Abstract
Recent studies have suggested that extracellular matrix rigidity regulates cancer invasiveness, including the formation of cellular invadopodial protrusions; however, the relevant mechanical range is unclear. Here, we used a combined analysis of tissue-derived model basement membrane (BM) and stromal matrices and synthetic materials to understand how substrate rigidity regulates invadopodia. Urinary bladder matrix-BM (UBM-BM) was found to be a rigid material with elastic moduli of 3-8 MPa, as measured by atomic force microscopy and low-strain tensile testing. Stromal elastic moduli were ∼6-fold lower, indicating a more compliant material. Using synthetic substrates that span kPa–GPa moduli, we found a peak of invadopodia-associated extracellular matrix degradation centered around 30 kPa, which also corresponded to a peak in invadopodia/cell. Surprisingly, we observed another peak in invadopodia numbers at 2 GPa as well as gene expression changes that indicate cellular sensing of very high moduli. Based on the measured elastic moduli of model stroma and BM, we expected to find more invadopodia formation on the stroma, and this was verified on the stromal versus BM side of UBM-BM. These data suggest that cells can sense a wide range of rigidities, up into the GPa range. Furthermore, there is an optimal rigidity range for invadopodia activity that may be limited by BM rigidity.
Introduction
Invasion by epithelial cancer cells across the basement membrane (BM) is considered to be a critical rate-limiting step in cancer metastasis (1). The BM is a thin, dense extracellular matrix (ECM) that is composed of a highly ordered and cross-linked type IV collagen network, along with laminin, nidogen/entactin, and various proteoglycans and glycoproteins (2). Once malignant cells penetrate this barrier, they must navigate the adjacent stroma and enter the vasculature for metastasis to occur.
The tensile properties of the ECM (i.e., stiffness or rigidity) have been implicated in the malignant transformation of the breast through activation of cellular mechanotransduction signaling pathways (3). This relationship is consistent with findings from both mouse tumor (4,5) and clinical (6,7) studies that showed a strong correlation between tissue density and cancer development and invasiveness. On the cellular level, our group previously linked mechanosensing of rigid substrates in vitro to the formation and activity of invadopodia, which are punctate, actin-rich structures with associated cell-surface proteinases that degrade the ECM and have been implicated in cancer invasion and metastasis (8,9). Although it is evident that the mechanical nature of tumor-associated ECM can drive an invasive phenotype, the relevant rigidity range with respect to the BM and stroma is unclear.
A significant challenge that exists in the field is to recapitulate in vitro physiologically relevant in vivo characteristics (10). For example, although biological hydrogels such as collagen and Matrigel are extremely useful for mimicking the stromal and BM environments, they lack many of the physical characteristics of in vivo tissues (1,11) that contribute to the mechanical properties of those tissues. Specifically, both pepsinized collagen gels and Matrigel are uncross-linked, have very low elastic moduli, and provide little barrier to cellular migration and invasion (11,12). Several recent studies have used processed (13,14) and native (11,12) biological tissues as ex vivo organotypic models to recapitulate the in vivo ECM environment experienced by invasive cells. Similarly, tissue scaffolds prepared from naturally occurring ECMs for tissue engineering and clinical applications have resulted in biological materials that have been thoroughly tested to ensure that they maintain their in vivo physical and mechanical properties (15). For example, urinary bladder matrix (UBM), which has an intact BM with an adjacent fibrous stroma (16), is well characterized and can be readily handled for mechanical testing (17) and ex vivo culturing (16).
Despite the availability of these tissue surrogates, regulation of behavior by tissue rigidity is usually explored in vitro with artificial substrates that are easily synthesized and manipulated to yield specific mechanical properties. One of the most common approaches is to graft ECM molecules onto polyacrylamide (PAA) gels of different rigidities. These hydrogels have been used to explore a host of biological processes, including migration (18) and stem cell differentiation (19), because their mechanical properties are elastic and tunable and their optical properties allow for favorable microscopic imaging. In a previous study using these substrates, we found that the number and degradative ability of invadopodia increased when the rigidity was increased by one order of magnitude, from E = 1 to 10 kPa (8). However, PAA gels are limited because they can be synthesized with elastic moduli that span just a few orders of magnitude (typically ∼0.1–30 kPa) (19,20), in contrast to biological tissues that have elastic moduli spanning up to nine orders of magnitude (0.1 kPa–10 GPa) (21,22). Alternatively, rubber-like polymers such as polyurethane (PUR) elastomers can be synthesized to reach much larger moduli values in the high MPa–GPa region (23,24).
In this study, we focused on defining how breast cancer cells respond to a wide range of substrate rigidities, and how that response corresponds to physiological ECM microenvironments that might be encountered in a developing or metastatic tumor. To determine the mechanical influence of relevant in vivo environments, we used intact UBM as our model of stroma or a thin delaminated version in which the majority of the stroma had been removed from the BM (UBM-BM). We first characterized the physical and mechanical properties of UBM-BM and compared them with UBM. Because both UBM-BM and UBM had much higher elastic moduli than observed in our previous in vitro invadopodia study (8), we developed synthetic invadopodia substrates (PAA and PUR) that span the kPa–GPa rigidity range, and determined the corresponding degradative capabilities of breast cancer cells. Surprisingly, we found that the breast cancer cells could sense a wide range of rigidities, as measured by ECM degradation and invadopodia formation. We further validated this observation by evaluating the expression of several genes that were found to peak at either relatively low or high moduli. Furthermore, there was an optimal peak of ECM degradation on the 30 kPa substrate, which was closer to the rigidity of the stroma than to that of the BM. Consistent with the notion that rigidity plays a role in vivo, the breast cancer cells formed significantly more invadopodia when cultured on the stromal side of UBM-BM compared with the BM side. Experiments repeated with 804G rat bladder carcinoma cells on synthetic substrates and UBM-BM yielded similar results, indicating a common response among cell types of different tissue origin.
Materials and Methods
UBM preparations
UBM and UBM-BM were kindly provided by Dr. Stephen Badylak (University of Pittsburgh). UBM-BM is prepared from porcine bladders similarly to UBM (25), except that the tissue is further mechanically delaminated close to the BM such that the remaining tissue is extremely thin and appears translucent.
Immunohistochemistry
Formalin-fixed, agar-coated samples were sectioned for either hematoxylin and eosin staining (Richard Allan Scientific, Kalamazoo, MI) or immunostaining with rabbit type IV collagen (Abcam, Cambridge, MA) or anti-laminin (Sigma, St. Louis, MO) antibodies, and visualized with the Envision+ HRP/DAB system (Dako, Carpinteria, CA) and Mayer's hematoxylin.
Electron microscopy
Samples were fixed in 2.5% glutaraldehyde, serially dehydrated, critical-point dried, and sputter-coated with 60% gold and 40% palladium for visualization with a Hitachi S4200 field emission scanning electron microscope. Samples were either mounted with double-sided tape onto stages or anchored onto CellCrown inserts (Scaffdex, Tampere, Finland). To visualize the type IV collagen network using a Philips CM-12 transmission electron microscope, samples were salt-extracted to remove noncollagenous molecules from the BM (26), fixed in 2.5% glutaraldehyde, serially dehydrated, resin-embedded, sectioned, and stained with 1% uranyl acetate and 1% lead citrate.
Swelling
The swelling ratio, Q, was calculated based on the sample mass before and after hydration with phosphate-buffered saline overnight (see the Supporting Material).
Dynamic mechanical analysis and rheology
UBM or UBM-BM samples (15×6.5 mm) were mounted in a tension submersion clamp of a TA Instruments Q800 DMA, allowed to equilibrate at room temperature for 30 min, and stretched at a strain rate of 1%/min. Stress, defined as force divided by the initial cross-sectional area (measured with calipers; see Fig. 2 B), was plotted versus strain. T3000 and T900 PURs were tested at a rate of 3 N/min, whereas T300 PURs were loaded at a rate of 5 mm/min using a three-point bending apparatus. Elastic moduli were determined by linear regression of the stress-strain curves. We previously tested soft and hard PAAs using rheometry (8), and therefore used a similar method to test the rigid PAA in the study presented here. In our previous study, we showed that the mechanical properties of ECM layers on synthetic substrates were not substantially altered (8).
Figure 2.
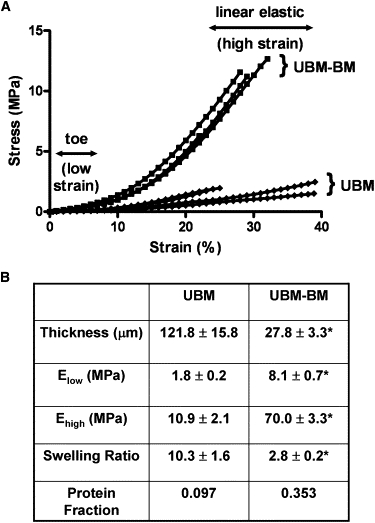
Stress-strain curves and elastic moduli for UBM-BM and UBM. (A) To determine the elastic moduli of our BM and stromal models, we performed uniaxial tensile DMA of UBM-BM and UBM. Both materials exhibited classic strain stiffening; however, UBM-BM exhibited steeper slopes in both the low-strain toe region and the high-strain linear elastic region, indicating stiffer mechanical behavior. One data point per % strain was reported for graphical representation. (B) Low- and high-strain elastic moduli were calculated from the slopes of the toe and linear elastic regions, respectively. Consistent with the UBM-BM being stiffer, this BM model had a smaller swelling ratio and larger protein fraction. Data are presented as mean ± SE, and ∗ indicates p < 0.05 for UBM-BM versus UBM comparisons. n = 4 for the thickness, Elow, and Ehigh values, and n ∼ 20 for the swelling ratios, respectively.
Atomic force microscopy of stromal and BM surfaces
We measured the moduli of the BM and stromal sides of UBM-BM using a Bioscope Catalyst atomic force microscope (Veeco Instruments,Plainview, NY) operated in peak force quantitative nanomechanical mode (see Supporting Material).
Invadopodia assay on synthetic substrates and tissue-derived scaffolds
Mechanically tunable substrates (∼75 μm thick) were made from PAA hydrogels and PUR elastomers according to previously established methods (18,27). In brief, they were cast on activated glass coverslips of 35 mm MatTek dishes and conjugated with 1% gelatin (cross-linked with 0.5% glutaraldehyde) and FITC-labeled fibronectin (8,28). The rigid PAA was formed from 12% acrylamide and 0.6% bis-acrylamide. PURs were prepared from lysine diisocyanate and different equivalent weights of polyester polyalcohols synthesized from a glycerol starter and a 70%/30% mixture of caprolactone/glycolide for the T3000 (3000 Da) and T900 (900 Da) PURs, and 100% caprolactone for the T300 (300 Da) PUR (29). PURs were soaked with 100 μg/mL poly-D-lysine at 37°C for 1 h and then coated as described above. UBM-BM samples were secured in Cell Crown inserts until they were taut, but without significant strain (i.e., low strain). MCF10A CA1d breast carcinoma cells at low density (∼3000 cells/cm2) were incubated on all substrates for 18 h.
Immunofluorescence
For identification of invadopodia, cells were fixed and stained with Alexa Fluor 546 phalloidin (Invitrogen) and 4F11 antibody (Upstate Biotechnology, Lake Placid, NY) to identify F-actin and cortactin, respectively. Fluorescent images were captured on a Nikon Eclipse TE2000-E microscope with a 40X Plan Fluor oil immersion lens or on a Zeiss LSM 510 confocal microscope with a Plan Apo 63X oil immersion lens (0.2 μm Z-sections). For the synthetic substrates, areas of FITC-fibronectin degradation were thresholded and quantified with Metamorph software based on the loss of FITC signal. Active and total invadopodia were manually counted and cell size was quantified as previously described (8).
Statistics
Data were evaluated for normality with the Shapiro-Wilk or Kolmogorov-Smirnov test. Data that passed the normality test were analyzed by means of Student's t-test or one-way analysis of variance. Data that did not pass the normality test were analyzed by a Mann-Whitney or Kruskal-Wallis test. If significance was determined within a group, a Tukey or Tamhane post hoc test was used for pairwise comparisons, with p < 0.05 considered statistically significant. All statistical analyses were performed with PASW Statistics 18 (SPSS, Chicago, IL).
Results
Our goal in this study was to test how mechanical properties corresponding to those found in tissues affect the formation and activity of cancer cell invadopodia. To isolate rigidity effects while also relating them to relevant in vivo physical properties, we used a combination of synthetic invadopodia substrates of tunable rigidity and tissue-derived ECM scaffolds that model the BM and stromal environments.
Characterization of model BM and stromal matrices
To obtain the relevant rigidities of stroma and BM, we began by characterizing the properties of the model ECM scaffolds UBM and UBM-BM, respectively. We chose to use bladder-derived scaffolds because the large BM surface in bladder can easily be isolated and used for both mechanical testing and experimental studies. By contrast, it is difficult to isolate the BM from ductal tissues, such as breast, for in vitro studies. UBM is a well-characterized, decellularized ECM scaffold that is predominantly composed of stromal connective tissue of the tunica propria underlying a thin BM layer (16); therefore, we used it as a model of stroma. As a model of BM, we used UBM-BM, which is created from the same precursor tissue as UBM but is further mechanically delaminated to remove the majority of the connective tissue layer and leave a thin layer containing the BM. To verify the presence and structural integrity of the BM in UBM-BM after this additional processing, we performed immunohistochemistry (IHC) staining and EM.
In contrast to the thick stromal layer in UBM (Fig. 1 A, double-headed arrow), UBM-BM contained a thin, supporting layer of connective tissue (Fig. 1 B, double-headed arrow). On the top side, UBM-BM maintained a smooth and contoured surface (Fig. 1 B, asterisk) consistent with the BM ultrastructure as observed in UBM (16). After salt extraction, UBM-BM retained a filamentous meshwork composed of 100–200 nm pores (Fig. 1 C) as previously described for BMs from a variety of tissues (26,30). Dense, positive IHC staining for both type IV collagen (Fig. 1 D) and laminin (Fig. 1 E) was observed in a continuous pattern localized to the luminal side that was also consistent with the presence of an intact BM in UBM-BM.
Figure 1.
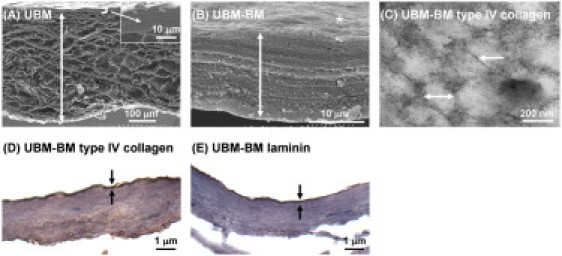
Characterization of UBM-BM and UBM. As model BM and stromal matrices, we characterized two ex vivo tissue scaffolds: UBM-BM and UBM. (A) Scanning EM image (×180) of a UBM cross-section reveals a thick stromal layer of connective tissue (double-headed arrow) underlying a thin layer that includes the BM (bracket and inset). (B) UBM-BM was created by mechanically delaminating porcine bladders to a further extent than UBM to obtain a very thin layer of connective tissue (double-headed arrow) underlying the BM (∗) that was equivalent to the top portion of UBM (bracket and inset of A) as shown in a cross-sectional scanning EM image (×8000). (C) After salt extraction of noncollagenous components, transmission EM imaging (×110,000) revealed that UBM-BM contains filaments (single-headed arrow) and 100–200 nm diameter pores (double-headed arrow) consistent with the polygonal type IV collagen network of a BM. The integrity of the BM of UBM-BM was confirmed histologically with continuous positive staining for (D) type IV collagen and (E) laminin (between the arrows).
UBM-BM is mechanically rigid
We first determined the mechanical properties of UBM-BM and UBM as BM and stroma models, respectively, using dynamic mechanical analysis (DMA). Because collagen is a well-known tensile load-bearing protein (5,31) and UBM is a fairly isotropic material (17), we generated stress-strain data by uniaxial tensile mechanical loading. Although collagen-rich biological tissues exhibit a complex nonlinear viscoelastic behavior, their biomechanical response can be approximated as pseudoelastic to yield repeatable stress-strain curves once they are preconditioned to reach a steady-state mechanical response (31). Previous studies have shown that ECM scaffolds do not necessarily require preconditioning to reach repeatable stress-strain responses (32,33). In our preliminary experiments, successive loading cycles did not yield significant shifts in the stress-strain curves; therefore, preconditioning was assumed.
During mechanical loading, both UBM-BM and UBM exhibited strain stiffening (Fig. 2 A), a typical phenomenon observed in collagenous tissues that is characterized by a transition from a compliant response at low strain (with the toe region of the curve representing low physiologic loading) to a stiffer response at high strain (with the linear elastic region of the curve representing high physiologic loading). UBM-BM exhibited steeper toe and linear elastic regions of the stress-strain curves, indicating stiffer mechanical behavior than UBM that was verified with linear regressions to determine the elastic moduli at both low and high strain (Fig. 2 B). For both scaffolds, the moduli were in the MPa range. The elastic moduli for UBM-BM in the low- and high-strain regions were respectively 4.5- and 6.4-fold larger than for UBM. UBM-BM experienced a higher degree of strain stiffening, with an increase in modulus from the toe to the linear elastic region by 8.6- versus 6.1-fold for UBM.
To further quantitate the rigidity of the stroma and BM, we performed atomic force microscopy (AFM) on the stromal and BM sides of UBM-BM (Fig. 3). Note that AFM measurements are on the nanometer–micrometer scale, which is similar to the subcellular–cellular scale (21,34). Interestingly, although there was a range of BM moduli (Fig. 3 B), the weighted average BM side modulus measured by AFM was ∼3 MPa (Fig. 3 C), which is similar to moduli measured for retinal BM by AFM (35,36) and >2-fold lower than the modulus measured by DMA tensile testing. The stromal side weighted average modulus measured by AFM was ∼0.4 MPa (Fig. 3 C), which was also lower than that measured by tensile testing (Fig. 2 B). In similarity to the DMA test results, however, the stromal side average elastic modulus was 7.5-fold lower than that of the BM side average modulus.
Figure 3.
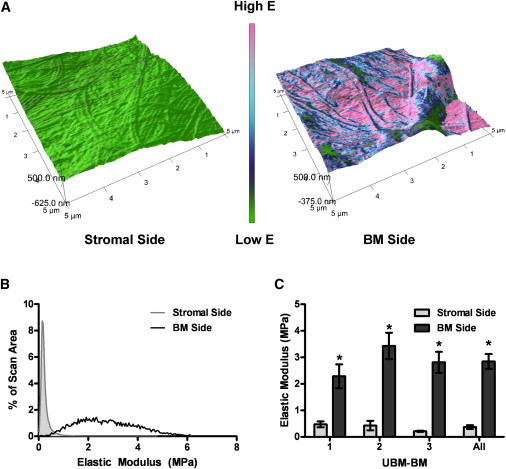
AFM measurements of the stromal and BM sides of UBM-BM. To determine whether rigidity differences between the stroma and BM were conserved at the nano- to microscale on each side of UBM-BM, (A) the stromal and BM sides of UBM-BM were scanned to determine (B) the distribution of elastic moduli in a 5×5 μm sample area (representative examples shown). (C) Weighted averages were calculated for each sample and showed significantly larger elastic moduli for the BM side. Data are presented as mean ± SE, and ∗ indicates p < 0.05 for stromal versus BM side comparisons. n = 9 total for each side from three independent UBM-BM specimens.
UBM-BM is highly cross-linked and dense
BM is thought to be a highly dense and cross-linked material. To indirectly measure the degree of cross-linking in each material, we determined the swelling ratio, Q, of UBM-BM and UBM. Q represents the ratio of wet to dry volume (or mass) of a polymer network, such as type I pepsinized collagen gels (37). As expected, UBM-BM had a significantly smaller Q than UBM (almost fourfold smaller), indicating a much more highly cross-linked network (Fig. 2 B). This finding is consistent with the dynamic mechanical properties of these materials, and with the fact that BM is the predominant component of UBM-BM but not the stromal-dominated UBM (Fig. 1). The inverse of Q yields the protein fraction, ν2, which is likewise fourfold larger for UBM-BM than for UBM (Fig. 2 B).
Development of invadopodia substrates that span eight orders of magnitude in rigidity
The elastic moduli of our model BM and stromal matrices, as measured by DMA or AFM, were considerably larger than those of the PAA gels that we previously tested for rigidity regulation of invadopodia (8). Therefore, we developed synthetic substrates for invadopodia testing that span a wider range of rigidities. These substrates included three PAA and three PUR substrates of defined rigidity that were polymerized in a thin layer on top of MatTek glass dishes and overlaid with 1% gelatin/FITC-fibronectin. Because PAA is fairly elastic, with a loss tangent much less than one (estimated as ∼0.2 from our previous rheometric measurements), the soft (storage modulus G′∼360 Pa) and hard (G′∼3300 Pa) PAA gels (8) were assumed to be incompressible and elastic such that the elastic modulus E = 3G′ (3). We synthesized an additional rigid PAA gel according to published protocols (38), which had a measured storage modulus of G′ = 9248 ± 598 Pa. Therefore, the elastic moduli of the soft, hard, and rigid PAA gels were calculated to be E = 1071, 9929, and 28,283 Pa, respectively. Because elastomeric PURs have larger moduli, we tested them by uniaxial tensile loading (T3000 and T900 PURs) or three-point bending (T300 PUR) using DMA, which yielded moduli values of 3.07, 5.58, and 1853 MPa, respectively. Glass was used as the ceiling of the rigidity spectrum with a known modulus of 69 GPa (39). The elastic moduli of these materials spanned eight orders of magnitude (Fig. 4), included MPa rigidities, and were used as substrates for invadopodia assays.
Figure 4.
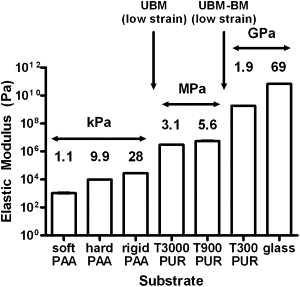
PAA, PUR, and glass substrates span eight orders of magnitude in rigidity. To isolate the effects of rigidity on invadopodia activity and span the MPA range of our stromal and BM model substrates, we synthesized PAA and PUR substrates with tunable rigidities. PAA elastic moduli were based on measurements of the storage modulus obtained by rheometry for soft, hard (8), or rigid PAAs. PUR elastic moduli were calculated by DMA. The glass elastic modulus was obtained from the literature (39). Data are presented as mean ± SE. Arrows indicate the rigidity regions relevant to the UBM and UBM-BM scaffolds. All elastic moduli were statistically significant from each other except between the T900 and T300 PUR substrates (p = 0.28; significances not shown on graph). n = 5 and 4 for rigid PAA and all PUR substrates, respectively.
ECM degradation as a function of rigidity exhibits a positively skewed distribution with a maximum at 30 kPa
To assess the regulation of invadopodia activity by substrate rigidity, we cultured CA1d breast carcinoma cells overnight on the FITC-fibronectin/gelatin/synthetic substrates, followed by fixation and immunostaining for the invadopodia markers actin and cortactin (Fig. 5), and quantitation of invadopodia-associated ECM degradation and activity (Fig. 6). As in our previous study (8), we observed that the CA1d cells degraded more ECM on hard PAA (10 kPa) surfaces than on soft PAA (1 kPa) surfaces. Of interest, however, there was a peak of ECM degradation activity, with the highest ECM degradation/cell occurring on the rigid PAA (E = 30 kPa) substrates, followed by the hard PAA (E = 10 kPa) substrates (Fig. 6 A). Contrary to expectation, there was significantly less ECM degradation/cell on the more-rigid PURs and glass substrates (Fig. 6 A). We observed a similar trend with 804G bladder carcinoma cells in which degradation/cell peaked in the kPa range but decreased in the GPa range (Fig. S1). In addition, quantitation of the total invadopodia/cell gave two separate peaks centered around the rigid PAA (E = 30 kPa) and the most rigid PUR (T300, E = 1.8 GPa; Fig. 6 B). These data suggest not only that cells sense a much wider rigidity range than previously thought (40,41) but that rigidity affects two different contributing processes to invadopodia activity: invadopodia formation and ECM degrading capability. We also saw differences in gene expression across a wide range of rigidities (Fig. S2), consistent with cellular sensing of rigidity in both the kPa and MPa–GPa range. The number of degrading invadopodia/cell (Fig. 6 C) paralleled the ECM degradation/cell curve (Fig. 6 A), with a major peak of degradation activity on the rigid PAA (E = 30 kPa) substrates. Cell size was not significantly altered and could not account for the observed differences in invadopodia activity (Fig. 6 D). Full statistical comparisons are reported in Table S1.
Figure 5.

Optimal peak of invadopodia-associated ECM degradation on 10 and 30 kPa substrates. Culture of breast cancer cells on synthetic substrates gave a surprising peak of ECM degradation activity on the hard and rigid PAA substrates, with reduced degradation at higher rigidities. Shown are representative wide-field immunofluorescence images of CA1d breast carcinoma cells after overnight culture on each of the rigidity substrates. Active degrading invadopodia are identified by colocalization of actin and cortactin-positive puncta with black degraded areas of the 1% gelatin/FITC-fibronectin matrix (arrows).
Figure 6.
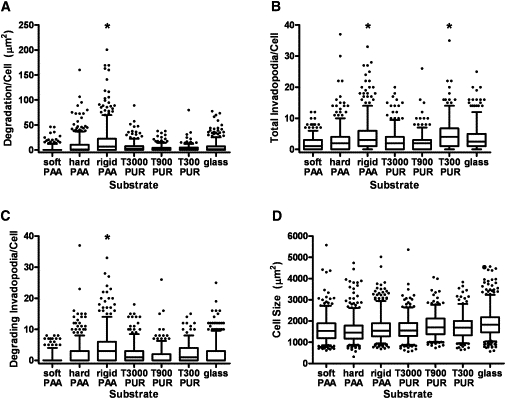
Quantitation of invadopodia numbers and activity on synthetic substrates. (A) Degradation area/cell peaked statistically on the rigid PAA substrates (i.e., significantly different from all other substrates) with a median value of 6.68 μm2. (B) Number of total invadopodia/cell (actively degrading and nondegrading) peaked statistically on both the rigid PAA (30 kPa) and T300 PUR (2 GPa) substrates with respective median values of 3 and 4. (C) The number of invadopodia actively degrading ECM/cell (as identified by colocalization of actin and cortactin over black areas only) peaked statistically only on the rigid PAA substrates with a median value of 3. (D) Differences in cell size were significant between some substrates but not between the majority of comparisons (except for glass, which was significantly different from all other substrates except T900 PUR; significances not shown on graph). Data are presented as box and whisker plots with solid lines indicating medians, whiskers representing 95% confidence intervals, and dots representing outliers. For comparisons depicted on the graphs, ∗ indicates p < 0.05 as described above for specific comparisons. For all statistical comparisons between groups, refer to Table S1. n ∼ 300–500 cells for each substrate, from four to six independent experiments.
Invadopodia formation is enhanced on the stromal side of UBM-BM versus the BM side
Our data up to this point indicate that optimal invadopodia activity occurs in cells somewhere between 10 kPa and 3 MPa (Figs. 5 and 6 A). For comparison, the mechanical analyses of stromal and BM tissue yielded E = 0.4 MPa (AFM) or 2 MPa (low-strain DMA), and E = 3 MPa (AFM) or 8 MPa (low-strain DMA), respectively, suggesting that from the rigidity standpoint, cells might have more invadopodia activity in stromal tissue than on BM. To test this hypothesis, CA1d breast carcinoma cells were seeded on either the BM or stromal side of UBM-BM overnight, and fixed and stained for actin and cortactin as markers of invadopodia (Fig. 7, A–D). Wide-field imaging was used for invadopodia quantification in CA1d cells (Fig. 7, A and B), and confocal imaging confirmed colocalization of the markers within the invadopodia structures and their formation on both surfaces (Fig. 7, C and D). The vast majority of cells (88% and 93% on the BM and stromal sides, respectively) exhibited at least one invadopodium. Of interest, cells seeded on the stromal side exhibited significantly more invadopodia/cell compared with those on the BM side (Fig. 7 E) and were also larger in size (Fig. 7 F), suggesting that the softer substrate is indeed conducive to formation of invadopodia. In addition, we tested 804G bladder carcinoma cells and again found that more invadopodia/cell were formed on the stromal side than on the BM side (Fig. S3) and that the majority of cells (87% and 77% on the stromal and BM surfaces, respectively) formed at least one invadopodium. The enhanced formation of invadopodia on the softer stromal side of the UBM-BM is consistent with the optimal-rigidity regime in the 10 kPa to 3 MPa range identified in our synthetic substrate studies (Fig. 6), although we cannot rule out other contributing factors, such as differences in ECM composition or topology.
Figure 7.
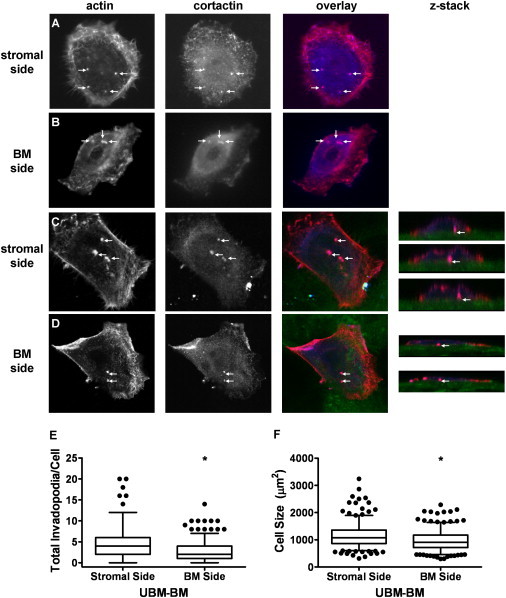
Invadopodia formation is enhanced on the stromal side of UBM-BM. To determine whether the stroma or BM is more permissive for formation of invadopodia, CA1d breast cancer cells were cultured overnight on the stromal or BM side of UBM-BM. Invadopodia (arrows) were identified by colocalization of actin (red) and cortactin (blue) for quantification on the (A) stroma or (B) BM using wide-field fluorescence imaging and confirmed with confocal imaging (C and D, z-stacks). The matrix surfaces were identified by collagen autofluorescence (green). (E) Quantitation of invadopodia formation on the stromal and BM sides of UBM-BM reveals a statistically significant increase in the total invadopodia/cell in cells cultured on the stroma. (F) Cell size is also statistically greater on the stroma. Data are presented as box and whisker plots with solid lines indicating medians, whiskers representing 95% confidence intervals, and dots representing outliers. ∗p < 0.05 for BM and stromal side comparisons; n ∼ 300 cells from two independent experiments.
Discussion
Mechanical signals are known to regulate a myriad of biological phenomena, including stem cell differentiation (19), cellular motility (18), tissue morphogenesis (3,42), and invadopodia activity (8). However, because we and others have used a limited range of substrate rigidities to test cellular responses (generally 0.1–30 kPa (3,18,19)), it is unclear what the rigidity ceiling is for mechanosensing. It is also unclear how in vivo tissue rigidities correspond to in vitro cellular responses. In this study, we sought to determine the optimal rigidity range that promotes ECM degradation by invasive cancer cells, and to connect our findings to tissue mechanics. Using both bulk tensile testing and nanoindentation methods, we characterized the mechanical properties of both the stromal and BM components of urinary bladder-derived tissue scaffolds and found that BM had a rigidity of 3–8 MPa at low strains and stromal tissue was ∼6-fold less rigid. Using synthetic substrates that span a wide range of rigidities, we found that breast cancer cells optimally degraded ECM on the ∼30 kPa substrates, with lower activity on substrates of higher rigidity (3 MPa to 69 GPa). Of interest, there were two peaks of invadopodia formation located around ∼30 kPa and 1.8 GPa, suggesting separate regulation of invadopodia formation and acquisition of proteolytic activity, and indicating a very wide rigidity range that elicits cellular responses. Gene expression data also support a wide range of rigidity sensing by cells. Consistent with our synthetic substrate data, the stromal side of the tissue scaffold UBM-BM supported better formation of invadopodia by breast cancer cells than the BM side of UBM-BM. A similar trend was observed with bladder carcinoma cells. Overall, our data suggest that the high rigidity of BM can serve as a restraining factor for invadopodia-associated ECM degradation, and that cells can sense differences in rigidity even in the MPa–GPa range.
Tissue rigidity has recently been implicated as a microenvironmental factor that promotes the development and progression of breast cancers. In humans, mammographically dense breast tissue is associated with the development of invasive breast carcinomas (6,7). In mouse tumor studies, the accumulation and cross-linking of stromal collagen fibers was shown to directly promote the formation and invasion of mammary tumors (4,43). Mechanotransduction signaling is thought to be critical for all of these effects (3,5,42,43). With regard to the rigidity of breast tissue, which represents a mixture of adipose, collagenous stromal, and ductal epithelial components, various elastic moduli have been reported. In similarity to our tensile DMA data, which showed elastic moduli in the low-MPa range for the stromal-dominated UBM, tensile loading of breast tissue in a previous study (44) yielded an apparent peak elastic modulus of 2.2 MPa. However, indentation testing by several groups (3,45–47) using 4–5 mm diameter tips yielded much softer moduli, with a range of 167 Pa to 30 kPa for normal tissue and 10–90 kPa for carcinomatous tissue. Using AFM (25–40 nm tips), we obtained an intermediate average elastic modulus of 400 kPa for the stromal component of UBM-BM. These differences may be due to the different modes of deformation used, the percent strain that was tested, and mechanical differences in bladder versus breast stroma. Our data do not resolve these differences, nor can our tests indicate what cells feel in the heterogenous local breast microenvironment; however, our data do indicate that the stroma is less rigid than the BM. Furthermore, stromal tissue is much closer in rigidity to the optimal rigidity range for invadopodia activity as defined using uniform synthetic substrates of tunable rigidity. Although the length scale at which matrix rigidity is probed has a significant effect on the measured mechanical properties of stromal tissue, test results for BM have been more consistent between nanoindentation versus macroscopic testing methods. Both techniques yield elastic moduli in the 1–20 MPa range (35,36,48), likely because the tissue properties do not change greatly across the nano-, micro-, and macroscale due to the high degree of cross-linking, small pore size, and uniformity of the matrix composition (1,2).
In this study we used two different experimental systems—native tissue scaffolds and synthetic substrates—to compare the effects of rigidity on invadopodia formation. The synthetic-substrate data were much more controlled because we were able to tune the rigidity without changing other parameters. However, both the mechanical testing and invadopodia experiments with the tissue scaffolds give context to our findings with the synthetic substrates. According to the elastic moduli measurements, the total invadopodia numbers for breast and bladder cancer cells cultured on the BM and stromal sides of UBM-BM fit the trend seen for the polymer substrates. Thus, more invadopodia were formed on the stromal component, which is closer in elastic modulus to the peak seen on the 30 kPa rigid PAA than is the BM substrate. If we correlate the invadopodia numbers with degradation, these results suggest that the stroma may be more conducive to degradation than the BM when rigidity is considered as the predominant factor. If we theoretically extrapolate from this finding and consider the early events in invasion as the BM is broken down by proteolysis at the primary tumor site, it appears that the BM begins to weaken, causing a decrease in elastic modulus and, according to our data, a shift to the left of the degradation curve, suggesting an increase in degradation. However, we must emphasize that the topology and chemical components of the ECM will also have an impact on cellular phenotype (49). In addition, ECM degradation is not the only factor that can affect invasion in a three-dimensional matrix (50). Nonetheless, substrate stiffness has been shown to be more important for determining cell shape than adhesive ligand density (51), implying that rigidity has an extremely strong influence on cellular behavior. In addition, chemical components could synergize with rigidity to limit invasiveness on BM substrates; for example, the BM component Laminin-332 was recently shown to limit invadopodia formation (52).
One of the most interesting findings of this study is that cells apparently can sense a wide range of rigidities, from kPa to GPa. This conclusion is supported by three major pieces of data. First, we find a peak of invadopodia-associated ECM degradation activity at ∼30 kPa, with a reduction in activity at higher rigidities. More convincingly, we observe two significant peaks of total invadopodia formation (one at ∼30 kPa and the other at 1.8 GPa) associated with substrate rigidity. Finally, we find regulation of gene expression across this same range (kPa–GPa). Thus, although it is thought that above a certain rigidity (e.g., 100 kPa) cells are performing isometric exercise (40,41) and may not feel differences from contraction, our data indicate that cells can sense rigidity differences even on highly rigid substrates. Considering that some physiologic substrates, such as BM and bone, are in the MPa–GPa range (35,48,54), it seems likely that mechanosensing across the full range could appropriately regulate behavior. Consistent with this idea, two recent studies using substrates with kPa–GPa moduli showed that preosteoblastic cells differentially sense and respond to moduli > 100 kPa by changing gene and protein expression (55,56). Likewise, breast and other cancer cell types that metastasize to bone may also experience low GPa rigidities, since calcified bone has an elastic modulus of 10–30 GPa as revealed by both nanoindentation and tensile testing methods (21,57). We speculate that differential sensing between MPa and GPa rigidities, combined with other contextual cues, might allow cells to distinguish between BM and bone. Nonetheless, future studies should investigate whether other cellular responses are altered in high-rigidity regimes and how the cell type of origin affects rigidity sensing at different matrix elasticities.
Acknowledgments
We thank Bryan Brown, Christopher Medberry, and Scott Johnson in Dr. Stephen Badylak's laboratory at the University of Pittsburgh for preparing the UBM and UBM-BM, and Thomas Gilbert for discussions on the mechanical characterization of ECM scaffolds.
This study was supported by National Institutes of Health (NIH) grants 1R01GM075126 to A.M.W., U54CA113007 (PI, Quaranta), a pilot project on 2 P50 CA098131-06 (Arteaga) to S.A.G., HL094707 to W.D.M., and 1K25CA143412 to A.P. We also thank the Vanderbilt University Medical Center Immunohistochemistry Core (CCSG 5P30 CA068485) and Cell Imaging Shared Resource (supported by NIH grants CA68485, DK20593, DK58404, HD15052, DK59637, and EY08126) for their assistance. Rheology experiments were performed at the Center for Nanophase Materials Science at Oak Ridge National Laboratory.
Supporting Material
References
- 1.Rowe R.G., Weiss S.J. Breaching the basement membrane: who, when and how? Trends Cell Biol. 2008;18:560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 3.Paszek M.J., Zahir N., Weaver V.M. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Provenzano P.P., Inman D.R., Keely P.J. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Provenzano P.P., Inman D.R., Keely P.J. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28:4326–4343. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vacek P.M., Geller B.M. A prospective study of breast cancer risk using routine mammographic breast density measurements. Cancer Epidemiol. Biomarkers Prev. 2004;13:715–722. [PubMed] [Google Scholar]
- 7.Boyd N.F., Rommens J.M., Paterson A.D. Mammographic breast density as an intermediate phenotype for breast cancer. Lancet Oncol. 2005;6:798–808. doi: 10.1016/S1470-2045(05)70390-9. [DOI] [PubMed] [Google Scholar]
- 8.Alexander N.R., Branch K.M., Weaver A.M. Extracellular matrix rigidity promotes invadopodia activity. Curr. Biol. 2008;18:1295–1299. doi: 10.1016/j.cub.2008.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weaver A.M. Invadopodia. Curr. Biol. 2008;18:R362–R364. doi: 10.1016/j.cub.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 10.Parekh, A., and A. M. Weaver. 2009. Regulation of cancer invasiveness by the physical extracellular matrix environment. Cell Adh. Migr. 3:288–292. [DOI] [PMC free article] [PubMed]
- 11.Sabeh F., Shimizu-Hirota R., Weiss S.J. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hotary K., Li X.Y., Weiss S.J. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 2006;20:2673–2686. doi: 10.1101/gad.1451806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akgül B., García-Escudero R., Storey A. The E7 protein of cutaneous human papillomavirus type 8 causes invasion of human keratinocytes into the dermis in organotypic cultures of skin. Cancer Res. 2005;65:2216–2223. doi: 10.1158/0008-5472.CAN-04-1952. [DOI] [PubMed] [Google Scholar]
- 14.McDaniel S.M., Rumer K.K., Schedin P. Remodeling of the mammary microenvironment after lactation promotes breast tumor cell metastasis. Am. J. Pathol. 2006;168:608–620. doi: 10.2353/ajpath.2006.050677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badylak S.F. Xenogeneic extracellular matrix as a scaffold for tissue reconstruction. Transpl. Immunol. 2004;12:367–377. doi: 10.1016/j.trim.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Brown B., Lindberg K., Badylak S.F. The basement membrane component of biologic scaffolds derived from extracellular matrix. Tissue Eng. 2006;12:519–526. doi: 10.1089/ten.2006.12.519. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert T.W., Wognum S., Badylak S.F. Collagen fiber alignment and biaxial mechanical behavior of porcine urinary bladder derived extracellular matrix. Biomaterials. 2008;29:4775–4782. doi: 10.1016/j.biomaterials.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelham R.J., Jr., Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engler A.J., Sen S., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 20.Engler A.J., Rehfeldt F., Discher D.E. Microtissue elasticity: measurements by atomic force microscopy and its influence on cell differentiation. Methods Cell Biol. 2007;83:521–545. doi: 10.1016/S0091-679X(07)83022-6. [DOI] [PubMed] [Google Scholar]
- 21.Moore S.W., Roca-Cusachs P., Sheetz M.P. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev. Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemir S., West J.L. Synthetic materials in the study of cell response to substrate rigidity. Ann. Biomed. Eng. 2010;38:2–20. doi: 10.1007/s10439-009-9811-1. [DOI] [PubMed] [Google Scholar]
- 23.Guelcher S.A. Biodegradable polyurethanes: synthesis and applications in regenerative medicine. Tissue Eng. Part B Rev. 2008;14:3–17. doi: 10.1089/teb.2007.0133. [DOI] [PubMed] [Google Scholar]
- 24.Oertel G. Hanser Publications; Munich: 1994. Polyurethane Handbook. [Google Scholar]
- 25.Freytes D.O., Badylak S.F., Rundell A.E. Biaxial strength of multilaminated extracellular matrix scaffolds. Biomaterials. 2004;25:2353–2361. doi: 10.1016/j.biomaterials.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Yurchenco P.D., Ruben G.C. Basement membrane structure in situ: evidence for lateral associations in the type IV collagen network. J. Cell Biol. 1987;105:2559–2568. doi: 10.1083/jcb.105.6.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey R.F., Wiggins J.S., Puckett A.D. Hydrolyzable poly(ester-urethane) networks from L-lysine diisocyanate and D,L-lactide/ɛ-caprolactone homo- and copolyester triols. J. Polym. Sci. A Polym. Chem. 1994;32:2345–2363. [Google Scholar]
- 28.Kandow C.E., Georges P.C., Beningo K.A. Polyacrylamide hydrogels for cell mechanics: steps toward optimization and alternative uses. Methods Cell Biol. 2007;83:29–46. doi: 10.1016/S0091-679X(07)83002-0. [DOI] [PubMed] [Google Scholar]
- 29.Guelcher S., Srinivasan A., Hollinger J. Synthesis, in vitro degradation, and mechanical properties of two-component poly(ester urethane)urea scaffolds: effects of water and polyol composition. Tissue Eng. 2007;13:2321–2333. doi: 10.1089/ten.2006.0395. [DOI] [PubMed] [Google Scholar]
- 30.Inoue S. Basic structure of basement membranes is a fine network of “cords,” irregular anastomosing strands. Microsc. Res. Tech. 1994;28:29–47. doi: 10.1002/jemt.1070280105. [DOI] [PubMed] [Google Scholar]
- 31.Fung Y. Springer-Verlag; New York: 1997. Biomechanics: Mechanical Properties of Living Tissues. [Google Scholar]
- 32.Gilbert T.W., Sacks M.S., Chancellor M.B. Fiber kinematics of small intestinal submucosa under biaxial and uniaxial stretch. J. Biomech. Eng. 2006;128:890–898. doi: 10.1115/1.2354200. [DOI] [PubMed] [Google Scholar]
- 33.Freytes D.O., Stoner R.M., Badylak S.F. Uniaxial and biaxial properties of terminally sterilized porcine urinary bladder matrix scaffolds. J. Biomed. Mater. Res. B Appl. Biomater. 2008;84:408–414. doi: 10.1002/jbm.b.30885. [DOI] [PubMed] [Google Scholar]
- 34.Machalek, A. 2005. An owner's guide to the cell. In Inside the Cell. National Institute of General Medical Sciences, Bethesda, MD. http://publications.nigms.nih.gov/insidethecell/index.html.
- 35.Candiello J., Balasubramani M., Lin H. Biomechanical properties of native basement membranes. FEBS J. 2007;274:2897–2908. doi: 10.1111/j.1742-4658.2007.05823.x. [DOI] [PubMed] [Google Scholar]
- 36.Candiello J., Cole G.J., Halfter W. Age-dependent changes in the structure, composition and biophysical properties of a human basement membrane. Matrix Biol. 2010;29:402–410. doi: 10.1016/j.matbio.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Chan B.P., So K.F. Photochemical crosslinking improves the physicochemical properties of collagen scaffolds. J. Biomed. Mater. Res. A. 2005;75:689–701. doi: 10.1002/jbm.a.30469. [DOI] [PubMed] [Google Scholar]
- 38.Yeung T., Georges P.C., Janmey P.A. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 39.Callister W. John Wiley & Sons; Somerset, NJ: 2000. Fundamentals of Material Science and Engineering: An Interactive E-Text. [Google Scholar]
- 40.Discher D.E., Janmey P., Wang Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 41.Wells R.G., Discher D.E. Matrix elasticity, cytoskeletal tension, and TGF-β: the insoluble and soluble meet. Sci. Signal. 2008;1:pe13. doi: 10.1126/stke.110pe13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wozniak M.A., Desai R., Keely P.J. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J. Cell Biol. 2003;163:583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levental K.R., Yu H., Weaver V.M. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edsberg L.E., Cutway R., Natiella J.R. Microstructural and mechanical characterization of human tissue at and adjacent to pressure ulcers. J. Rehabil. Res. Dev. 2000;37:463–471. [PubMed] [Google Scholar]
- 45.Krouskop T.A., Wheeler T.M., Hall T. Elastic moduli of breast and prostate tissues under compression. Ultrason. Imaging. 1998;20:260–274. doi: 10.1177/016173469802000403. [DOI] [PubMed] [Google Scholar]
- 46.Samani A., Zubovits J., Plewes D. Elastic moduli of normal and pathological human breast tissues: an inversion-technique-based investigation of 169 samples. Phys. Med. Biol. 2007;52:1565–1576. doi: 10.1088/0031-9155/52/6/002. [DOI] [PubMed] [Google Scholar]
- 47.Van Houten E.E.W., Doyley M.M., Paulsen K.D. Initial in vivo experience with steady-state subzone-based MR elastography of the human breast. J. Magn. Reson. Imaging. 2003;17:72–85. doi: 10.1002/jmri.10232. [DOI] [PubMed] [Google Scholar]
- 48.Welling L.W., Zupka M.T., Welling D.J. Mechanical properties of basement membrane. News Physiol. Sci. 1995;10:30–35. [Google Scholar]
- 49.Geiger B., Spatz J.P., Bershadsky A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 50.Brábek J., Mierke C.T., Fabry B. The role of the tissue microenvironment in the regulation of cancer cell motility and invasion. Cell Commun. Signal. 2010;8:22. doi: 10.1186/1478-811X-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Engler A.J., Griffin M.A., Discher D.E. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J. Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu S., Yamashita H., Quaranta V. Laminin-332-β1 integrin interactions negatively regulate invadopodia. J. Cell. Physiol. 2010;223:134–142. doi: 10.1002/jcp.22018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reference deleted in proof.
- 54.Nyman J.S., Leng H., Wang X. Differences in the mechanical behavior of cortical bone between compression and tension when subjected to progressive loading. J. Mech. Behav. Biomed. Mater. 2009;2:613–619. doi: 10.1016/j.jmbbm.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khatiwala C.B., Peyton S.R., Putnam A.J. The regulation of osteogenesis by ECM rigidity in MC3T3-E1 cells requires MAPK activation. J. Cell. Physiol. 2007;211:661–672. doi: 10.1002/jcp.20974. [DOI] [PubMed] [Google Scholar]
- 56.Smith K.E., Hyzy S.L., Boyan B.D. The dependence of MG63 osteoblast responses to (meth)acrylate-based networks on chemical structure and stiffness. Biomaterials. 2010;31:6131–6141. doi: 10.1016/j.biomaterials.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chevalier Y., Pahr D., Zysset P. Validation of a voxel-based FE method for prediction of the uniaxial apparent modulus of human trabecular bone using macroscopic mechanical tests and nanoindentation. J. Biomech. 2007;40:3333–3340. doi: 10.1016/j.jbiomech.2007.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


