Abstract
The Escherichia coli dihydroxyacetone (Dha) kinase is an unusual kinase because (i) it uses the phosphoenolpyruvate carbohydrate: phosphotransferase system (PTS) as the source of high-energy phosphate, (ii) the active site is formed by two subunits, and (iii) the substrate is covalently bound to His218K* of the DhaK subunit. The PTS transfers phosphate to DhaM, which in turn phosphorylates the permanently bound ADP coenzyme of DhaL. This phosphoryl group is subsequently transferred to the Dha substrate bound to DhaK. Here we report the crystal structure of the E. coli Dha kinase complex, DhaK–DhaL. The structure of the complex reveals that DhaK undergoes significant conformational changes to accommodate binding of DhaL. Combined mutagenesis and enzymatic activity studies of kinase mutants allow us to propose a catalytic mechanism for covalent Dha binding, phosphorylation, and release of the Dha-phosphate product. Our results show that His56K is involved in formation of the covalent hemiaminal bond with Dha. The structure of H56NK with noncovalently bound substrate reveals a somewhat different positioning of Dha in the binding pocket as compared to covalently bound Dha, showing that the covalent attachment to His218K orients the substrate optimally for phosphoryl transfer. Asp109K is critical for activity, likely acting as a general base activating the γ-OH of Dha. Our results provide a comprehensive picture of the roles of the highly conserved active site residues of dihydroxyacetone kinases.
Keywords: DhaK–DhaL complex, kinase mechanism
Dihydroxyacetone phosphate (Dha-P), an intermediate for the synthesis of pyruvate, can be generated in bacteria through glycerol fermentation by the sequential actions of glycerol dehydrogenase forming dihydroxyacetone and Dha kinase generating Dha-P. Dha kinases are divided into two classes, those using ATP as the phosphoryl donor or those using the PTS-dependent DhaM protein to provide the phosphoryl group (1). The ATP-dependent Dha kinases are present in animals, plants, and some bacteria and consist of a single chain containing two domains, whereas the PTS-dependent Dha kinases exist only in bacteria and are composed of two subunits. The most studied representative of the PTS-dependent family is the Escherichia coli Dha kinase, which is composed of DhaK and DhaL subunits. DhaM, the phosphotransferase subunit of Dha kinase, is phosphorylated by the small phospho-carrier protein HPr of the PTS (1). Phosphorylated DhaM then forms a complex with the ADP-bound DhaL subunit and transiently phosphorylates ADP to ATP (1). The ATP-loaded DhaL subsequently associates with the DhaK subunit containing the Dha substrate covalently bound to His218K* through a hemiaminal bond (2). Finally, ATP bound to DhaL, serving as a coenzyme (3), transfers phosphate to Dha yielding Dha-P.
Crystal structures for the two individual subunits of the E. coli PTS-dependent Dha kinase—DhaK (2) and DhaL (4)—have been previously determined, as well as the structure of an ATP-dependent single chain, two-domain Dha kinase from Citrobacter freundii (5), which has 30% sequence identity to the E. coli kinase. Both ATP- and PTS-dependent Dha kinases display high structural similarity of their corresponding domains/subunits and are structurally distinct from other kinases. The active site residues are highly conserved between bacteria and eukaryotes reflecting the unique phosphotransfer mechanism. The structure of the homodimeric, ATP-dependent C. freundii Dha kinase shows swapped K- and L-domains, with bound Dha and an ATP analogue separated by approximately 14 Å (5). It is therefore unlikely that this structure represents a phosphotransfer-competent state of this enzyme, leaving the question of the mechanism unanswered. In addition to the structures of DhaK and DhaL, the structure of the Lactococcus lactis DhaM domain has been reported as well as its complex with DhaL, providing insight into phosphotransfer from phosphohistidine to ADP (6).
We have determined the crystal structure of the E. coli DhaK–DhaL complex bound to ADP and Dha at 2.2-Å resolution, providing a snapshot of the final step of phosphoryl transfer from ATP to Dha. Site-directed mutagenesis, activity measurements, and structural data indicate that His56K is important for formation of the covalent hemiaminal bond. We show further that the covalent attachment of Dha to His218K optimally positions the substrate for phosphoryl transfer but is likely dispensable for enzyme activity. Together, these data allow us to propose a general catalytic mechanism for dihydroxyacetone kinases, consistent with the available structural and biochemical data.
Results and Discussion
Structure of the DhaK–DhaL Complex Reveals Conformational Rearrangements upon Complex Formation.
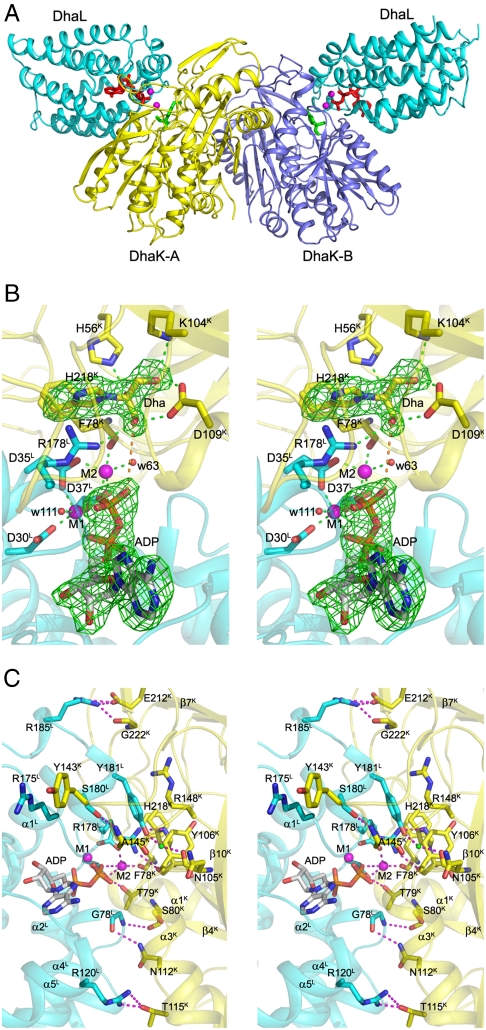
Each asymmetric unit contains one molecule each of DhaK and DhaL with all residues well defined in the electron density map. The biological unit of the complex is a heterotetramer containing a central DhaK dimer and two molecules of DhaL (2∶2 stoichiometry). Each DhaL subunit interacts with only one DhaK subunit of the dimer, with the two DhaL subunits being approximately 37 Å apart (Fig. 1A). An ADP molecule and two magnesium ions are bound to DhaL, whereas DhaK contains a Dha molecule covalently bound to His218K (Fig. 1B).
Fig. 1.
Crystal structure of the E. coli DhaK–DhaL complex. (A) Cartoon representation of the DhaK–DhaL heterotetramer. The DhaL molecules are shown in cyan with the two DhaK subunits A and B colored yellow and mauve, respectively. The Dha molecules covalently bound to His218K are colored green. The ADP molecules and magnesium ions bound to DhaL are shown as red sticks and magenta spheres, respectively. (B) Active site of the DhaK–DhaL complex. The DhaK (yellow) and DhaL (cyan) molecules are shown in cartoon representation. The ADP molecule and residues described in the text are shown in stick mode. The two magnesium ions are shown as magenta spheres. Interactions involving the Dha molecule and the two metal ions are indicated by green dashed lines. The distance between the γ-OH group of Dha and the oxygen atom of the ADP β-phosphate is 4.8 Å, which is shown as an orange dashed line. The difference (Fo - Fc) electron density map for Dha covalently linked to His218K as well as ADP (omitted from refinement) contoured at 4σ is shown in green. (C) Binding interface and interactions between DhaK and DhaL. The color scheme is the same as B. Hydrogen bonding interactions are shown as magenta dashed lines. The water molecule mediating the interaction between Y181L and N105K is shown as a green sphere.
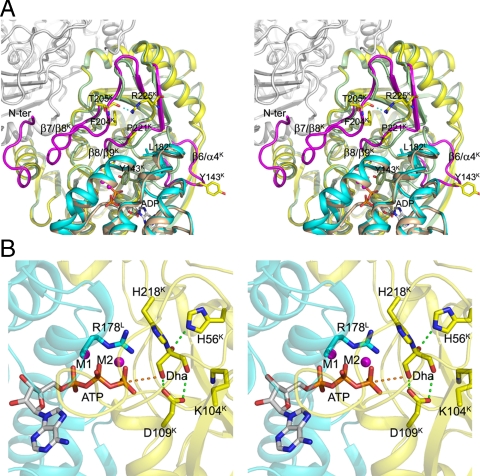
DhaL binds to DhaK through the narrower end of an eight-helix barrel that harbors the ADP. This is consistent with the role of DhaL-ATP as the phosphoryl donor to the DhaK-bound Dha. The structure of DhaL in the complex is very similar to that of free DhaL (4), with an rmsd of 0.42 Å for all Cα atoms. Binding to DhaK involves residues in loops α1L/α2L, α3L/α4L, α7L/α8L, and portions of helices α1L, α2L, α4L, and α5L (Fig. 1C). In contrast, DhaK undergoes significant conformational changes in several regions as compared to the free DhaK previously reported [PDB ID code 1OI2, (2)] (Fig. 2A). The rmsd for the Cα atoms of free and complexed DhaK is 1.38 Å (for 336 aligned residues). The loop β6K/α4K undergoes a significant relocation to accommodate the narrow tip of DhaL, with the Cα atoms of Leu142K and Tyr143K moving by approximately 13 Å. Removing this loop from the comparison results in an rmsd of 0.5 Å for 328 Cα atoms. The loop β8K/β9K moves by approximately 3 Å at Pro221K to accommodate Leu182L. A consequence of the adjustment of loop β8K/β9K is the ordering of the neighboring loop β7K/β8K, which is disordered in the free DhaK (2). This ordering effect also includes the N-terminal segment of DhaK (residues 2–9), which is now sandwiched between the β7K/β8K loop and the β9K′/α6K′ loop of the other DhaK subunit and forms part of the dimerization interface. DhaK residues involved in the interface are found in loops β4K/α2K, β6K/α4K, β7K/β8K, β8K/β9K and in helix α3K (for more details see SI Text).
Fig. 2.
Conformational changes in the DhaK subunit induced by complex formation. (A) Uncomplexed forms of DhaK (PDB ID code 1OI2, shown in pale green) and DhaL (PDB ID code 2BTD, shown in wheat) superimposed onto their respective subunits from the DhaK–DhaL complex (yellow and cyan, respectively). The four regions in DhaK that undergo significant conformational changes are colored in magenta. The second subunit of DhaK is shown in white. (B) Model of the DhaK–DhaL(ATP) complex refined by molecular dynamics. The ATP molecule, two magnesium ions, and active site residues are shown. In this model, the γ-phosphoryl atom of ATP is located 3.9 Å away (indicated by the orange dashed line) from the γ-OH group of Dha. The interactions between the Dha molecule and the residues His56K and Asp109K, which we show are important in catalysis, are shown as green dashed lines.
Although the overall structure of both DhaK and DhaL subunits in the E. coli complex are similar to the corresponding domains of the ATP-dependent C. freundii Dha kinase, with rmsd values of 1.4 Å (307 Cα atoms) and 1.7 Å (162 Cα atoms), respectively, our crystal structure reveals that the binding interface between these two subunits/domains is quite different as a result of domain swapping in C. freundii (Fig. S1A). Importantly, and in contrast to the C. freundii Dha kinase structure, in the E. coli structure, the orientation of DhaL relative to DhaK is consistent with a direct transfer of the γ-phosphoryl moiety from ATP to the γ-OH group of Dha (Fig. 1B).
We have also crystallized E. coli DhaK in a different crystal form (Table S1) than that previously reported (2). Interestingly, in our free DhaK structure, the loop β6K/α4K (residues 138–145) displays a conformation intermediate between that of free DhaK (PDB ID code 1OI2) and that of DhaL-bound DhaK (see above), providing a snapshot of the movement of this loop (Fig. S2). The flexibility of the loop β6K/α4K is pivotal for the ability of DhaK to bind DhaL, because conformations of this loop in both free DhaK structures are not compatible with complex formation. The difference between the two structures of free DhaK is likely caused by changes in packing environment.
The Dha Kinase Active Site Is Formed at the Interface of DhaL-DhaK Subunits.
In order to bring DhaL-bound ADP in proximity to DhaK-bound Dha, the tip of the DhaL helical barrel containing loop α7L/α8L (residues 175–185) that caps the nucleotide-binding site, dips into the cavity formed between the β6K/α4K and β8K/β9K loops, displacing both loops from their locations observed in free DhaK (Fig. 2A). The distance between the β-phosphate oxygen of ADP and the γ-OH group of Dha is 4.8 Å, sufficient to accommodate the missing ATP γ-phosphate in our DhaK–DhaL(ADP) complex as shown by our model of DhaK–DhaL(ATP) (Fig. 2B).
Both subunits contribute residues to form the active site. The key residues in the Dha binding pocket, including His218K covalently bound to Dha, Gly53K, His56K, Lys104K, and Asp109K, are in almost identical positions as in the free DhaK structure. Dha is further anchored by Arg178L, whose guanidinium group hydrogen bonds to the γ-OH of Dha and likely stabilizes the trigonal bipyramidal transition state during phosphoryl transfer. All of these residues are highly conserved in bacterial Dha kinases. Mutations of His218K in E. coli DhaK or Arg161L in L. lactis DhaL (corresponding to Arg178L in E. coli) result in inactive enzymes (2, 6).
Despite the lack of conformational changes in DhaL upon binding to DhaK, the adenine ring of ADP undergoes a syn to anti conversion (Fig. S3) when compared with the free DhaL structure (4). Our molecular dynamics (MD) simulations indicate that in the syn conformation, ADP has a potential steric clash of its adenine ring with Thr107K (Fig. S4). Flipping of the adenine ring is accompanied by formation of three additional water-mediated H bonds, two between its purine N1 and the carbonyl group of Val117L and the amide group of Ala123L and the third between its purine N6 and the OH group of Thr107K. In addition, one of the two Mg2+ ions (M2) in the ADP binding site of DhaL moves by approximately 1.0 Å compared to free DhaL and becomes a bridging element at the DhaK–DhaL interface (Fig. S3). Its coordination number increases from three to five with the addition of the carbonyl of Phe78K and a water molecule.
His56K and Asp109K Are Important for Dha Kinase Activity.
In order to evaluate the contributions of residues in the active site for activity, we performed site-directed mutagenesis and activity measurements for several mutant enzymes, focusing on the conserved residues His56K, Asp109K, His218K, and Arg178L. Gel filtration chromatography revealed that all of the DhaK mutant enzymes (H56AK, H56NK, D109AK, D109NK, and H218KK) were able to form complexes with DhaL, while no complex formation between DhaK and R178EL was observed (see SI Text). Two variants of His56K, H56AK and H56NK, show a moderate decrease in  but at least a 40- to 300-fold increase in
but at least a 40- to 300-fold increase in 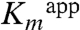 (Table 1). Both D109AK and D109NK mutant enzymes showed no measurable activity (Fig. S5). Mutation of His218K to either lysine (Fig. S5) (2) or alanine (2) also abolishes activity, consistent with its role in binding and orienting the substrate. In addition to the coupled enzyme assay, similar results were obtained by NMR spectroscopy (Fig. S6).
(Table 1). Both D109AK and D109NK mutant enzymes showed no measurable activity (Fig. S5). Mutation of His218K to either lysine (Fig. S5) (2) or alanine (2) also abolishes activity, consistent with its role in binding and orienting the substrate. In addition to the coupled enzyme assay, similar results were obtained by NMR spectroscopy (Fig. S6).
Table 1.
Kinetic constants of wild-type DhaK and His56K mutants for Dha substrate
| WT | H56AK | H56NK | |
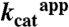 (min-1) (min-1) |
507 ± 25 | 96.1 ± 2.0 | 223 ± 4.3 |
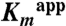 (mM) (mM) |
< 0.008* | 0.302 ± 0.005 | 2.20 ± 0.15 |
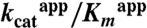 (mM-1 min-1) (mM-1 min-1) |
> 6.3 × 104 | 318 | 101 |
Standard deviations were calculated from three independent experiments.
*Due to the limited sensitivity of the glycerol-3-phosphate dehydrogenase coupled assay for WT at low substrate concentrations the  could not be accurately determined.
could not be accurately determined.
Structures of DhaK H56NK and H56AK Reveal the Ability of Dha to Bind Noncovalently.
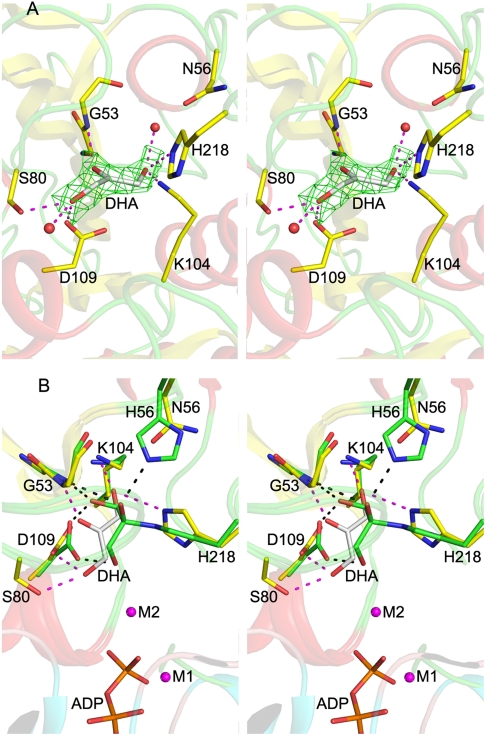
To gain further insight into the reduced activity exhibited by the H56NK and H56AK mutant enzymes we determined their crystal structures. The overall structure of either the H56NK or H56AK enzyme is almost identical to that of the previously reported structure of wild-type DhaK (2), although they crystallized in a different packing environment (Table S1) with four subunits in the asymmetric unit. However, in contrast to the structures of wild-type DhaK, no electron density corresponding to the Dha substrate was observed in any of the subunits in the structures of these mutant enzymes (Fig. S7). These structures were immediately suggestive of a critical role for His56K in hemiaminal bond formation with Dha. The H56NK and H56AK structures reveal very small conformational changes in the active site as compared to wild-type DhaK with covalently bound Dha. These differences include an approximatley 35° rotation of the imidazole ring of His218K, possibly resulting from the shorter Asn56K and Ala56K side chains in the mutant enzyme structures. The carboxylate group of Asp109K also rotates approximatley 35°, possibly due to the loss of its hydrogen bonding interaction with the hydroxyl groups of Dha.
To better understand how the H56NK mutant enzyme could retain some activity in the absence of covalent substrate binding, we determined the crystal structure of H56NK in the presence of Dha by supplementing the crystallization buffer with 10 mM Dha. These crystals contained four molecules in the asymmetric unit (Table S1). In one subunit there is clear electron density showing the presence of Dha bound in the active site (Fig. 3A). Although this non-covalently bound Dha molecule occupies the same binding pocket as in the covalently bound structure, its C2 and O2 atoms are displaced by approximately 2 Å and approximately 3 Å, respectively, as compared with covalently bound Dha. The absence of the covalent bond results in somewhat different interactions between Dha and the surrounding residues. The Dha carbonyl group is anchored via hydrogen bonds with the main chain amide group of Gly53K; the hydroxyl group (α-OH) buried in the binding pocket is within H-bonding distance of Lys104K(NZ) and His218K(NE2), while the other exposed hydroxyl group (γ-OH) forms a hydrogen bond to Asp109K(OD1) and Ser80K(OG) (Fig. 3A). Although this γ-OH is still orientated toward the β-phosphate group of ADP, it is shifted approximately 1.7 Å away from its position in the covalently bound substrate (Fig. 3B). We surmise that this rearrangement contributes to the observed reduced catalytic efficiency of the mutants (Table 1). Another contribution to the lower activity of the H56NK and H56AK mutants is their weaker affinity for the substrate, as reflected by a significant increase in the 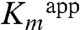 (Table 1). Although our upper estimate of
(Table 1). Although our upper estimate of 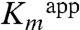 for Dha in wild-type DhaK is 8 μM, the corresponding
for Dha in wild-type DhaK is 8 μM, the corresponding 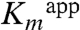 is 302 μM and 2.2 mM for H56AK and H56NK, respectively (Table 1). Despite the different positioning of Dha in the H56NK active site, the γ-OH remains within H-bonding distance (∼2.9 Å) of Asp109K(OD) therefore retaining the ability of Asp109K to act as a catalytic base by deprotonating Dha.
is 302 μM and 2.2 mM for H56AK and H56NK, respectively (Table 1). Despite the different positioning of Dha in the H56NK active site, the γ-OH remains within H-bonding distance (∼2.9 Å) of Asp109K(OD) therefore retaining the ability of Asp109K to act as a catalytic base by deprotonating Dha.
Fig. 3.
Crystal structure of the DhaK H56NK mutant. (A) Active site of the DhaK H56NK mutant bound noncovalently to Dha. The DhaK molecule is shown in cartoon representation. Asn56K and the residues involved in substrate binding are shown in stick mode. Hydrogen bonding interactions involving Dha are indicated by magenta dashed lines. Red spheres represent two water molecules. The difference (Fo - Fc) electron density map for Dha (omitted from refinement) contoured at 2.5σ is shown in green. (B) Substrate binding pocket of the DhaK H56NK–Dha complex superimposed onto that of the DhaK–DhaL complex. The carbon atoms of the labeled residues in the DhaK H56NK mutant and in the DhaK–DhaL complex are colored in yellow and green, respectively. The phosphate moieties of ADP as well as the two magnesium ions in the DhaK–DhaL complex are also shown. Hydrogen bonds involving the noncovalently and covalently bound Dha molecules are represented by magenta and black dash lines, respectively.
Mechanistic Insight into Phosphotransfer.
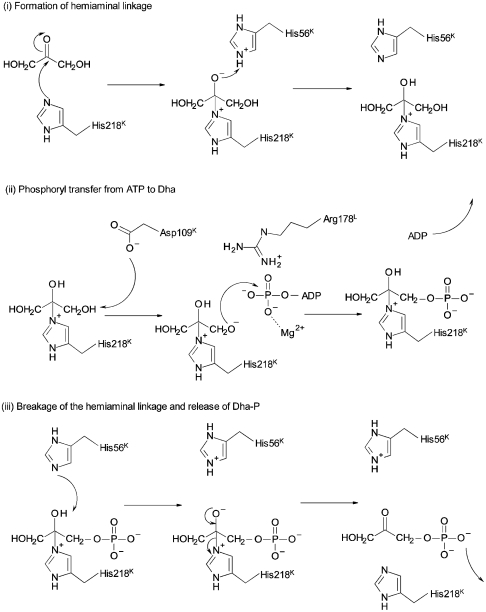
The chemical mechanisms of phosphoryl transfer in general, and with respect to kinases in particular, have been well studied (reviewed in ref. 7). However, little has been proposed regarding how DhaK forms the covalent adduct, catalyzes phosphoryl transfer, and ultimately releases its product. We divide the overall reaction into three stages: (i) binding of Dha to DhaK and formation of the covalent hemiaminal linkage; (ii) phosphoryl transfer from ATP to the covalently bound Dha, generating Dha-P and ADP; and (iii) release of Dha-P from DhaK, setting the stage for a new catalytic cycle. We propose here a plausible chemical mechanism for the overall reaction (Fig. 4). With the exception of the demonstrated roles for His218K of DhaK (2) and Arg178L of DhaL (6), the remaining residues in the active site have not been previously implicated as having a role in catalysis.
Fig. 4.
Proposed catalytic mechanism for the DhaK–DhaL kinase, consisting of (i) initial formation of the hemiaminal linkage between His218K and Dha; (ii) phosphoryl transfer from ATP to Dha, yielding Dha-P; and (iii) breaking of the hemiaminal linkage and release of Dha-P.
Consistent with our structural data, we propose that formation of the initial covalent complex with Dha (Fig. 4i) involves nucleophilic attack of His218K on the electrophilic carbonyl Cβ atom of Dha. For this, the NE2 atom of His218K needs to be deprotonated. The proximal protonated His56K acts subsequently as a general acid, donating its proton to form the Dha β-OH moiety (Fig. 4i). Although in general a water molecule could also play the role of general acid, there is no suitably located water molecule in any of the available crystal structures of wild-type DhaK. Formation of the covalent enzyme–substrate complex allows for precise orientation of the substrate with respect to other active site residues, including Gly53K, His56K, Lys104K, and Asp109K.
Following formation of the covalent DhaK–Dha complex, the next step is transfer of the γ-phosphoryl group of ATP to the γ-OH of Dha (Fig. 4ii). This reaction requires activation of the γ-OH through removal of a proton, in order to generate an oxyanion suitable for attack at the ATP γ-phosphorous atom. This activation requires a residue acting as a general base, which we identify as Asp109K, within H-bonding distance of Dha γ-OH. This role is supported by the loss of activity observed for the Asp109K mutant enzymes (Fig. S5). In the DhaK–DhaL complex, the leaving oxygen (O2B of ADP) and the entering oxygen (Oγ of Dha) are separated by 4.8 Å (Fig. 1B). Based on molecular dynamics simulations on the DhaK–DhaL–ATP model, the Pγ-Oγ(Dha) distance in the Michaelis complex is estimated to be 3.9 Å (Fig. 2B). The locations of negative charges developing during phosphoryl transfer may be inferred from the location of stabilizing, positively charged groups provided by the enzyme. An invariant residue, Arg178L, is located at the active site of the DhaK–DhaL complex and has been suggested to be essential for activity by mutagenesis (6). This residue has been postulated to stabilize the γ-phosphoryl group during phosphoryl transfer. In the crystal structure, the guanidinium group of Arg178L is within H-bonding distance of the γ-OH group of Dha while at the same time does not directly interact with the β-phosphate of ADP (3.8 Å away). As previously stated, the second magnesium ion (M2) in the complex is coordinated by five oxygens, one of which is from a water molecule located between the β-phosphate of ADP and the substrate γ-OH group and likely mimics one of the oxygen atoms in the γ-phosphate group of ATP. Such an arrangement indicates that both Arg178L and M2 are important for stabilizing the negative charges accumulated on the γ-phosphoryl group (γ-phosphate) during phosphoryl transfer. In the DhaK–DhaL complex, no positively charged residue other than Arg178L could be found at the interface.
The final step in catalysis involves dissociation of Dha-P from DhaK, requiring breaking the hemiaminal linkage. Based on the structure of the active site, His56K is well positioned to act as a general base, abstracting the proton from the β-OH of Dha-P, with concomitant formation of the carbonyl group at C2 and breakage of the Dha-P-His218NE2 bond (Fig. 4iii). The only other amino acid that could potentially act as a general base, Asp109K, which interacts directly with the primary hydroxyl groups of Dha via its γ-carboxyl oxygens, is located too far to interact with the β-OH of Dha. Here, the oxygen–oxygen distance is 5.4 Å as opposed to 2.6 Å between NE2 of His56K and the β-OH of Dha. As suggested previously (8), the γ-carboxyl oxygen of Asp109K loses the ability to form an H bond with the primary hydroxyl group of Dha after the phosphoryl-ester is formed. Unfavorable interactions between the phosphate group of Dha-P and the carboxylate group of Asp109K may help release the product from the enzyme. Together our data allow us to propose a chemical mechanism for DhaK supported by several structural and biochemical studies, consistent with involvement of His56K and Asp109K in addition to the previously characterized His218K (2) and Arg178L (4). These residues are highly sequence conserved in both bacterial and eukaryotic Dha kinases suggesting their common mechanism.
Our results indicating some activity for the H56AK and H56NK mutants require explanation. The first possibility is that covalently linked substrate is not essential for catalysis. Noncovalent binding of Dha, as reflected by our crystal structure, leads to reduced affinity as indicated by increased 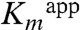 as well as to a suboptimal orientation of γ-OH, affecting
as well as to a suboptimal orientation of γ-OH, affecting  . If the covalent linking is not essential for activity, why are H218KK and H218AK mutants dead? We believe that because the lysine mutant introduces a more bulkly side chain it does not allow for the binding of Dha. Furthermore, the small Dha molecule could not be well positioned in the absence of the histidine imidazole ring in the H218AK mutant. The second possibility is that catalysis in H56K mutants is achieved through the covalently bound Dha, assuming that His56K determines the equilibrium between the covalently and noncovalently bound states of Dha. The covalent bond might be formed without participation of His56K; for example, a water molecule could replace His56K as general acid in the H56AK and H56NK mutants. The fact that
. If the covalent linking is not essential for activity, why are H218KK and H218AK mutants dead? We believe that because the lysine mutant introduces a more bulkly side chain it does not allow for the binding of Dha. Furthermore, the small Dha molecule could not be well positioned in the absence of the histidine imidazole ring in the H218AK mutant. The second possibility is that catalysis in H56K mutants is achieved through the covalently bound Dha, assuming that His56K determines the equilibrium between the covalently and noncovalently bound states of Dha. The covalent bond might be formed without participation of His56K; for example, a water molecule could replace His56K as general acid in the H56AK and H56NK mutants. The fact that 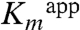 rather than
rather than 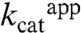 is more affected by the His56K mutation provides some support for this idea. However, we could not identify appropriate water molecule in any of the His56K mutant structures. Moreover, despite having analyzed 40 independent subunits of the crystal structures of DhaK H56AK or H56NK mutants, we were not capable of capturing the covalently bound substrate even at 40 mM Dha. Therefore, it is more likely that covalent binding of Dha is not essential for activity.
is more affected by the His56K mutation provides some support for this idea. However, we could not identify appropriate water molecule in any of the His56K mutant structures. Moreover, despite having analyzed 40 independent subunits of the crystal structures of DhaK H56AK or H56NK mutants, we were not capable of capturing the covalently bound substrate even at 40 mM Dha. Therefore, it is more likely that covalent binding of Dha is not essential for activity.
Functional Implications of Covalent vs. Noncovalent Substrate Binding by Dha Kinases.
Formation of a transient, covalent bond between substrate and enzyme usually directly contributes to catalysis (9, 10, 11); however, covalent substrate binding by dihydroxyacetone kinases is distinctively different. Previous studies have suggested that the primary role of the hemiaminal bond formed between DhaK and Dha or Dha-P is to provide the enzyme with a way of specifically selecting for short chain ketoses and aldoses vs. polyalcohols, because the enzyme displays remarkable selectivity for Dha in comparison with the closely related molecule, glycerol (8). This is in contrast to glycerol kinase, which is able to effectively utilize both Dha and glycerol as substrates for phosphorylation (12, 13). Quantum-mechanical calculations did not provide any evidence for an influence of the hemiaminal bond on activation of the Dha γ-O atom involved in nucleophilic attack of the γ-phosphate of ATP (8). In our hands, addition of glycerol (10 mM–1 M) to phosphotransferase reactions showed no decrease in kinase activity for wild-type and both H56K mutant enzymes, indicating that the specificity of DhaK for Dha is not a consequence of hemiaminal bond formation per se. The apparent inability of glycerol to compete with Dha for binding to the DhaK active site may be a consequence of subtle structural differences between the two molecules, due to the sp2 β-C in Dha vs. the sp3 β-C of glycerol, resulting in steric conflicts of the latter within the quite rigid DhaK active site (based on comparison of all available crystal structures).
Our structural and biochemical results show that covalent bond formation does contribute, in part, to orientation of the substrate for optimal phosphoryl transfer. An additional potential role for hemiaminal bond formation to that in catalysis is for it to serve as an on/off switch, thereby exerting a regulatory function. A covalent bond between Dha and DhaK would support the enzyme existing in either a “Dha-loaded” or “Dha-unloaded” form, as dictated by (i) the inherent affinity of the enzyme for substrate and (ii) the effective concentration of Dha within the cell. The hemiaminal bond, once formed, commits DhaK to the “loaded” state, regardless of further changes in intracellular Dha concentration. It also provides a form of DhaK already primed for catalysis, once an ATP-loaded form of DhaL is available. In this manner, Dha binding and phosphoryl transfer are effectively uncoupled, allowing, for example, variations in the intracellular PEP pool independent of the particular state of DhaK. In a similar manner, the covalent bond would allow for an on/off switch with regards to transcriptional activation or repression that has been shown to occur via interactions with the transcription regulator DhaR (14). In either case, one would predict that a DhaK enzyme incapable of forming the hemiaminal bond in vivo would exhibit an attenuated response as opposed to an on/off behavior, given that the substrate could bind/unbind to the enzyme active site. Importantly, His56K is as strongly sequence conserved as is the catalytic Asp109K in Dha kinases across various species, including higher eukaryotes. This suggests a physiological importance for covalent substrate binding for this class of kinase in both PEP and ATP-dependent branches. The precise nature of the physiological consequences on cellular Dha metabolism resulting from loss of hemiaminal bond formation is the subject of future studies.
Materials and Methods
Protein Expression, Purification, and Characterization.
Details for cloning, mutagenesis, expression, and purification are available in SI Text. Briefly, DhaK and DhaL N-terminal His8-fusion proteins were purified by nickel affinity chromatography and the tags on DhaK removed by cleavage with tobacco etch virus (TEV) protease. Enzyme I and DhaM GST-fusion proteins were purified by glutathione affinity chromatography and cleaved from the column by TEV. HPr was purified by solubilizing inclusion bodies in urea and refolding the protein on nickel resin. For crystallization, the His8-tag was cleaved from DhaL and the DhaK–DhaL complex was further purified on a Superdex 200 size exclusion column equilibrated with 50 mM Tris pH 8, 10 mM NaCl, 1 mM magnesium acetate, 1 μM AMP-PNP, and 1 mM DTT. Complex formation of kinase mutants was analyzed by size exclusion chromatography in the same manner except that 1 μM ADP was substituted for AMP-PNP.
Crystallization.
Crystals of the DhaK–DhaL complex were obtained using the Anion Suite screen (Qiagen). Optimization of conditions led to the best crystals from hanging drop vapor diffusion at 19 °C by equilibrating 2 μL of protein (10 mg/ml) with 1 μL reservoir solution (0.1 M HEPES pH 7.5, 3.5 M sodium formate) over 1 mL of reservoir solution. Crystals belong to space group P41212 with unit cell dimensions a = b = 74.6, c = 268.8 Å. Crystals of wild-type DhaK and mutant enzymes were also obtained by hanging drop vapor diffusion by equilibrating 1 μL of protein (2.5 mg/ml of DhaK and 1.7 mg/ml of DhaL-his) with 1 μL reservoir solution (0.1 M sodium citrate pH 5.6, 20% [w/v] PEG 8000) over 1 mL of reservoir solution. Crystals of the wild-type DhaK belong to space group P21 with a = 49.8, b = 91.5, c = 73.2 Å, β = 89.9°. Crystals of H56AK and H56NK belong to space group P1 with unit cell dimensions a = 59.7, b = 82.6, c = 92.9 Å, α = 77.9, β = 78.1, γ = 71.1°, whereas that of the DhaK-H56NK-Dha complex form in space group P21 with a = 82.2, b = 101.1, c = 99.3 Å, β = 89.95°. For data collection, crystals were transferred to reservoir solution supplemented with 12% (v/v) ethylene glycol and flash cooled in a nitrogen stream at 100 K (Oxford Cryosystems).
X-ray Data Collection, Structure Solution, and Refinement.
Diffraction data for free DhaK and the DhaK–DhaL complex were collected to 2.2 Å at the 31-ID beamline (LRL-CAT), Advanced Photon Source, Argonne National Laboratory. Data integration and scaling were performed with the program HKL2000 (15). Structure determination was performed by molecular replacement using the program Phaser (16) from the CCP4 suite with the previously reported E. coli DhaK (PDB 1OI2) (2) and DhaL (PDB ID code 2BTD) (4) structures as the search models. Refinement was carried out with the program Refmac5 (17) with final Rwork/Rfree of 0.172/0.210 and 0.189/0.225 for DhaK and DhaK–DhaL, respectively. Diffraction data for crystals of DhaK mutant enzymes were collected to 2.55 Å (H56AK), 1.97 Å (H56NK) and 2.2 Å (H56NK-Dha) at the CMCF1 beamline at the Canadian Light Source. Data processing and structure determination was done similarly as described above. All models have good geometry as analyzed with PROCHECK (18). Final data collection and refinement statistics are shown in Table S1.
Molecular Dynamics Simulations.
The DhaK–DhaL–ATP, DhaK–DhaL–syn–ADP, and DhaK–DhaL–anti–ADP complexes were sampled by 5-ns MD simulations using the AMBER10 suite of programs (19) together with the AMBER ff03 force field for the proteins and a modified force field for ATP and ADP (20). Details of the calculations are available in SI Text.
Phosphotransferase Assay.
Phosphorylation of Dha was measured in a coupled assay similar to that described previously (1). To remove residual endogenous DhaK that could copurify with recombinant DhaL, the latter was further purified by size exclusion (Superdex 200) in buffer containing 20 mM Tris pH 8, 300 mM NaCl, 1 mM DTT, 1 mM MgCl2, and 0.01 mM ADP. Phosphotransferase reactions for specific activity measurements contained 1 μM Enzyme I, 1 μM HPr, 0.5 μM DhaM, 0.5 μM DhaL, 0.5 μM DhaK, 50 mM potassium phosphate pH 7.5, 16 mM Dha, 2.5 mM DTT, 2.5 mM MgCl2, 2 units glycerol-3-phosphate dehydrogenase (Sigma Chemical Co.), and 1 mM NADH and were initiated by the addition of 2 mM PEP. The glycerol-3-phosphate dehydrogenase coupling enzyme uses NADH to convert Dha-P to glycerol-3-phosphate. The production of Dha-P in the reaction was followed by monitoring oxidation of NADH at 340 nm in a Spectramax 250 plate reader at room temperature. The effect of glycerol on phosphotransferase activity for wild-type, H56AK and H56NK was examined for reactions in the presence of 10 mM, 100 mM and 1 M glycerol. 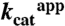 was measured for wild-type enzyme from coupled phosphotransferase reactions (0–1.6 mM Dha) containing 0.5 μM Enzyme I, 0.5 μM HPr, 0.25 μM DhaM, 0.25 μM DhaL, 0.25 μM DhaK, 50 mM potassium phosphate pH 7.5, 2.5 mM DTT, 2.5 mM MgCl2, 2 units glycerol-3-phosphate dehydrogenase (Sigma Chemical Co.), and 1 mM NADH, initiated by addition of 2 mM PEP. These conditions are similar to those reported previously (8), and kinetic constants were derived by varying concentration of Dha only, leading to apparent values of kinetic parameters. Due to the limited sensitivity of the assay for wild-type enzyme seen at low substrate concentrations an accurate value for
was measured for wild-type enzyme from coupled phosphotransferase reactions (0–1.6 mM Dha) containing 0.5 μM Enzyme I, 0.5 μM HPr, 0.25 μM DhaM, 0.25 μM DhaL, 0.25 μM DhaK, 50 mM potassium phosphate pH 7.5, 2.5 mM DTT, 2.5 mM MgCl2, 2 units glycerol-3-phosphate dehydrogenase (Sigma Chemical Co.), and 1 mM NADH, initiated by addition of 2 mM PEP. These conditions are similar to those reported previously (8), and kinetic constants were derived by varying concentration of Dha only, leading to apparent values of kinetic parameters. Due to the limited sensitivity of the assay for wild-type enzyme seen at low substrate concentrations an accurate value for 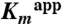 could not be determined. For H56AK and H56NK,
could not be determined. For H56AK and H56NK, 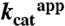 and
and 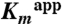 were determined from reactions containing 1 μM Enzyme I, 1 μM HPr, 0.5 μM DhaM, 0.5 μM DhaL, and 0.5 μM DhaK at a range from 0–16 mM and 0–64 mM Dha, respectively. Values were calculated by nonlinear regression fit to a hyperbola.
were determined from reactions containing 1 μM Enzyme I, 1 μM HPr, 0.5 μM DhaM, 0.5 μM DhaL, and 0.5 μM DhaK at a range from 0–16 mM and 0–64 mM Dha, respectively. Values were calculated by nonlinear regression fit to a hyperbola.
Supplementary Material
Acknowledgments.
We thank Dr. Shaunivan Labiuk (Canadian Light Source; CLS) for collecting several datasets; John Wagner, Ming-Ni Hung, and Linhua Zhang for cloning; and Drs. Traian Sulea, Enrico O. Purisima, and Stephan Grosse for helpful discussions. This research was supported by Canadian Institutes of Health Research (CIHR) Grant MOP-48370 (I.E., A.M., and M.C.). X-ray diffraction data for this study were measured at Lilly Research Laboratory Collaborative Access Team (LRL-CAT) at the Advanced Photon Source, Argonne National Laboratory and at CMCF1 at CLS. Use of the Advanced Photon Source at Argonne National Laboratory was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. Use of the LRL-CAT beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly & Company, which operates the facility. The Canadian Macromolecular Crystallography Facility is supported by the Canadian Foundation for Innovation, the Natural Sciences and Engineering Research Council of Canada, and CIHR. This is NRCC publication 50686.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012596108/-/DCSupplemental.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3PNK (DhaK), 3PNL (DhaK–DhaL complex), 3PNM (DhaK H56A), 3PNO (DhaK H56N), and 3PNQ ( DhaK H56N-Dha)].
*Superscript letter K (or L) denotes kinase subunit.
References
- 1.Gutknecht R, Beutler R, Garcia-Alles LF, Baumann U, Erni B. The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J. 2001;20:2480–2486. doi: 10.1093/emboj/20.10.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siebold C, Garcia-Alles LF, Erni B, Baumann U. A mechanism of covalent substrate binding in the x-ray structure of subunit K of the Escherichia coli dihydroxyacetone kinase. Proc Natl Acad Sci USA. 2003;100:8188–8192. doi: 10.1073/pnas.0932787100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bächler C, Flükiger-Brühwiler K, Schneider P, Bähler P, Erni B. From ATP as substrate to ADP as coenzyme: Functional evolution of the nucleotide binding subunit of dihydroxyacetone kinases. J Biol Chem. 2005;280:18321–18325. doi: 10.1074/jbc.M500279200. [DOI] [PubMed] [Google Scholar]
- 4.Oberholzer AE, Schneider P, Baumann U, Erni B. Crystal structure of the nucleotide-binding subunit DhaL of the Escherichia coli dihydroxyacetone kinase. J Mol Biol. 2006;359:539–545. doi: 10.1016/j.jmb.2006.03.057. [DOI] [PubMed] [Google Scholar]
- 5.Siebold C, Arnold I, Garcia-Alles LF, Baumann U, Erni B. Crystal structure of the Citrobacter freundii dihydroxyacetone kinase reveals an eight-stranded alpha-helical barrel ATP-binding domain. J Biol Chem. 2003;278:48236–48244. doi: 10.1074/jbc.M305942200. [DOI] [PubMed] [Google Scholar]
- 6.Zurbriggen A, et al. X-ray structures of the three Lactococcus lactis dihydroxyacetone kinase subunits and of a transient intersubunit complex. J Biol Chem. 2008;283:35789–35796. doi: 10.1074/jbc.M804893200. [DOI] [PubMed] [Google Scholar]
- 7.Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: Evolution of a catalytic scaffold. Trends Biochem Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Alles LF, et al. Phosphoenolpyruvate- and ATP-dependent dihydroxyacetone kinases: Covalent substrate-binding and kinetic mechanism. Biochemistry. 2004;43:13037–13045. doi: 10.1021/bi048575m. [DOI] [PubMed] [Google Scholar]
- 9.Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN. The pentacovalent phosphoryl transfer reaction. Science. 2003;299:2067–2071. doi: 10.1126/science.1082710. [DOI] [PubMed] [Google Scholar]
- 10.Verschueren KH, Seljée F, Rozeboom HJ, Kalk KH, Dijkstra BW. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature. 1993;363:693–698. doi: 10.1038/363693a0. [DOI] [PubMed] [Google Scholar]
- 11.Heine A, et al. Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science. 2001;294:369–374. doi: 10.1126/science.1063601. [DOI] [PubMed] [Google Scholar]
- 12.Jin RZ. Glycerol kinase as a substitute for dihydroxyacetone kinase in a mutant of Klebsiella pneumoniae. J Bacteriol. 1982;152:1303–1307. doi: 10.1128/jb.152.3.1303-1307.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson EA, Burke SK, Forage RG, Lin EC. Purification and properties of dihydroxyacetone kinase from Klebsiella pneumoniae. J Bacteriol. 1984;160:55–60. doi: 10.1128/jb.160.1.55-60.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bächler C, Schneider P, Bähler P, Lustig A, Erni B. Escherichia coli dihydroxyacetone kinase controls gene expression by binding to transcription factor DhaR. EMBO J. 2005;24:283–293. doi: 10.1038/sj.emboj.7600517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 18.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 19.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meagher KL, Redman LT, Carlson HA. Development of polyphosphate parameters for use with the AMBER force field. J Comput Chem. 2003;24:1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.