-specific kinase-defective isoform. Mol Cell Biol. 2003;23:9208–9221. doi: 10.1128/MCB.23.24.9208-9221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Reilly M, Marshall E, Speirs HJ, Brown RW. WNK1, a gene within a novel blood pressure control pathway, tissue-specifically generates radically different isoforms with and without a kinase domain. J Am Soc Nephrol. 2003;14:2447–2456. doi: 10.1097/01.asn.0000089830.97681.3b. [DOI] [PubMed] [Google Scholar]
- 13.Piechotta K, Lu J, Delpire E. Cation chloride cotransporters interact with the stress-related kinases Ste20-related proline-alanine-rich kinase (SPAK) and oxidative stress response 1 (OSR1) J Biol Chem. 2002;277:50812–50819. doi: 10.1074/jbc.M208108200. [DOI] [PubMed] [Google Scholar]
- 14.Gagnon KB, England R, Delpire E. Volume sensitivity of cation-chloride cotransporters is modulated by the interaction of two kinases: SPAK and WNK4. Am J Physiol Cell Physiol. 2006;290:C134–C142. doi: 10.1152/ajpcell.00037.2005. [DOI] [PubMed] [Google Scholar]
- 15.Moriguchi T, et al. WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J Biol Chem. 2005;280:42685–42693. doi: 10.1074/jbc.M510042200. [DOI] [PubMed] [Google Scholar]
- 16.Vitari AC, Deak M, Morrice NA, Alessi DR. The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J. 2005;391:17–24. doi: 10.1042/BJ20051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anselmo AN, et al. WNK1 and OSR1 regulate the Na+, K+, 2Cl− cotransporter in HeLa cells. Proc Natl Acad Sci USA. 2006;103:10883–10888. doi: 10.1073/pnas.0604607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delpire E, Gagnon KB. Genome-wide analysis of SPAK/OSR1 binding motifs. Physiol Genomics. 2007;28:223–231. doi: 10.1152/physiolgenomics.00173.2006. [DOI] [PubMed] [Google Scholar]
- 19.Villa F, et al. Structural insights into the recognition of substrates and activators by the OSR1 kinase. EMBO Rep. 2007;8:839–845. doi: 10.1038/sj.embor.7401048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moniz S, Jordan P. Emerging roles for WNK kinases in cancer. Cell Mol Life Sci. 2010;67:1265–1276. doi: 10.1007/s00018-010-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong C, et al. Epigenome scans and cancer genome sequencing converge on WNK2, a kinase-independent suppressor of cell growth. Proc Natl Acad Sci USA. 2007;104:10974–10979. doi: 10.1073/pnas.0700683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choe KP, Strange K. Evolutionarily conserved WNK and Ste20 kinases are essential for acute volume recovery and survival after hypertonic shrinkage in Caenorhabditis elegans. Am J Physiol Cell Physiol. 2007;293:C915–C927. doi: 10.1152/ajpcell.00126.2007. [DOI] [PubMed] [Google Scholar]
- 23.Björklund M, et al. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 2006;439:1009–1013. doi: 10.1038/nature04469. [DOI] [PubMed] [Google Scholar]
- 24.Sun X, Gao L, Yu RK, Zeng G. Down-regulation of WNK1 protein kinase in neural progenitor cells suppresses cell proliferation and migration. J Neurochem. 2006;99:1114–1121. doi: 10.1111/j.1471-4159.2006.04159.x. [DOI] [PubMed] [Google Scholar]
- 25.Conery AR, Harlow E. High-throughput screens in diploid cells identify factors that contribute to the acquisition of chromosomal instability. Proc Natl Acad Sci USA. 2010;107:15455–15460. doi: 10.1073/pnas.1010627107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veríssimo F, Silva E, Morris JD, Pepperkok R, Jordan P. Protein kinase WNK3 increases cell survival in a caspase-3-dependent pathway. Oncogene. 2006;25:4172–4182. doi: 10.1038/sj.onc.1209449. [DOI] [PubMed] [Google Scholar]
- 27.Zagórska A, et al. Regulation of activity and localization of the WNK1 protein kinase by hyperosmotic stress. J Cell Biol. 2007;176:89–100. doi: 10.1083/jcb.200605093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King RW. When 2+2=5: The origins and fates of aneuploid and tetraploid cells. Biochim Biophys Acta. 2008;1786:4–14. doi: 10.1016/j.bbcan.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gromley A, et al. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 30.Lee BH, et al. WNK1 phosphorylates synaptotagmin 2 and modulates its membrane binding. Mol Cell. 2004;15:741–751. doi: 10.1016/j.molcel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 31.Xu B, et al. WNK1 activates SGK1 to regulate the epithelial sodium channel. Proc Natl Acad Sci USA. 2005;102:10315–10320. doi: 10.1073/pnas.0504422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh E, Heise CJ, English JM, Cobb MH, Thurmond DC. WNK1 is a novel regulator of Munc18c-syntaxin 4 complex formation in soluble NSF attachment protein receptor (SNARE)-mediated vesicle exocytosis. J Biol Chem. 2007;282:32613–32622. doi: 10.1074/jbc.M706591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie J, et al. Endothelial-specific expression of WNK1 kinase is essential for angiogenesis and heart development in mice. Am J Pathol. 2009;175:1315–1327. doi: 10.2353/ajpath.2009.090094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: Regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Archambault V, Glover DM. Polo-like kinases: Conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 36.do Carmo Avides M, Tavares A, Glover DM. Polo kinase and Asp are needed to promote the mitotic organizing activity of centrosomes. Nat Cell Biol. 2001;3:421–424. doi: 10.1038/35070110. [DOI] [PubMed] [Google Scholar]
- 37.Donaldson MM, Tavares AA, Ohkura H, Deak P, Glover DM. Metaphase arrest with centromere separation in polo mutants of Drosophila. J Cell Biol. 2001;153:663–676. doi: 10.1083/jcb.153.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Luca M, Lavia P, Guarguaglini G. A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle. 2006;5:296–303. doi: 10.4161/cc.5.3.2392. [DOI] [PubMed] [Google Scholar]
- 39.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sunkel CE, Glover DM. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- 41.Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barr FA, Gruneberg U. Cytokinesis: Placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Chen W, Yazicioglu M, Cobb MH. Characterization of OSR1, a member of the mammalian Ste20p/germinal center kinase subfamily. J Biol Chem. 2004;279:11129–11136. doi: 10.1074/jbc.M313562200. [DOI] [PubMed] [Google Scholar]
- 44.Costes SV, et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.