Abstract
Glial cell line-derived neurotrophic factor (GDNF) is indispensable for ureteric budding and branching. If applied exogenously, GDNF promotes ectopic ureteric buds from the Wolffian duct. Although several downstream effectors of GDNF are known, the identification of early response genes is incomplete. Here, microarray screening detected several GDNF-regulated genes in the Wolffian duct, including Visinin like 1 (Vsnl1), which encodes a neuronal calcium-sensor protein. We observed renal Vsnl1 expression exclusively in the ureteric epithelium, but not in Gdnf-null kidneys. In the tissue culture of Gdnf-deficient kidney primordium, exogenous GDNF and alternative bud inducers (FGF7 and follistatin) restored Vsnl1 expression. Hence, Vsnl1 characterizes the tip of the ureteric bud epithelium regardless of the inducer. In the tips, Vsnl1 showed a mosaic expression pattern that was mutually exclusive with β-catenin transcriptional activation. Vsnl1 was downregulated in both β-catenin-stabilized and β-catenin-deficient kidneys. Moreover, in a mouse collecting duct cell line, Vsnl1 compromised β-catenin stability, suggesting a counteracting relationship between Vsnl1 and β-catenin. In summary, Vsnl1 marks ureteric bud tips in embryonic kidneys, and its mosaic pattern demonstrates a heterogeneity of cell types that may be critical for normal ureteric branching.
The differentiation of the permanent kidney or metanephros is initiated at embryonic day (E) 10.5 by sprouting of a ureteric bud (UB) from the Wolffian duct (WD). The UB then undergoes several cycles of branching to form the collecting duct system, whereas the UB tips induce secretory nephrons in the surrounding metanephric mesenchyme (MM).1
Both genetic and tissue culture studies have pinpointed glial cell line-derived neurotrophic factor (GDNF) as an essential regulator of ureteric budding and branching.2 GDNF is synthesized by the cap condensate cells of the MM.3 A dimeric complex of GDNF and the coreceptor GDNF family receptor α1 (GFRα1) binds to and phosphorylates Ret receptor tyrosine kinase in the UB tips.4–6 Mouse ablation of Gdnf, Ret, or GFRα1 disrupts either primary ureteric budding or subsequent branching, resulting in renal agenesis or severe hypodysplasia.7–9 On the other hand, forced expression of GDNF by the ureteric epithelium in vivo or GDNF supplementation in vitro promotes supernumerary budding from WD.10,11 GDNF upregulates Ret, Sox9, Wnt11, Etv4, and Etv5 in the UB tips.12,13 The tip identity of the ureteric epithelium, characterized by the above mentioned genes for instance, has been suggested to be critical for UB budding and branching.14
A substantial number of mice lacking Gdnf (27%)15 or Ret (40 to 45%)9 show rudimentary kidneys, suggesting that pathways partially redundant with GDNF contribute to UB branching. In vitro and in vivo studies have revealed roles for members of fibroblast growth factor (FGF) family in primary ureteric budding. A combination of FGF7 and follistatin, an inhibitor of activin A, promotes supernumerary UBs from the WD in vitro,16 and FGF10 alone has the same effect.17 Genetic models support these results, because mice lacking either Fgf7, Fgf10,18,19 or Fgfr2 deleted specifically in the ureteric epithelium20 show renal hypoplasia. The disruption of renal differentiation in these mice is far less severe than in mice lacking Gdnf or Ret, indicating that FGF signaling has a secondary role in vivo. However, FGF10 can partially substitute many renal functions of GDNF in the absence of Sprouty.17
Wnt signaling regulates cell functions by activating the canonical β-catenin pathway, the planar cell polarity pathway, or the calcium pathway. Two Wnt reporter mouse lines, TCF-gal21 and BAT-gal,22 have been used in assessing renal β-catenin activity. Both lines show high 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) labeling in the UBs.23,24 β-Catenin acts upstream of essential genes required for ureteric branching, because mice lacking β-catenin in the ureteric epithelium display renal agenesis or severe hypodysplasia.25,26
To search for early response genes to GDNF in ureteric budding, we performed a genome-wide analysis of mRNA expression of WDs cultured for 16 hours with or without GDNF. Vsnl1 was among the genes upregulated by GDNF in the Wolffian duct.
In situ hybridization on wild-type and Gdnf -deficient embryonic kidneys was used to validate the results. Vsnl1 has been previously identified as a UB molecule,27 but there are no reports on its expression pattern or possible function in kidney development. Vsnl1 was specifically expressed in the UB tip and was lacking in the E11 Gdnf−/− kidneys. Vsnl1 is one of the neuronal calcium sensor proteins and is highly expressed in the brain, but low levels are detectable also in the adult heart, testis, ovary, and colon.28 Binding of calcium to Vsnl1 leads to a conformational change (calcium-myristoyl switch), exposing hydrophobic stretches and the myristoyl residues at the protein surface, thereby making these domains available for interaction with the cell membrane and/or target molecules.29,30 Vsnl1 modulates the activity of guanylyl cyclase B through clathrin-dependent cycling31 and enhances insulin secretion by pancreatic β cells.32
We now demonstrate that Vsnl1 mRNA characterizes the UB tips, and the protein shows a unique mosaic pattern in the UB. Besides Gdnf knockouts, Vsnl1 is downregulated in several other genetic models with disrupted UB branching and is upregulated by all known UB inducers. Vsnl1 expression pattern is mutually exclusive with β-catenin activity in the UB tips of BAT-gal reporter mice. Moreover, Vsnl1 colocalizes in the tips with high intracellular calcium levels and compromises β-catenin stability in mIMCD3 cells, suggesting that Vsnl1 modulates β-catenin activity in the ureteric epithelium.
RESULTS
Microarray Analysis of GDNF-regulated Genes in the Wolffian Duct
To identify new GDNF target genes, we did a genome-wide transcriptional profiling of mouse WD cultured for 16 hours with or without GDNF (Supplemental Figure 1, A through C) by using GeneChip® Mouse Genome 430 2.0 Array containing more than 45,000 probe sets.
16 hours of GDNF exposure showed no morphologic responses (Supplemental Figure 1B); however, the treatment had a clear effect on gene expression in the microarray samples, as shown by principal component analysis (Supplemental Figure 1E), PCA2, which explained more than 16% of the total variation in the data, correlated with GDNF treatment (R2 = 0.79, P = 0.0389). A Volcano plot of the differentially expressed genes is shown in Supplemental Figure 1D.
Using a P value cutoff of 0.05 combined with a fold change of 1.890 (corresponding to the fold change of GFRα1, a known GDNF target) or more, resulted in 69 upregulated genes (Supplemental Table 1). A similar cutoff was used for the downregulated genes resulting in 50 downregulated candidate genes (Supplemental Table 2).
The microarray list of upregulated genes contained a number of known GDNF targets (Ret, Spry1, and Etv4)12,33,34 or genes that have been previously identified in kidneys (Crlf1, Ros1, Calb1, Arg2, Mtmr7, Wfdc2, Neu1, Tuba4, and Vsnl1) (Supplemental Table 1).27,35–38
The data were validated by in situ hybridization on kidneys of E11 wild-type, GDNF-treated, and Gdnf-deficient mice (Figure 1 and data not shown). Although the genes upregulated by GDNF showed ureteric epithelium expression, only few of them, such as Vsnl1, were absent in the Gdnf−/− kidney explants (Figure 1, middle column). This gene was therefore selected for further analysis. In situ hybridization showed that the downregulated genes were preferentially expressed by the surrounding mesenchyme and not by the tips of UB (data not shown). Therefore, we excluded the downregulated genes from other analysis.
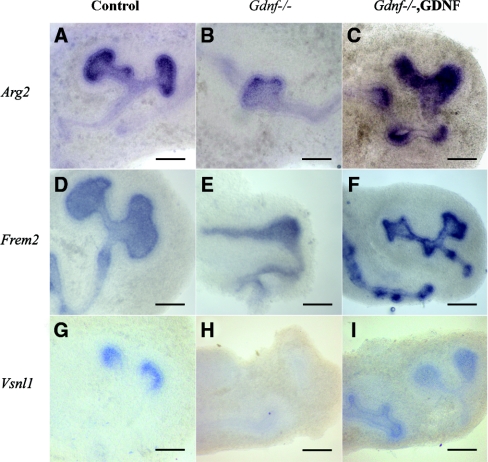
Figure 1.
In situ hybridization validates candidate GDNF targets. Shown are examples of whole-mount in situ hybridization of E11 wild-type (left column), Gdnf-deficient (middle column), and GDNF-treated Gdnf-null kidney explants (right column). (A through C) Arg2. (D through F) Frem2. (G through I) Vsnl1. All of them are expressed in the UBs of the control kidneys and in the ectopic buds of GDNF-treated explants. Unlike Arg2 (B) and Frem2 (E), Vsnl1 is missing in the Gdnf−/− kidney explants (H). Scale bars, 20 μm.
Vsnl1 Is a New Ureteric Bud Tip Marker
Vsnl1 mRNA and protein showed an expression pattern similar to Ret, first in the caudal segment of the WD at E10 and then in the tips of UB at E11, E12, and E13 (Figure 2, A through D). At E15, Vsnl1 protein expression was high in the UB tips, gradually decreasing toward the medullary stalk region (Figure 2E), but the mRNA remained UB tip specific (Figure 2S). At all of the stages analyzed, Vsnl1 protein showed a mosaic expression pattern in the ureteric epithelium (Figure 2, D and E, and Supplemental Figure 2).
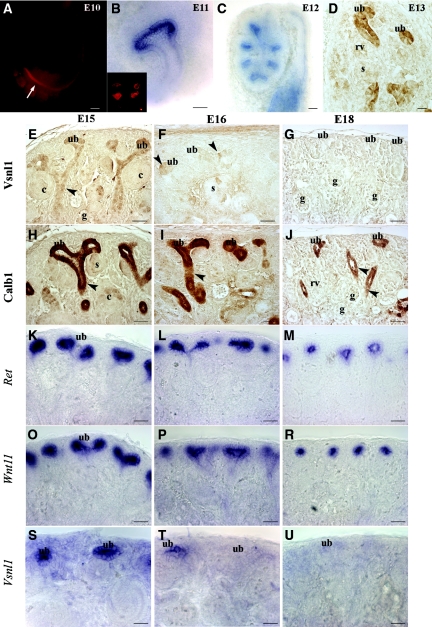
Figure 2.
Vsnl1 is required for early ureteric branching and is not required for the late stage of kidney development. Whole-mount antibody staining of Vsnl1 at E10 shows high levels at the caudal segment of WD close to cloaca (arrow in A). In situ hybridization (B) of E10.5 kidneys and protein labeling (B, inset) show Vsnl1 expression in the tips of the UB. (C) Vsnl1 mRNA in E12 kidneys is tip specific. (D) At E13, Vsnl1 protein levels are high in the UB tips and terminal branches but absent from renal vesicles (rv) and S-shaped bodies (s). (E) At E15, Vsnl1 protein is highly expressed in the tips of the UBs, and gradually lowering expression is seen throughout the terminal stalk region (arrowhead). No expression is observed in comma shaped bodies (c) or glomeruli (g). (S) In contrast to the protein, the Vsnl1 mRNA remain specifically expressed in the UB tips at E15. (F and T) At E16, Vsnl1 starts to decline, and few UB cells are still expressing at low levels the protein (F, arrowheads) and mRNA (T). (G and U) By E18 Vsnl1 protein and transcript expression are absent from the UB tips. (H through R) For comparison, the expression of Calb1 (H through J), Ret (K through M), and Wnt11 (O through R) is shown at E15, E16, and E18. Scale bars, 50 μm.
We compared Vsnl1 expression with another calcium-binding molecule, Calb1 expressed throughout the ureteric epithelium (Figure 2, H through J), with Ret (Figure 2, K through M) and Wnt11 (Figure 2, O through R), which are exclusively found in the UB tips. In contrast to these genes, Vsnl1 mRNA (Figure 2T) and protein expression started to decline by E16 (Figure 2F), and at E18 Vsnl1 staining was not detectable any more (Figure 2, G and U). At all stages of nephrogenesis, no Vsnl1 labeling was seen in pretubular aggregates, secretory nephrons, or stromal or vascular cells (Supplemental Figure 2).
Vsnl1 Is Induced during Ureteric Budding and Branching
Vsnl1 was next analyzed in cultured kidneys and in different genetic models with disrupted UB morphogenesis. Like the transcript (Figure 1, G through I), Vsnl1 protein was upregulated in the supernumerary buds induced by exogenous GDNF (Figure 3A). No Vsnl1 was detected in the E11 Gdnf−/− mouse kidney rudiments (Figure 3B), but its expression was restored by exogenous GDNF (Figure 3C).
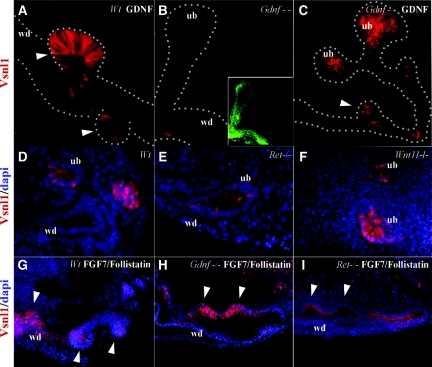
Figure 3.
Vsnl1 is induced during ureteric budding and branching. (A) Vsnl1 (red) is expressed in GDNF-induced ectopic buds. (B) Vsnl1 is absent in E11 Gdnf−/− kidney rudiments where UB is labeled with pan-cytokeratin antibody (green). (C) Vsnl1 expression is rescued by exogenous GDNF (50 ng, 32 hours) in these explants. (D through I) Vsnl1 in Wt (D), Ret−/− (E), and Wnt11−/− (F) E11 kidney sections and in wt (G), Gdnf−/− (H), and Ret−/− (I) kidney explants grown with FGF7/follistatin combination (125/500 ng, 48 hours). The white arrowheads in (A, C, G, H, and I) mark ectopic buds. DAPI staining (blue) marks the nuclei. wd, Wolffian duct; ub, ureteric bud. Scale bars, 50 μm.
To analyze whether Vsnl1 is specifically regulated by GDNF signaling or rather serves as a marker for ureteric tips, Vsnl1 expression was analyzed in different genetic models with disrupted UB branching. Vsnl1 was expressed at low levels in E11 Ret-deficient UB (Figure 3E). In Wnt11-deficient mice, which display a mild defect in ureteric branching,39 Vsnl1 was present at the UB tips (Figure 3F) at levels comparable with the wild-type kidneys (Figure 3D).
FGFs modulate renal branching morphogenesis in vivo and in vitro.17–19 E11 wild-type Gdnf−/− and Ret−/− urogenital block explants were therefore exposed to the combination of FGF7 and follistatin for 48 hours. This combination upregulated Vsnl1 in the supernumerary buds formed along the WD in all explants, irrespective of the genotype (Figure 3, G through I).
The Mosaic Expression Pattern of Vsnl1 Does Not Correlate with Cell Cycle or ERK Phosphorylation
Because Vsnl1 showed a mosaic pattern in the UB tips, we searched for other molecules with uneven or mosaic pattern in these cells. The diphosphorylated form of ERK (dp-ERK1/2) is an established indicator of receptor tyrosine kinase signaling,40,41 with a heterogeneous expression pattern in the UB epithelium.42 Double labeling with antibodies against dp-ERK1/2 and Vsnl1 showed no positive or negative correlation in the UB cells because dp-ERK1/2-positive cells contained variable levels of Vsnl1 (Figure 4, A through C). Double labeling with antibodies against cell cycle markers and Vsnl1 showed that Vsnl1 levels were low during the S phase (Figure 4, D through F) and often high at mitosis (Figure 4, G through L). However, no clear correlation between Vsnl1 and the cell cycle phases could be verified.
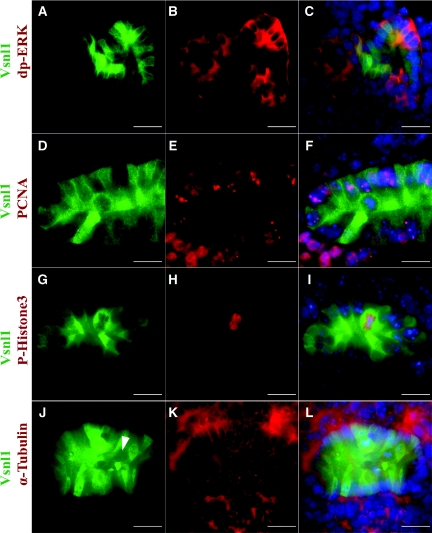
Figure 4.
The mosaic expression pattern of Vsnl1 protein does not correlate with cell cycle or ERK activity. (A through C) Double immunolabeling of E12 kidney sections with anti-dp-ERK1/2, a proliferation marker, and vsnl1 shows no correlation between the two molecules. (D through F) To visualize the synthesis phase of the cell cycle, double labeling with antibodies for Vsnl1 (D and F) and proliferating cell nuclear antigen (E and F) was performed. (G through L) The mitotic cells were visualized by double immunostaining with antibodies against Vsnl1 (G, I, J, and L) and P-His3 (H and I). Vsnl1 expression is high in P-His3-positive cells but also in some P-His3-negative cells. α-Tubulin marks the mitotic spindles (K and L). The arrowhead in J marks a cell in mitosis showing low levels of Vsnl1. DAPI (blue) marks the nuclei in C, F, I, and L). Scale bars, 10 μm.
Cultured Ureteric Bud Tips Show High Vsnl1 Expression and Fluo4 Labeling of Ca2+
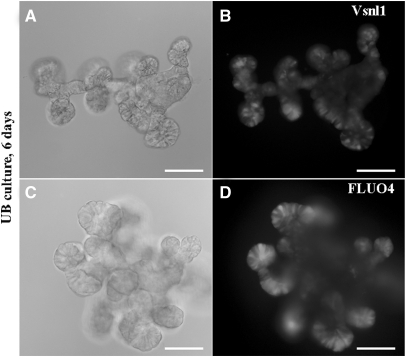
Vsnl1 is a calcium-binding protein, and calcium enables Vsnl1 to interact with the cell membrane by a mechanism known as the calcium-myristoyl switch.30 Vsnl1 was highly expressed at the UB tips of isolated UBs growing on GDNF containing Matrigel43 (Figure 5, A and B). The tips of the UB cultures also contain elevated intracellular calcium levels demonstrated by the strong signal intensity in Fluo4 assay (Figure 5, C and D).
Figure 5.
Vsnl1 expression and Ca2+ activity are high in cultured UB tips. Isolated UB epithelia from E12 kidneys were cultured in GDNF-containing Matrigel for 6 days. (A and B) Whole-mount staining with anti-Vsnl1 antibody reveals high Vsnl1 expression in the tip-like structures in the periphery of the explants. (C and D) Fluo4 staining of the UBs reveals high intracellular Ca2+ release also in the UB tips. Scale bars, 20 μm.
Possible Interaction between β-Catenin and Vsnl1 in Collecting Duct Cells In Vitro and in the Ureteric Epithelium In Vivo
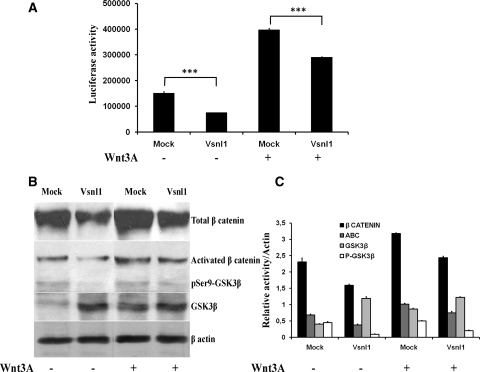
Canonical β-catenin activity has been shown to be antagonized by the calcium sensors.44,45 To explore the possible relationship between Vsnl1 and stabilized β-catenin, we used the mIMCD3 cell line, transiently cotransfected with the TCF reporter TOPFLASH and full-length murine Vsnl1 cDNA (pCAb-Vsnl1-IRES-EGFPm) or with a control vector (pCAb-IRES-EGFPm). The transfected cells were cultured in control or in Wnt3A conditioned medium. Luciferase assay showed reduced reporter activity in Vsnl1-transfected cells cultured in control (approximately 50%; P < 0.005) and in Wnt3A conditioned medium (approximately 30%; P < 0.005) (Figure 6A). Vsnl1 transfection diminished the levels of stabilized β-catenin in both control and stimulated cells (Figure 6B). GSK3β is a key inhibitor of the β-catenin stability. Quantification of the Western blot results (Figure 6C) revealed a decrease of the total amount of β-catenin, about 30% (P < 0.005) in control medium and respectively 24% (P < 0.005) in Wnt3A-stimulated cells. Vsnl1 increased the levels of total GSK3β (2.9-fold in control and 1.4-fold in Wnt3A-stimulated cells) and lowered the levels of inactivated GSK3β (ser9-phospho-GSK3β) (5-fold in control and 2.5-fold in Wnt3A-stimulated cells) as compared with mock-transfected cells (Figure 6C).
Figure 6.
Vsnl1 suppresses canonical WNT signaling activity in mIMCD3 cells. (A) Luciferase reporter assay in mIMCD3 cells. The cells were transiently cotransfected with TOPFLASH and Vsnl1 cDNA or empty vector and cultured in Wnt3A conditioned medium or control medium for 2 hours. Luciferase activity was reduced in Vsnl1-transfected cells (about 50%; P < 0.005) grown in both control and Wnt3A-conditioned medium (about 30%; P < 0005). (B) Western blot analysis. Western blotting of mIMCD3 cells lysates was done for phosphoSer9-GSK3β, total GSK3β, activated β-catenin, and total β-catenin. β-Actin served as a loading control. Vsnl1 increased the total GSK3β levels and decreased the levels of inactive phosphoSer9-GSK3β when compared with controls. (C) Quantitative analysis. (relative activity/β-actin) of the Western blots results revealed a decrease of the total level of β-catenin in both control (about 30%, P < 0005) and Wnt3A medium (24%, P < 0005). In Vsnl1 transfected cells, total GSKβ level was increased in non-Wnt3A-treated cells by 2.9-fold and by 1.4-fold in Wnt3A-stimulated cells. Respectively, the inactivated ser9-phospho GSK3β shows a 5-fold decrease in control medium and a 2.5=fold decrease in Wnt3A-stimulated cells, when compared with mock transfections (B and C). The fold changes represent the averages ± SE in three independent experiments. ***P < 0.005.
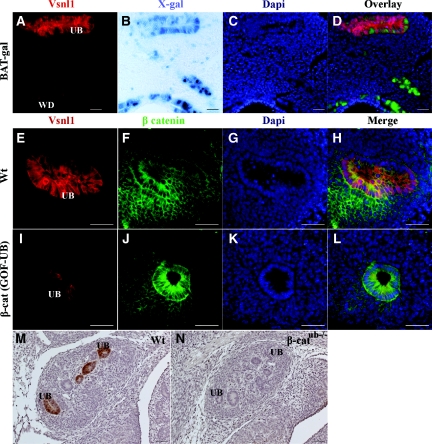
BAT-gal reporter mice22 were used to analyze β-catenin activity and Vsnl1 expression in the ureteric epithelium (Figure 7, A through D). X-gal staining of the E11.5 BAT-gal kidneys revealed mosaic labeling in the ureteric epithelium but with more intense labeling in the WD than in the UB tips (Figure 7, B through D). In the same sections, high levels of Vsnl1 protein were detected in the UB tips, and in individual UB tip cells, the expression of Vsnl1 was mutually exclusive with X-gal labeling in BAT-gal mice (Figure 7, A and D). Vsnl1 expression was then studied in the ureteric epithelium in a mouse model with conditionally stabilized β-catenin in the ureteric epithelium (β-catGOF-UB) (D.B. Bridgewater, V. Di Giovanni, J.E. Cain, B. Cox, B. Jakobson, K. Sainio, and N.D. Rosenblum, manuscript submitted),24 and in β-catenin-deficient mice (β-catub−/−).25 Vsnl1 was downregulated in E12.5 β-catGOF-UB kidneys compared with wild-type kidneys (Figure 7, E through L). Also, in E12.5 β-catub−/− kidney explants, Vsnl1 expression was missing from the UBs (Figure 7, M and N).
Figure 7.
Vsnl1 is downregulated in the ureteric epithelium of β-catenin mutant mice. (A) Vsnl1 staining in BAT-gal kidney explants. (B) Whole-mount X-gal staining of E11.5 BAT-gal reporter kidney. Note the higher β catenin activity in the WD than in the UB. (C) DAPI staining. (D) Overlay. Note that, a cellular level, Vsnl1 expression is nonoverlapping with Xgal labeling in UBs of BAT-gal mice. (E through L) Double staining for Vsnl1 (red) and β catenin (green) in E12.5 wt (E through H) and β-catenin stabilized (β-catGOF-UB) kidneys (I through L) Vsnl1 is downregulated in the ureteric epithelium of β-catenin mutant mice. (M and N) Vsnl1 expression in sections of E12.5 wt kidney (M) and kidneys deficient for β-catenin (β-catub−/−) in ureteric epithelium (N). DAPI in (C, G, and K) marks the nuclei (blue). Scale bars, 20 μm in A through D, M, and N; 50 μm in E through L.
DISCUSSION
To identify genes regulated by GDNF before ureteric budding from Wolffian duct, we conducted a microarray screen. In addition to previously described GDNF targets in the UB (Ret, Etv4, and Spry1), we identified a new putative target gene, Vsnl1. Vsnl1 was identified previously in the UB of the embryonic murine kidneys,27 but its expression pattern and possible function in the kidney have never been approached.
During early steps of kidney morphogenesis, Vsnl1 expression dynamically overlaps with that of the GDNF receptor Ret. Although Vsnl1 protein shows the highest expression in the UB tips, it is also seen at low levels in the medullary ureteric stalks, in contrast to its mRNA. This may indicate that the protein turnover is slow, but currently no data are available on the stability of Vsnl1.
Vsnl1 levels start to decline at late stages of kidney development clearly before other tip markers, such as Ret and Wnt11, are downregulated. At E16, only few cells in the cortical UBs express Vsnl1. By E18, Vsnl1 cannot be detected in the UB. The expression of Ret and Wnt11 remains, and the formation of nephrons continues through the first postnatal days, suggesting that Vsnl1 is not involved in the late stages of kidney development.
Vsnl1 is downregulated or reduced in several genetic models with disrupted ureteric branching (Gdnf- and Ret-deficient mice, and mice with stabilized or deficient β-catenin in the ureteric epithelium) but is expressed at normal levels in Wnt11-deficient mice that exhibit only mild renal hypoplasia. Thus, Vsnl1 levels seem to correlate with the degree of ureteric branching defect. In Gdnf-deficient kidneys, Vsnl1 expression and UB branching are restored by exogenous GDNF but also by alternative bud inducers FGF7/follistatin. This combination also upregulates Vsnl1 in Ret-deficient kidneys. Hence, Vsnl1 characterizes the tip of the ureteric bud epithelium regardless of the inducer, also in the absence of Ret.
Vsnl1 shows a unique mosaic expression pattern in the UB tips. One possible explanation for the mosaic pattern of Vsnl1 could be its differential expression during cell cycle progression. We could not verify this in our analysis with different cell cycle markers, but further studies are needed to completely exclude any correlation between Vsnl1 levels and cell cycle phases. Vsnl1 belongs to the neuronal calcium sensor protein family that modulates calcium-dependent cell signaling events.46 Activation of ERK-MAPK pathway by GDNF/Ret is a calcium-dependent process essential to cell proliferation.47,48 The dp-ERK is an established reporter of Ret signaling,40,41 with an uneven expression in the UB.42 However, no correlation between Vsnl1 mosaicism and the heterogeneous UB expression of dp-ERK could be established.
On the other hand, increased intracellular Ca2+ and activated calcium sensors have been shown to antagonize β-catenin activity.44,45 Indeed, the expression of Vsnl1 is mutually exclusive with the mosaic X-gal labeling in the UBs of BAT-gal reporter mice (Figure 7). Although the mouse line is artificial, this prompted us to study whether β-catenin and Vsnl1 are counteracting.
First, the calcium activity in UBs was analyzed. As revealed by Fluo4 labeling of cultured UBs, the intracellular calcium release is most prominent in the tip cells, where Vsnl1 expression is also the highest. However, the low resolution of vital dye labeling excluded any attempts to analyze calcium activity at a cellular level.
Second, to explore more in detail the possible antagonism between β-catenin stability and Vsnl1, we used the mIMCD3 cell line transiently expressing Vsnl1. In mIMCD3 cells, Vsnl1 suppresses β-catenin activity. In accordance with a previous study of pancreatic β cells,32 Vsnl1 upregulates GSK3β in mIMCD3 cells. Because activation of β-catenin depends on the inhibition of GSK3β,49 our data suggest that Vsnl1 promotes β-catenin degradation in mIMCD3 cells by controlling GSK3β levels. Conversely, in a molecular screening of DLD1 adenocarcinoma cell line, Vsnl1 was found to be directly regulated by β-catenin.50
To address the possibility of Vsnl1 regulation by β-catenin in the kidney in vivo, we used genetic models with β-catenin loss (β-catub−/−) and gain of function (β-catGOF-UB) in the ureteric epithelium. In the UBs of β-catGOF-UB kidneys, Vsnl1 expression was downregulated. Therefore, high activity of the canonical Wnt signaling may inhibit Vsnl1 expression in the UB. Likewise, also β-catub−/− kidneys demonstrated a loss of Vsnl1 expression in the UB. These results would suggest regulation of Vsnl1 by β-catenin in the kidney. Nevertheless, in β-catub−/− kidneys, Emx2, Lim1, and Pax2 transcription factors and consequently Ret and Wnt11 are also downregulated.25 Therefore, a loss of Vsnl1 might be secondary to the inhibition of ureteric branching or may actually reflect the possible absence of tip identity in this genetic model. In β-catGOF-UB kidneys few cells are still expressing Vsnl1 in the UB, suggesting that Vsnl1 expression is not completely dependent on activated β-catenin in the ureteric epithelium.
In conclusion, Vsnl1 mRNA and protein are new markers for UB tips in embryonic kidneys. The mosaic pattern of Vsnl1 shows that the tip has molecularly different cell populations that could respond differently to mesenchyme-derived inductive signals or differ in their ability to promote nephrogenesis. This raises the intriguing possibility that the UB tip has functionally different cell types, some forming new tips and the others becoming stalks. This hypothesis is supported by the exclusion of Ret-deficient cells from UB tips in mosaic Ret null-wild-type mice.42 Taking these results together, the future analysis of Vsnl1 deficient mice will bring more insights about the possible correlation of Vsnl1 with β-catenin and the real function of Vsnl1 in ureteric branching morphogenesis.
CONCISE METHODS
Affymetrix Array Analysis
Urogenital blocks including gonadal ridge, mesonephros, WD, and the MM were dissected from C57Bl mouse embryos at E10.5 and cultured in tissue culture media (DMEM, 10% FBS, 2 mM l-glutamine, 100 units/ml streptomycin, and 100 units/ml penicillin). Half of the urogenital block was exposed to exogenous GDNF (50 ng/ml; R & D Systems) for 16 hours, and the contralateral half served as a control without GDNF supplementation. The 16-hour time point was selected because it is just before the first morphologic response to GDNF to be seen. From the cultured urogenital blocks, a segment of WD between mesonephric and metanephric rudiments was microdissected for microarray analysis. The WD segment used for microarray contained a minimal amount of attached mesenchymal cells. Gonadal ridge, mesonephros, and metanephros were discharged.
The tissues were snap frozen in Trizol (Invitrogen) and taken to RNA extraction. Total RNA from four sample pairs was extracted using TRIzol reagent (Invitrogen) and purified with an RNeasy Cleanup Kit (Qiagen). The RNA integrity was confirmed with 2100 Bioanalyzer (Agilent Technologies). 50 ng of purified RNA was used for cDNA synthesis and amplification (Roche's Microarray Target Amplification Kit), followed by in vitro transcription to biotin labeled aRNA (Roche's Microarray RNA Target Synthesis Kit). The labeled aRNA was fragmented and hybridized to Affymetrix GeneChip® mouse genome 430 2.0 microarrays. The microarrays were scanned with Affymetrix GeneChip scanner, using streptavidin-conjugated fluorochromes to visualize the bound aRNA.
The data were analyzed using R/Bioconductor51 packages affy, limma, and ade4. Raw intensity measures were normalized using RMA52 with a custom Chip Definition File, with probes reannotated for 16331 Entrez genes (version 12) obtained from BRAINARRAY.53 Expression differences were quantified using linear fitting for the paired design, with sample pairs and GDNF treatment as factors, using the limma package.54
Using a P value cutoff of 0.05 (Q value 0.722) revealed 1102 differentially expressed genes. Because the number of expected false positives at this threshold was too high, we decided to use a fold change of 1.890 as an additional cutoff. The level of fold change was determined by the genes known to be affected by GDNF treatment (Ret and Gfrα1). This resulted in a list of 69 upregulated and 50 downregulated candidate genes to validate.
A principal component analysis of the normalized intensities was done using the program dudi.pca of the R package ade4,55 with centering and scaling the values. Correlation of PCA2 with GDNF treatment was tested using a linear model with PCA2 explained by treatment (fixed factor) and sample pair (random factor).
Mice, Genotyping, and In Vitro Morphogenesis Assay
Wild-type embryos were from C57Bl and NMRI strains maintained at the Research Animal Center of the University of Helsinki, and Gdnf-deficient,8 Wnt11-deficient,39 and Ret-deficient56 embryos were generated by crossing heterozygous mice. Hoxb7-Cre:EGFP mice20 were crossed with mice containing loxP sites flanking exon 3 of the β-catenin allele (β -catΔ3/Δ3),57 to generate β-catenin gain-of-function mutant mice, termed β-catGOF-UB (D.B. Bridgewater, V. Di Giovanni, J.E. Cain, B. Cox, B. Jakobson, K. Sainio, and N.D. Rosenblum, manuscript submitted), or with mice with a β-catenin allele containing LoxP sites flanking exons two through six,58 to generate Hoxb7-Cre:EGFP; β-catenin-deficient mice, termed β -catub−/−.25 BAT-gal reporter mice22 were used to determine β-catenin transcriptional activation.
X-gal staining for β-galactosidase was performed as described previously.59 For in vitro experiments, E11.5 urogenital blocks were microdissected in sterile Dulbecco's PBS as described previously.10 For all of the studies, the urogenital blocks were cut into two halves; one was manipulated, and the other served as a control for the experiment. The culture medium was DMEM supplemented with 10% FBS, GlutaMAX, and penicillin-streptomycin (Lonza, Belgium). To induce ectopic ureter buds, either 50 ng/ml GDNF (R & D Systems) or FGF7/follistatin 125 ng/500 ng/ml (R & D Systems) was added to the culture medium for 24, 32, or 48 hours. The use of all of the mouse lines was licensed by the local authorities.
Immunohistochemistry
E13–E18 kidneys were fixed overnight with 4% paraformaldehyde and processed for paraffin embedding and sectioning. Whole-mount immunohistochemistry was performed as described earlier.60 Rabbit anti-vsnl1 (1:1000 dilution), and mouse anti-calbindin (1:3000 dilution; Sigma) immunohistochemistry was performed as described previously.61 Biotinylated secondary antibodies (1:2000 dilution; Leica Microsystems) were used in combination with the Vectastain ABC Elite Kit (Vector Laboratories). For vibratome sectioning of the whole kidneys, the samples were embedded into 5% agarose and cut with Leica VT 1000S Vibratome (Leica) into 8-μm sections. Immunofluorescence on vibratome sections was performed using the following antibodies: rabbit anti-vsnl1 (1:1000 dilution), rat anti-NCAM (1:250; Chemicon International), proliferating cell nuclear antigen (1:500; BD Biosciences-Pharmingen), rabbit anti-phospho-Histone-H3 (1:1000; Upstate), mouse anti-α-tubulin (Sigma; 1:2000), mouse anti-dp-Erk2 (1:100; Sigma), and mouse monoclonal anti-β-catenin (1:1000 dilution; BD Transduction Laboratories). 4′,6′-Diamino-2-phenylindole (DAPI) (Vector Laboratories) was used for the nuclear staining. The secondary antibodies were Alexa 563 and Alexa 488 in 1:250 dilutions (Invitrogen, St. Louis, MO). The samples were photographed with a Zeiss Axioplan II microscope with epifluorescence. Whole-mount staining was done on MeOH fixed kidney explants using monoclonal anti-pan-cytokeratin (1:1000, Sigma) and rabbit anti-vsnl1 (1:750).
In Situ mRNA Hybridization
Whole-mount in situ hybridization was performed with InSituPro Automate (Intavis, Cologne, Germany) as described previously.62 The samples were hybridized with digoxigenin-labeled cRNA probes to Vsnl1. Vsnl1 full-length cDNA was subcloned into pYX-Asc vector. The plasmid was then linearized with SalI and transcribed with T3 RNA polymerase for the antisense probe. The sense probe was transcribed with T7 RNA polymerase after MfeI linearization. The samples were photographed with a Leica stereomicroscope equipped with a DC300 camera and the IM1000 software. The Ret and Wnt11 probes have been described earlier,39,63 as well as Arg235 and Frem2.64
Isolated UB Culture
As described previously,43 UBs were microdissected from E12 mouse embryonic kidneys. The UBs were then suspended within extracellular matrix gels (1:2 mixture of growth factor-reduced Matrigel and culture medium; BD Biosciences). UBs were cultured in DMEM/F12 with 30 ng of GDNF. After 6 days of culture, the UBs were fixed and stained for Vsnl1.
Fluo4 Staining
UB cultures were washed in Hanks' balanced salt solution (HBSS; Invitrogen) and resuspended in HBSS supplemented with 25 mM HEPES buffer (Invitrogen) containing Fluo-4 NW. UB cultures were incubated at 37°C for 30 minutes and then at room temperature for an additional 30 minutes and then cleared in HBSS. The samples were photographed with Zeiss Axioplan II microscope with epifluorescence. The Fluo4 NW Calcium Assay Kit was obtained from Invitrogen.
Cell Culture, DNA Reagents, and Luciferase Assay
The mIMCD3 (mouse inner medullary collecting duct) cell line (ATCC, CRL-2123) was grown in DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 units/ml streptomycin, and 100 units/ml penicillin. Expression plasmids for the transient assays include: TCF/LEF luciferase reporter constructs (TOPflash) from Upstate Biotechnology, pCAb-IRES-EGFPm empty vector (a gift from Jonathan Gilthorpe) used as a control (Mock), and Vsnl1 full-length cDNA cut out with NotI and EcoRI from pYX-Asc vector cloned into a pCAb-IRES-EGFPm vector. Transient transfections were performed using LipofectAMINE Plus reagent (Invitrogen) following the manufacturer's protocol.
For luciferase assay cotransfections (TOPFLASH and pCAb-Vsnl1-IRES-EGFPm or the empty vector) performed in triplicate in 24-well plates, each experiment was performed three times. After 48 hours of transfection, the cells were starved for 12 hours and stimulated with Wnt3A conditioned medium versus Wnt3A control medium for 2 hours. The firefly luciferase reporter activity was determined using a Luciferase Assay System (Promega, Madison, WI) following the manufacturer's instructions. Reporter activity was expressed as a ratio of luciferase to protein quantification. The statistical analyses were performed using t test.
Western Blotting
Subconfluent mIMCD3 cells were transiently transfected with an empty expression plasmid pCAb-IRES-EGFPm (Mock) or Vsnl1-expressing plasmid (pCAb-Vsnl1-IRES-EGFPm). After 48 hours, the cells were starved for 12 hours, stimulated by Wnt3A conditioned or control medium for 2 hours, lysed in lysis buffer supplemented with 1 mM sodium orthovanadate, and analyzed on Western blots as described.65 The blots were probed with the indicated antibodies and developed with ECL reagents (Amersham Biosciences). The antibodies used for Western blotting were against phospho-GSK3β (Ser9; Cell Signaling, Danvers, MA), pan-GSK3β (Cell Signaling, Danvers, MA), β-catenin (Sigma), β-actin (Santa Cruz Biotechnology), activated β-catenin (Millipore), goat α-mouse horseradish peroxidase, and goat α-rabbit horseradish peroxidase (Bio-Rad Laboratories).
DISCLOSURES
None.
Acknowledgments
We thank Ms. Agnes Viherä, Ms. Lea Armassalo, and Ms. Iulia Diaconu for their outstanding technical assistance; Mr. Fares Zeidan Chulia for the Wnt conditioned and control medium; Dr. Jonathan Gilthorpe for the pCAb-IRES-EGFPm plasmid; Professor Irma Thesleff for BAT-gal reporter mice; and Professor Seppo Vainio for the Wnt11 null mice. This work was supported by the Marie Curie Early Stage Research Training program at the University of Helsinki, Finland; Sigrid Juselius Foundation; Nona and Kullervo Väre Foundation; and the Academy of Finland.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental information for this article is available online at http://www.jasn.org/.
REFERENCES
- 1. Dressler GR: The cellular basis of kidney development. Annu Rev Cell Dev Biol 22: 509–529, 2006 [DOI] [PubMed] [Google Scholar]
- 2. Costantini F, Shakya R: GDNF/Ret signaling and the development of the kidney. Bioessays 28: 117–127, 2006 [DOI] [PubMed] [Google Scholar]
- 3. Suvanto P, Hiltunen JO, Arumae U, Moshnyakov M, Sariola H, Sainio K, Saarma M: Localization of glial cell line-derived neurotrophic factor (GDNF) mRNA in embryonic rat by in situ hybridization. Eur J Neurosci 8: 816–822, 1996 [DOI] [PubMed] [Google Scholar]
- 4. Jing S, Wen D, Yu Y, Holst PL, Luo Y, Fang M, Tamir R, Antonio L, Hu Z, Cupples R, Louis JC, Hu S, Altrock BW, Fox GM: GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell 85: 1113–1124, 1996 [DOI] [PubMed] [Google Scholar]
- 5. Sanicola M, Hession C, Worley D, Carmillo P, Ehrenfels C, Walus L, Robinson S, Jaworski G, Wei H, Tizard R, Whitty A, Pepinsky RB, Cate RL: Glial cell line-derived neurotrophic factor-dependent RET activation can be mediated by two different cell-surface accessory proteins. Proc Natl Acad Sci U.S.A. 94: 6238–6243, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Treanor JJ, Goodman L, de Sauvage F, Stone DM, Poulsen KT, Beck CD, Gray C, Armanini MP, Pollock RA, Hefti F, Phillips HS, Goddard A, Moore MW, Buj-Bello A, Davies AM, Asai N, Takahashi M, Vandlen R, Henderson CE, Rosenthal A: Characterization of a multicomponent receptor for GDNF. Nature 382: 80–83, 1996 [DOI] [PubMed] [Google Scholar]
- 7. Cacalano G, Farinas I, Wang LC, Hagler K, Forgie A, Moore M, Armanini M, Phillips H, Ryan AM, Reichardt LF, Hynes M, Davies A, Rosenthal A: GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron 21: 53–62, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pichel JG, Shen L, Sheng HZ, Granholm AC, Drago J, Grinberg A, Lee EJ, Huang SP, Saarma M, Hoffer BJ, Sariola H, Westphal H: Defects in enteric innervation and kidney development in mice lacking GDNF. Nature 382: 73–76, 1996 [DOI] [PubMed] [Google Scholar]
- 9. Schuchardt A, D'Agati V, Larsson-Blomberg L, Costantini F, Pachnis V: Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367: 380–383, 1994 [DOI] [PubMed] [Google Scholar]
- 10. Sainio K, Suvanto P, Davies J, Wartiovaara J, Wartiovaara K, Saarma M, Arumae U, Meng X, Lindahl M, Pachnis V, Sariola H: Glial-cell-line-derived neurotrophic factor is required for bud initiation from ureteric epithelium. Development 124: 4077–4087, 1997 [DOI] [PubMed] [Google Scholar]
- 11. Shakya RE, Jho H, Kotka P, Wu Z, Kholodilov N, Burke R, D'Agati V, Costantini F: The role of GDNF in patterning the excretory system. Dev Biol 283: 70–84, 2005 [DOI] [PubMed] [Google Scholar]
- 12. Lu BC, Cebrian C, Chi X, Kuure S, Kuo R, Bates CM, Arber S, Hassell J, MacNeil L, Hoshi M, Jain S, Asai N, Takahashi M, Schmidt-Ott KM, Barasch J, D'Agati V, Costantini F: Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat Genet 41: 1295–1302, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pepicelli CV, Kispert A, Rowitch DH, McMahon AP: GDNF induces branching and increased cell proliferation in the ureter of the mouse. Dev Biol 192: 193–198, 1997 [DOI] [PubMed] [Google Scholar]
- 14. Shakya R, Watanabe T, Costantini F: The role of GDNF/Ret signaling in ureteric bud cell fate and branching morphogenesis. Dev Cell 8: 65–74, 2005 [DOI] [PubMed] [Google Scholar]
- 15. Moore MW, Klein RD, Farinas I, Sauer H, Armanini M, Phillips H, Reichardt LF, Ryan AM, Carver-Moore K, Rosenthal A: Renal and neuronal abnormalities in mice lacking GDNF. Nature 382: 76–79, 1996 [DOI] [PubMed] [Google Scholar]
- 16. Maeshima A, Sakurai H, Choi Y, Kitamura S, Vaughn DA, Tee JB, Nigam SK: Glial cell-derived neurotrophic factor independent ureteric bud outgrowth from the Wolffian duct. J Am Soc Nephrol 18: 3147–3155, 2007 [DOI] [PubMed] [Google Scholar]
- 17. Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D'Agati V, Licht JD, Martin GR, Costantini F: Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet 6: e1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, Itoh N: FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun 277: 643–649, 2000 [DOI] [PubMed] [Google Scholar]
- 19. Qiao J, Uzzo R, Obara-Ishihara T, Degenstein L, Fuchs E, Herzlinger D: FGF-7 modulates ureteric bud growth and nephron number in the developing kidney. Development 126: 547–554, 1999 [DOI] [PubMed] [Google Scholar]
- 20. Zhao H, Kegg H, Grady S, Truong HT, Robinson ML, Baum M, Bates CM: Role of fibroblast growth factor receptors 1 and 2 in the ureteric bud. Dev Biol 276: 403–415, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheon SS, Cheah AY, Turley S, Nadesan P, Poon R, Clevers H, Alman BA: beta-Catenin stabilization dysregulates mesenchymal cell proliferation, motility, and invasiveness and causes aggressive fibromatosis and hyperplastic cutaneous wounds. Proc Natl Acad Sci U.S.A. 99: 6973–6978, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S: Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U.S.A. 100: 3299–3304, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iglesias DM, Hueber PA, Chu L, Campbell R, Patenaude AM, Dziarmaga AJ, Quinlan J, Mohamed O, Dufort D, Goodyer PR: Canonical WNT signaling during kidney development. Am J Physiol Renal Physiol 293: F494–F500, 2007 [DOI] [PubMed] [Google Scholar]
- 24. Merkel CE, Karner CM, Carroll TJ: Molecular regulation of kidney development: Is the answer blowing in the Wnt? Pediatr Nephrol 22: 1825–1838, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bridgewater D, Cox B, Cain J, Lau A, Athaide V, Gill PS, Kuure S, Sainio K, Rosenblum ND: Canonical WNT/beta-catenin signaling is required for ureteric branching. Dev Biol 317: 83–94, 2008 [DOI] [PubMed] [Google Scholar]
- 26. Marose TD, Merkel CE, McMahon AP, Carroll TJ: Beta-catenin is necessary to keep cells of ureteric bud/Wolffian duct epithelium in a precursor state. Dev Biol 314: 112–126, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwab K, Patterson LT, Aronow BJ, Luckas R, Liang HC, Potter SS: A catalogue of gene expression in the developing kidney. Kidney Int 64: 1588–1604, 2003 [DOI] [PubMed] [Google Scholar]
- 28. Gierke P, Zhao C, Brackmann M, Linke B, Heinemann U, Braunewell KH: Expression analysis of members of the neuronal calcium sensor protein family: Combining bioinformatics and Western blot analysis. Biochem Biophys Res Commun 323: 38–43, 2004 [DOI] [PubMed] [Google Scholar]
- 29. Spilker C, Dresbach T, Braunewell KH: Reversible translocation and activity-dependent localization of the calcium-myristoyl switch protein VILIP-1 to different membrane compartments in living hippocampal neurons. J Neurosci 22: 7331–7339, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zozulya S, Stryer L: Calcium-myristoyl protein switch. Proc Natl Acad Sci U.S.A. 89: 11569–11573, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brackmann M, Schuchmann S, Anand R, Braunewell KH: Neuronal Ca2+ sensor protein VILIP-1 affects cGMP signalling of guanylyl cyclase B by regulating clathrin-dependent receptor recycling in hippocampal neurons. J Cell Sci 118: 2495–2505, 2005 [DOI] [PubMed] [Google Scholar]
- 32. Dai FF, Zhang Y, Kang Y, Wang Q, Gaisano HY, Braunewell KH, Chan CB, Wheeler MB: The neuronal Ca2+ sensor protein visinin-like protein-1 is expressed in pancreatic islets and regulates insulin secretion. J Biol Chem 281: 21942–21953, 2006 [DOI] [PubMed] [Google Scholar]
- 33. Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T, McMahon AP, Wilson PD, Costantini FD, Mason IJ, Licht JD: Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell 8: 229–239, 2005 [DOI] [PubMed] [Google Scholar]
- 34. de Graaff E, Srinivas S, Kilkenny C, D'Agati V, Mankoo BS, Costantini F, Pachnis V: Differential activities of the RET tyrosine kinase receptor isoforms during mammalian embryogenesis. Genes Dev 15: 2433–2444, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Caruana G, Cullen-McEwen L, Nelson AL, Kostoulias X, Woods K, Gardiner B, Davis MJ, Taylor DF, Teasdale RD, Grimmond SM, Little MH, Bertram JF: Spatial gene expression in the T-stage mouse metanephros. Gene Expr Patterns 6: 807–825, 2006 [DOI] [PubMed] [Google Scholar]
- 36. Schmidt-Ott KM, Yang J, Chen X, Wang H, Paragas N, Mori K, Li JY, Lu B, Costantini F, Schiffer M, Bottinger E, Barasch J: Novel regulators of kidney development from the tips of the ureteric bud. J Am Soc Nephrol 16: 1993–2002, 2005 [DOI] [PubMed] [Google Scholar]
- 37. Schwab K, Hartman HA, Liang HC, Aronow BJ, Patterson LT, Potter SS: Comprehensive microarray analysis of Hoxa11/Hoxd11 mutant kidney development. Dev Biol 293: 540–554, 2006 [DOI] [PubMed] [Google Scholar]
- 38. Sonnenberg E, Godecke A., Walter B., Bladt F., Birchmeier C.: Transient and locally restricted expression of the ros1 protooncogene during mouse development. EMBO J 10: 3693–3702, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Majumdar A, Vainio S, Kispert A, McMahon J, McMahon AP: Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development 130: 3175–3185, 2003 [DOI] [PubMed] [Google Scholar]
- 40. Corson LB, Yamanaka Y, Lai KM, Rossant J: Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development 130: 4527–4537, 2003 [DOI] [PubMed] [Google Scholar]
- 41. Lunn JS, Fishwick KJ, Halley PA, Storey KG: A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev Biol 302: 536–552, 2007 [DOI] [PubMed] [Google Scholar]
- 42. Chi X, Michos O, Shakya R, Riccio P, Enomoto H, Licht JD, Asai N, Takahashi M, Ohgami N, Kato M, Mendelsohn C, Costantini F: Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev Cell 17: 199–209, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Steer DL, Shah MM, Bush KT, Stuart RO, Sampogna RV, Meyer TN, Schwesinger C, Bai X, Esko JD, Nigam SK: Regulation of ureteric bud branching morphogenesis by sulfated proteoglycans in the developing kidney. Dev Biol 272: 310–327, 2004 [DOI] [PubMed] [Google Scholar]
- 44. Schneider I, Houston DW, Rebagliati MR, Slusarski DC: Calcium fluxes in dorsal forerunner cells antagonize beta-catenin and alter left-right patterning. Development 135: 75–84, 2008 [DOI] [PubMed] [Google Scholar]
- 45. Maye P, Zheng J, Li L, Wu D: Multiple mechanisms for Wnt11-mediated repression of the canonical Wnt signaling pathway. J Biol Chem 279: 24659–24665, 2004 [DOI] [PubMed] [Google Scholar]
- 46. Braunewell KH, Spilker C, Behnisch T, Gundelfinger ED: The neuronal calcium-sensor protein VILIP modulates cyclic AMP accumulation in stably transfected C6 glioma cells: Amino-terminal myristoylation determines functional activity. J Neurochem 68: 2129–2139, 1997 [DOI] [PubMed] [Google Scholar]
- 47. Nozaki C, Asai N, Murakami H, Iwashita T, Iwata Y, Horibe K, Klein RD, Rosenthal A, Takahashi M: Calcium-dependent Ret activation by GDNF and neurturin. Oncogene 16: 293–299, 1998 [DOI] [PubMed] [Google Scholar]
- 48. van Weering DH, Moen TC, Braakman I, Baas PD, Bos JL: Expression of the receptor tyrosine kinase Ret on the plasma membrane is dependent on calcium. J Biol Chem 273: 12077–12081, 1998 [DOI] [PubMed] [Google Scholar]
- 49. Huelsken J, Behrens J: The Wnt signalling pathway. J Cell Sci 115: 3977–3978, 2002 [DOI] [PubMed] [Google Scholar]
- 50. Major MB, Roberts BS, Berndt JD, Marine S, Anastas J, Chung N, Ferrer M, Yi X, Stoick-Cooper CL, von Haller PD, Kategaya L, Chien A, Angers S, MacCoss M, Cleary MA, Arthur WT, Moon RT: New regulators of Wnt/beta-catenin signaling revealed by integrative molecular screening. Sci Signal 1: ra12, 2008 [DOI] [PubMed] [Google Scholar]
- 51. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J: Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP: Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264, 2003 [DOI] [PubMed] [Google Scholar]
- 53. Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F: Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33: e175, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smyth GK: Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3, 2004 [DOI] [PubMed] [Google Scholar]
- 55. Charif D, Thioulouse J, Lobry JR, Perriere G: Online synonymous codon usage analyses with the ade4 and seqinR packages. Bioinformatics 21: 545–547, 2005 [DOI] [PubMed] [Google Scholar]
- 56. Schuchardt A, D'Agati V, Pachnis V, Costantini F: Renal agenesis and hypodysplasia in ret-k- mutant mice result from defects in ureteric bud development. Development 122: 1919–1929, 1996 [DOI] [PubMed] [Google Scholar]
- 57. Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM: Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J 18: 5931–5942, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R: Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128: 1253–1264, 2001 [DOI] [PubMed] [Google Scholar]
- 59. Godin RE, Takaesu NT, Robertson EJ, Dudley AT: Regulation of BMP7 expression during kidney development. Development 125: 3473–3482, 1998 [DOI] [PubMed] [Google Scholar]
- 60. Sariola H, Aufderheide E, Bernhard H, Henke-Fahle S, Dippold W, Ekblom P: Antibodies to cell surface ganglioside GD3 perturb inductive epithelial-mesenchymal interactions. Cell 54: 235–245, 1988 [DOI] [PubMed] [Google Scholar]
- 61. Sariola H, Holm K, Henke-Fahle S: Early innervation of the metanephric kidney. Development 104: 589–599, 1988 [DOI] [PubMed] [Google Scholar]
- 62. Kuure S, Sainio K, Vuolteenaho R, Ilves M, Wartiovaara K, Immonen T, Kvist J, Vainio S, Sariola H: Crosstalk between Jagged1 and GDNF/Ret/GFRalpha1 signalling regulates ureteric budding and branching. Mech Dev 122: 765–780, 2005 [DOI] [PubMed] [Google Scholar]
- 63. Pachnis V, Mankoo B, Costantini F: Expression of the c-ret proto-oncogene during mouse embryogenesis. Development 119: 1005–1017, 1993 [DOI] [PubMed] [Google Scholar]
- 64. Jadeja S, Smyth I, Pitera JE, Taylor MS, van Haelst M, Bentley E, McGregor L, Hopkins J, Chalepakis G, Philip N, Perez Aytes A, Watt FM, Darling SM, Jackson I, Woolf AS, Scambler PJ: Identification of a new gene mutated in Fraser syndrome and mouse myelencephalic blebs. Nat Genet 37: 520–525, 2005 [DOI] [PubMed] [Google Scholar]
- 65. Lindahl M, Poteryaev D, Yu L, Arumae U, Timmusk T, Bongarzone I, Aiello A, Pierotti MA, Airaksinen MS, Saarma M: Human glial cell line-derived neurotrophic factor receptor alpha 4 is the receptor for persephin and is predominantly expressed in normal and malignant thyroid medullary cells. J Biol Chem 276: 9344–9351, 2001 [DOI] [PubMed] [Google Scholar]