Abstract
The minor histocompatibility antigen 60 (H60a) is expressed in BALB/C and 129/Sv but not in C57BL/6 strains of mice. We recently found that IFNγ down-regulates H60a, but the mechanism of regulation is not known. To better understand the regulation of H60a, we examined the genomic locus of H60a in 129/Sv and C57BL/6 strains. We found that the upstream regulatory region of H60a was present and functional in both strains. Interestingly, IFNγ can down-regulate H60a transcripts in cell lines from 129/Sv but not C57BL/6 strains of mice, suggesting that IFNγ-dependent regulation of H60a proceeds through cis elements other than the conserved promoter region. We determined that the regulation of H60a by IFNγ proceeds through the 3′UTR of H60a, which is present in 129/Sv, but not C57BL/6 cells. We also found that the H60a 3′UTR and microRNAs can contribute to the level of constitutive expression of H60a in tumor cell lines. We conclude that in 129/Sv strain mice, H60a can be regulated by its 3′UTR through IFNγ and unknown microRNAs. Since H60a mediates NK cell target recognition, our studies identify a cis element that can regulate virus and tumor surveillance.
Keywords: NKG2D, H60, natural killer cells, interferon, microRNA
1. Introduction
Natural Killer Group 2D (NKG2D) is a receptor expressed on NK cells that mediates the detection of stressed cells that are infected by viruses or undergoing transformation (Cerwenka and Lanier, 2001; Raulet, 2003). It recognizes a ligand family that is heterogeneous and generally not expressed at functional levels in normal tissues, but can be up-regulated by certain stimuli, including DNA damage and viral infection (Gasser et al., 2005; Yokoyama, 2000). H60a is an NKG2D ligand that was originally identified as a minor histocompatibility antigen between BALB/b and C57BL/6 mice (Malarkannan et al., 1998). It is known to be expressed by hematopoietic cells and tumor cell lines from various strains, including BALB/C and 129/Sv (Bui et al., 2006a; Cerwenka et al., 2000; Diefenbach et al., 2000; Malarkannan et al., 1998), but is a pseudogene in C57BL/6 mice. The NKG2D ligands H60b and H60c were recently identified (Takada et al., 2008; Whang et al., 2009) and share 73 and 44% amino acid identity with H60a. H60b was shown to be expressed in BALB/C and C57BL/6 tissues, but its expression in the 129/Sv-strain has not been studied.
The expression of NKG2D ligands is regulated via complex pathways involving transcriptional, post-transcriptional, and post-translational mechanisms (Mistry and O’Callaghan, 2007; Nausch and Cerwenka, 2008; Yadav et al., 2009). Although these mechanisms have been described for many of the NKG2D ligands, surprisingly little is known about the signals that regulate H60a. For example, increased MULT1, RAE, and MICA/B transcripts can be found in cells undergoing DNA damage (Gasser et al., 2005), but it is not known whether H60a is induced by similar signals. Furthermore, virus infection seems to induce RAE (Lodoen et al., 2003) and H60b (Takada et al., 2008), but not H60a. Interestingly, the expression pattern of H60a, b, and c in normal cells is different, even though these molecules all can function as NKG2D ligands and activate NK cell recognition. H60c is expressed exclusively in keratinocytes (Whang et al., 2009), while H60b and H60a seem to be more broadly expressed (Takada et al., 2008).
The function of H60a as a recognition determinant for NK cells during tumor formation and viral infection is supported by multiple studies. H60a is induced during carcinogenesis (Girardi et al., 2001), and overexpression of H60a on tumor cells is sufficient to mediate tumor rejection by NK cells and prime adaptive immunity (Diefenbach et al., 2001). Nevertheless, little is known about other signals that induce its expression during carcinogenesis and maintains it on tumor cell lines. We have found that the cytokines IFNα/β and IFNγ potently down-regulate H60a on tumor cells, rendering them resistant to NK cell lysis (Bui et al., 2006a), but the mechanism by which IFNs regulate H60a in tumor cells is not known. In this study, we defined the H60a locus in the 129/Sv and C57BL/6 strains and found that the promoter region of H60a is identical and functional in both strains. We further show that IFNγ regulates H60a transcripts via its 3′UTR. Inhibition of DICER led to enhanced 3′UTR activity and H60a protein expression, thus suggesting that a mouse NKG2D ligand is regulated by microRNAs (miRNAs).
2. Materials and Methods
2.1. Shot-gun sequencing of the H60a locus in 129S6/SvEv Tac strain
We screened a bacterial artificial chromosome (BAC) library constructed from cells from a 129S6/SvEvTac mouse (http://bacpac.chori.org) for H60a sequences. Shot-gun sequencing (http://genome.wustl.edu/) of one clone generated a 44 kilobase (kB) contiguous sequence (contig) (Accession Number HM590820). This sequence was aligned with publicly available sequence using a BLAST program in NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi ).
2.2. Cell lines and stimulations
MCA-induced sarcomas were isolated and passaged in vitro as described (Bui et al., 2006b; Shankaran et al., 2001). Cell lines were maintained in RPMI 1640 supplemented with 10% FCS, L-glutamine, NEAA, sodium pyruvate, sodium bicarbonate, pen/strep, and β-mercaptoethanol. Recombinant murine IFNγ was obtained from eBioscience (San Diego, CA) and used at 100 U/ml.
2.3. Real-time PCR
RNA was generated using Trizol Reagent (Invitrogen, San Diego). cDNA was made using the Applied Biosystems (Foster City, CA) protocol. Real-time Taqman PCR reactions were performed using the following primers: H60a for, 5′GAG CCA CCA GCA AGA GCA A; H60a rev, 5′CCA GTA TGG TCC CCA GAT AGC T; H60a probe VIC-5′TTG CCT GAT TCT GAG CCT TTT CAT TCT GCT-TAMRA (Bui et al., 2006a); GAPDH for, 5′CTT AGC ACC CCT GGC CAA G; GAPDH rev, 5′TGG TCA TGA GTC CTT CCA CG; GAPDH probe, VIC-5′CAT CCA TGA CCA CCC CTG GCC AAG-MGB (Buchau et al.). Sybr green reactions were performed with the following primers: RAE1 for, 5′ATC AAC TTC CCC GCT TCC A; RAE1 rev, 5′AGA TAT GAA GAT GAG TCC CAC AGA GAT A (Bui et al., 2006a); H60b for, 5′AGC CTT TTG GTC CTG CTG AAT; H60b rev, 5′ATG TTT TTT ATC ACC AAA ATC AAG GAG T (Takada et al., 2008); H60a exon2-4 for, 5′CTG AGC TAT CTG GGG ACC AT; H60a exon2-4 rev, 5′AGA TTG TGT TGT GAC ATT CAA GG. The RAE1 primer recognizes all RAE1 family members. H60a exon2-4 sybr primers detect H60a transcripts from 129/Sv but not C57BL/6 mice. H60a taqman primers detect H60a transcripts from both strains. All experiments use at least duplicate conditions and duplicate real-time wells and are done at least twice.
2.4. Luciferase Assay
The H60a promoter region containing 527 bp of sequence upstream of the transcriptional start site and 501 bp of 5′UTR was amplified from a 129/SvEv BAC clone and ligated into a luciferase reporter plasmid (PGL3-basic, Promega) using pGLOW TOPO cloning protocol (Invitrogen) and the following primers: 5′AGA AGA AAA CCT GAG GGT GGG3′, 5′GGT CTT CCC TCA GAC CCT GCT3′. The H60a 3′UTR was amplified from a 129/SvEv BAC clone into a TOPO shuttle vector (Invitrogen) and subcloned into a reporter construct that contained both the firefly and renilla luciferase genes downstream of independent promoters (OmicsLink, Genecopoeia, Rockville, MD) using the following primers: 5′ TCA TGT GAC TCT TCA AC3′, 5′TAT GCA AGC TTA GTT C. The H60a 3′UTR was inserted downstream of the firefly luciferase gene. Reporter activity was measured by dividing firefly luciferase light values by renilla luciferase light values using a dual luciferase assay (Promega). Transfection was done through Lipofectamine 2000 (Invitrogen) in triplicate wells in a 96-well plate using four different DNA to cell ratios. Cells were left unstimulated or stimulated with IFNγ the next day, and activity was measured after 24 hours of IFNγ stimulation. In some experiments, siRNA control or siRNA specific for DICER were used (Dharmacon). All experiments were done at least twice.
2.5. Antibodies and FACS analysis
All cell stainings were done with 50-80% confluent cells. Cells were harvest with dPBS or HBSS supplemented with 2.5 mM EDTA. Trypsin was not used since it decreased H60a staining (unpublished observations, J.D.B.). Monoclonal antibodies to H60a were obtained from R&D (Minneapolis, MN). Secondary antibodies were obtained from Biolegend (San Diego, CA). Staining was conducted for 15-30 minutes at 4° in FACS tubes containing 0.5-2 million total cells, 0.5-1 μl of antibody, and 100 μl of FACS buffer (PBS+1% FCS+0.09% NaN3, Sigma). All analyses were done on live cells identified by forward and side scatter properties. All experiments were done at least twice.
2. Results
3.1. Alignment of H60a and H60b transcripts with 129/Sv genomic sequence
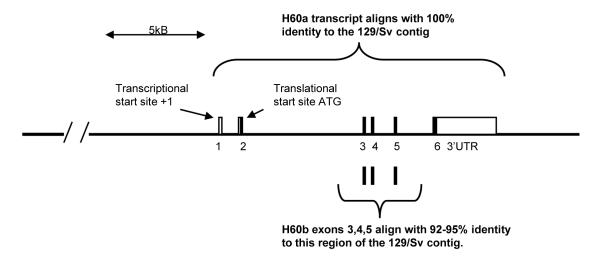
Most mouse genomic data is derived from the C57BL/6 strain, which does not have the intact H60a gene (Malarkannan et al., 1998; Samarakoon et al., 2009). To examine the H60a genomic locus, we screened a 129/Sv-strain BAC library and sequenced a putative clone that contains the H60a gene. We generated a 44 kb contig of the H60a genomic locus via this approach (Accession Number HM590820). This sequence was aligned with transcripts of H60a (NM_010400.2) and H60b (NM_001177775) (Figure 1). We found that when the H60a transcript was aligned to the 129/Sv genomic sequence, there were regions of 100% identity that corresponded to the six exons of H60a (Takada et al., 2008), thus confirming the accuracy of our sequence. In contrast, the H60b transcript did not align with 100% identity to our contig. There was 92-95% sequence identity between exons 3, 4, and 5 of H60a and H60b, but not between exons 1, 2, or 6. These results are concordant with previous studies showing high homology in the alpha 1, alpha 2, and transmembrane regions of the H60a and H60b proteins (Samarakoon et al., 2009; Takada et al., 2008) and further substantiates our approach. We conclude that our 129/Sv contig contains the entire H60a gene, including 30 kb of upstream sequence and 5 kb of downstream sequence, but does not contain any region of the H60b coding sequence.
Figure 1. Alignment of H60a and H60b transcripts with a 44 kb contig obtained by shotgun sequencing of a 129/Sv-strain BAC clone.
There is 100% identity in this alignment between H60a and the contig. The H60b gene is not present in this region, but there is 92-95% identity between exons 3,4,5 of H60b with the contig and with H60a. The other H60b exons (1, 2, 6) do not align with significant identity. Sequences used were H60a (NM_010400.2), H60b (NM_001177775), and 129/Sv contig (HM590820).
3.2. Alignment of H60a and H60b transcripts with C57BL/6 genomic sequence
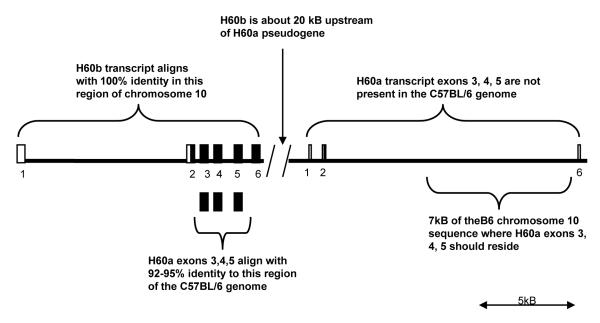
Next, we wished to elucidate the structure of the H60a locus in the C57BL/6 genome, where it is a pseudogene. Figure 2 shows that when the H60a transcript was aligned to C57BL/6 genomic sequences from chromosome 10 (NCBI Build 37.1), there was partial alignment of the first 2 exons and part of the 3′UTR; however, the C57BL/6 genome did not contain exons 3, 4, and 5 of H60a. In fact, when these exons were used to BLAST the C57BL/6 genome, they corresponded to exons 3, 4, and 5 of H60b with >90% identity. Interestingly, we found that the H60b gene locus lies approximately 20 kb upstream of the H60a putative transcriptional start site in the C57BL/6 strain (Figure 2), while the corresponding region in the 129/Sv strain does not contain H60b sequences (Figure 1). Similar to previous studies (Samarakoon et al., 2009; Takada et al., 2008), we found that H60b is encoded by six exons (similar to H60a) suggesting that the genes arose by a duplication or gene conversion event (Samarakoon et al., 2009; Takada et al., 2008; Whang et al., 2009). Both genes have a first exon that is 5′UTR. Both genes have an exon coding for the signal peptide region, three exons with high identity coding for the alpha 1, alpha 2, transmembrane, and part of the cytoplasmic region, and a final exon with the translation stop codon and 3′UTR. These results are consistent with previously published reports (Samarakoon et al., 2009; Takada et al., 2008).
Figure 2. Alignment of H60a and H60b transcript with chromosome 10 of the C57BL/6 strain.
There is 100% identity in this alignment between H60b and chromosome 10. The H60a exons align roughly 20 kB downstream of H60b exon 6. Only H60a exons 1 and 2 are present and align with 100% identity. There is intervening sequence in the H60a locus of the C57BL/6 strain that does not align with any H60a or H60b sequence. H60a exons 3,4,5 are not present in the C57BL/6 strain but align with 92-95% identity to the H60b exons.
3.3. Alignment of the 129/Sv and C57BL/6 genomic sequence at the H60a locus
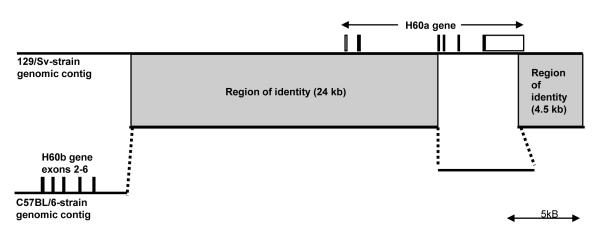
We next examined the similarities and differences between the H60a loci in 129/Sv versus C57BL/6 strains by aligning the respective contigs. Figure 3 shows that the two loci have 100% identity beginning with a region 20 kb upstream of the H60a transcriptional start site through to part of the third exon of H60a. This extensive region of identity upstream of the H60a transcriptional start site suggests that the regulation of H60a transcription may be similar between the two strains, and furthermore suggests that H60a transcripts may detected in C57BL/6 cells.
Figure 3. Alignment of the 44 kb contig from 129/Sv strain with the corresponding region of chromosome 10 of the C57BL/6 strain.
There is significant identity (shaded areas) in the central region encompassing a 24 kb of sequence that begins downstream of H60b and contains the entire upstream promoter region of H60a and exons 1 and 2 of H60a. The 129/Sv strain contig does not contain H60b sequences, while the C57BL/6 strain does not contain H60a exons 3,4,5.
We also found significant differences between the strains. First, the C57BL/6 genome does not have the complete H60a gene, as shown previously (Malarkannan et al., 1998; Samarakoon et al., 2009). The H60a pseudogene in the C57BL/6 genome lacks the 6.6 kb region that contains exons 3, 4, and 5 of H60a and instead has a 7.4 kb region that does not display sequence identity to any other NKG2D ligand or possess any similarity to the rattus norvegicus genome (data not shown). In addition, this region is not predicted to encode for any genes (data not shown). These results suggest that the events that rendered H60a a pseudogene in one strain and an intact gene in another strain are not simple deletional, insertional, or translocation events.
The second major difference between the strains is that the H60b locus was not present in the 129/Sv contig. Instead of the H60b gene, this region of the 129/Sv contig (approximately 9.1 kb in length about 25 kb upstream of the putative H60a transcriptional start site) has limited (70%) sequence identity to the RAET1L gene of rattus norvegicus (data not shown). We conclude that in the C57BL/6 strain, the H60b gene is located 20 kb upstream of H60a, whereas in the 129/Sv strain, H60b may be elsewhere or may not be present.
3.4. H60a transcripts can be detected in C57BL/6 cell lines
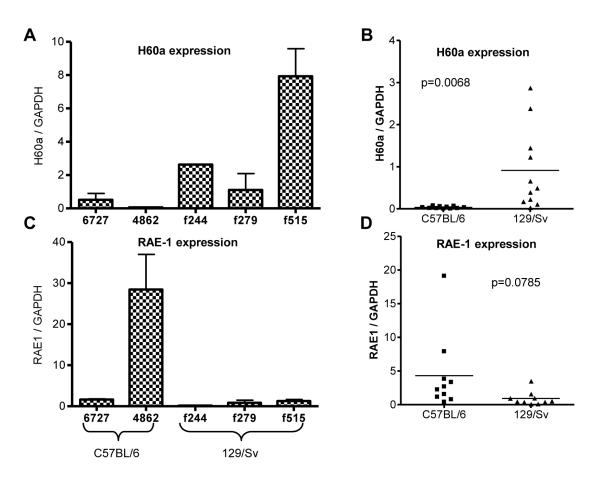
As shown in Figures 1-3, both 129/Sv and C57BL/6 strains possess the putative H60a promoter region and the entire exons 1 and 2. Since the entire promoter region up to 20 kb was present and identical between the strains, we hypothesized that H60a transcripts could be detected in C57BL/6 cells. To determine whether the H60a promoter region is functional in C57BL/6 cells, we used qRT-PCR to detect sequences from exon 2 of H60a in C57BL/6 and 129/Sv cell lines. We found that indeed, low levels of exon 2 (Figures 4A-B) but not exon 4 (data not shown) sequences can be detected in transcripts from C57BL/6 MCA sarcoma cell lines, suggesting that C57BL/6 cells produced a truncated H60a transcript. On average, these transcripts were 20-fold decreased in C57BL/6 cells compared to 129/Sv cells, indicating that perhaps the truncated transcript could not accumulate due to instability. In contrast, another NKG2D ligand, RAE1, displayed transcripts levels that were generally higher in C57BL/6 versus 129/Sv cells (Figure 4C-D), suggesting a compensatory mechanism for the lack for H60a expression. We did not detect appreciable levels of the NKG2D ligand MULT1 protein in cell lines from either strain (data not shown) and therefore did not examine MULT1 transcripts in these cell lines. We did not examine whether truncated H60a protein can be found in C57BL/6 mice, but the very low levels of transcript indicate that H60a protein, if translated, would be difficult to detect.
Figure 4. Truncated H60a transcripts can be detected in C57BL/6 tumor cell lines.
Quantitative real-time PCR was used to measure the level of (A, B) H60a or (C-D) RAE-1 transcript. Individual cell lines are shown in A, C, and multiple cell lines are shown in B, D, with each symbol representing a different cell line. Results are representative of 2 total experiments.
3.5. IFNγ does not regulate the H60a promoter region
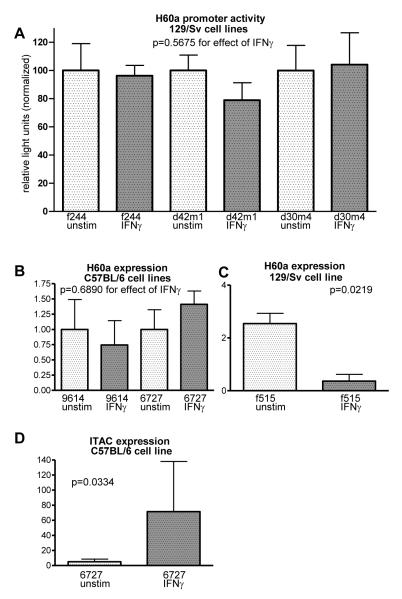
Previously, we showed that IFNs can down-regulate the level of H60a transcript in 129/Sv sarcomas cell lines (Bui et al., 2006a). This regulation could occur via the promoter region or the 3′UTR, as IFN is known to induce many different miRNAs (Pedersen et al., 2007), some of which can regulate NKG2D ligands (Yadav et al., 2009). To determine whether IFN-dependent regulation of H60a occurred via cis elements upstream of the H60a coding sequence, we constructed a luciferase reporter plasmid containing a 527 bp region upstream of the H60a transcription start site. We transfected this plasmid into several 129/SvEv cell lines and measured luciferase activity in IFNγ-stimulated cells compared to cells that were not stimulated. We did not find any change in normalized luciferase activity in cells treated with IFNγ (Figure 5A, p=0.5675).
Figure 5. IFNγ does not act on the H60a promoter region.
(A) H60a promoter activity was measured using a luciferase reporter assay. The 129/SvEv-strain cell lines f244, d42m1, and d30m4 were transfected with a reporter construct containing 527 bp of the H60a promoter region and were left unstimulated or stimulated with IFNγ. Reporter activity was normalized to renilla control and expressed as percentage of unstimulated cells. (B-D) Quantitative real-time PCR was used to measure H60a and ITAC transcripts in cells that were not stimulated or stimulated with IFNγ. H60a was measured in the (B) C57BL/6 cell lines 6727 and 9614 or (C)129/Sv cell line f515. (D) ITAC was used as a positive control for IFNγ responsiveness in the 6727 cell line. Results are representative of at least 2 total experiments for each section.
Since we only tested a short region of the promoter in this luciferase assay, we could not rule-out that the IFNγ-responsive element was outside of the tested region. To examine a larger region of the H60a promoter, we took advantage of the finding that C57BL/6 cells possessed 20 kb of H60a promoter region but not 3′UTR sequences (Figures 1-3). Even though the transcript from C57BL/6 mice was truncated, we postulated that IFNγ could still regulate transcript levels if it acted on the conserved 20 kb promoter region. Therefore, we compared the level of H60a transcripts in C57BL/6 and 129/Sv sarcoma cell lines with and without IFNγ treatment. We found that IFNγ did not down-regulate H60a transcripts in C57BL/6 cell lines (Figure 5B, p=0.6890), whereas it reduced H60a transcripts threefold in a 129/Sv cell line (Figure 5C, p=0.0219), as described previously (Bui et al., 2006a). We confirmed that the C57BL/6 cells did indeed respond to IFNγ by their upregulation of ITAC (IFN-γ-inducible α-chemoaatractant, CXCL11), a known downstream target of IFN (Uppaluri et al., 2008) (Figure 5D, p=0.0334).
3.6. IFNγ acts on the H60a 3′UTR
A notable mechanism by which IFNγ can regulate transcripts involves the induction miRNAs (Pedersen et al., 2007; Yadav et al., 2009) that in turn act on the 3′UTR of certain transcripts to inhibit translation or lead to their degradation (Bartel, 2004). Since IFNγ did not regulate H60a in C57BL/6 cells, which possess an intact H60a promoter region, we hypothesized that this cytokine would regulate H60a expression via the H60a 3′UTR. We amplified the entire 3.3 kb 3′UTR of the H60a gene from our 129/Sv contig and placed it downstream of the luciferase gene to generate a luciferase reporter for the H60a 3′UTR (http://www.genecopoeia.com/product/mirna/targets.php). Using this reporter plasmid, we found that IFNγ indeed acted on the 3′UTR of H60a by decreasing luciferase activity by twofold (Figure 6, p<0.0001). This was seen with three separate cell lines, and even occurred in cell lines that expressed very little H60a, thus providing evidence that IFNγ regulates H60a expression via its 3′UTR.
Figure 6. IFNγ acts on the H60a 3′UTR to decrease expression of a reporter construct.

The H60a 3′UTR was inserted downstream of a luciferase reporter gene that was tranfected into three different cell lines. The next day, cells were stimulated with IFNγ or left unstimulated, and relative light units were measured and normalized to renilla luciferase activity. Results are representative of 2 total experiments.
3.7. Correlation between basal H60a protein expression, H60a transcript levels, and H60a 3′UTR activity
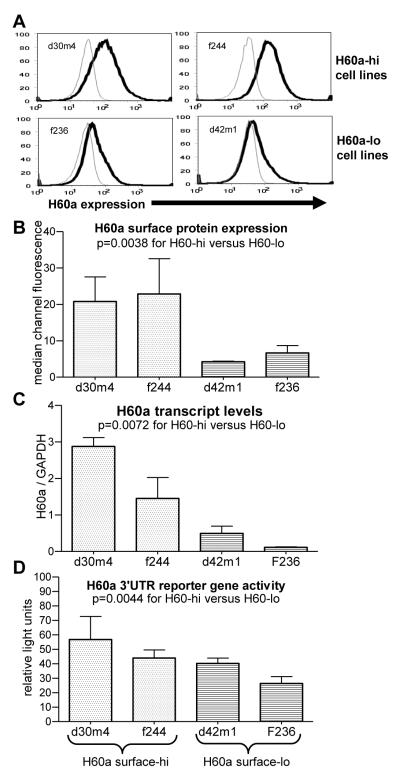
One interesting aspect of H60a expression is its heterogeneous expression among tumor cell lines of similar origin, indicating that its expression is actively regulated by tumor cells (Bui et al., 2006a). We next examined whether basal H60a surface protein expression could be correlated with H60a transcript levels and/or 3′UTR activity. Using flow cytometry, we identified two cell lines that had high (f244 and d30m4) and two that had low (d42m1 and f236) levels of surface H60a protein (Figure 7A-B, p=0.0038). We then examined the basal levels of H60a transcript (Figure 7C) in these cell lines. We found that the levels of H60a transcript correlated with the surface H60a expression, i.e., H60a-hi cell lines had high levels of H60a transcript, and H60a-lo cell lines had low levels of H60a transcript (p=0.0072). Since the transcript levels can be regulated by transcriptional or post-transcriptional mechanisms, we examined the basal activity of the H60a 3′UTR. We found that the repression of luciferase reporter activity by the H60a 3′UTR did not completely correlate with the level of H60a protein (Figure 7D), suggesting that the H60a 3′UTR functioned to limit H60a surface protein expression in some cell lines but not others. Nevertheless, the amount of 3′UTR activity was statistically greater in H60a-hi cells versus low cells (p=0.0044).
Figure 7. Constitutive H60a surface protein levels correlate with transcript and 3′UTR activity.
H60a-hi and H60a-lo cell lines were examined for H60a (A-B) protein by flow cytometry (C) transcript by real-time PCR, or (D) 3′UTR activity by luciferase reporter assay. Results are representative of at least 2 total experiments for each section.
3.8. Inhibition of DICER leads to increased 3′UTR activity
Next, we examined whether basal levels of miRNAs could regulate H60a protein levels and 3′UTR activity. Towards this end, we blocked the production of miRNAs by inhibiting DICER expression using siRNAs. When siRNAs specific for DICER were transfected into the H60a-lo cell line f236, we found two-fold increased H60a-3′UTR activity, suggesting that miRNAs present in f236 were acting on the H60a-3′UTR to limit H60a expression (p=0.0363). Correspondingly, there were increased levels of H60a surface protein in cells transfected with siRNA specific for DICER versus control transfection, but these findings were not statistically significant (p=0.1979). These results confirm that the basal expression of H60a is regulated by miRNAs, similar to recent findings on the regulation of the human NKG2D ligand MICA by miRNAs (Stern-Ginossar et al., 2008; Yadav et al., 2009).
4. Discussion
Since NKG2D ligands can mediate tumor surveillance and influence tumor immunotherapy (Groh et al., 2002; Guerra et al., 2008; Smyth et al., 2005), there is strong rationale for elucidating the complex regulatory pathways that control NKG2D ligand expression in tumor cells (Nausch and Cerwenka, 2008). We sought to understand the regulation of the NKG2D ligand H60a by studying its genomic structure.
We generated a 44 kb contig of the H60a locus in 129/Sv mice and used this sequence to study the cis elements that regulate H60a. We believe that our contiguous sequence is valid since we were able to generate a 6-exon gene structure of H60a similar to previous findings (Samarakoon et al., 2009; Takada et al., 2008). When we compared this sequence to the publicly available C57BL/6 sequence, we found that the H60a promoter region is conserved, with roughly 20 kB of identical sequence between 129/Sv and C57BL/6 strains of mice. We also found that the H60a promoter of C57BL/6 mice is functional and leads to the generation of a truncated transcript.
Previously, we showed that IFNs down-regulated H60a transcript via a mechanism that required the transcription factor STAT1 (Signal Transducers and Activators of Transcription 1) (Bui et al., 2006a), but the cis elements mediating this regulation are not known. Our studies showed that IFNγ did not regulate H60a through the promoter region based on two observations. First, a reporter construct containing 527 bp of the H60a promoter did not exhibit responsiveness to IFNγ. Second, IFNγ did not regulate H60a transcript in C57BL/6 cells even though there is 20 kb of identical promoter sequence to 129/Sv cells, which respond to IFNγ. It is possible that the very low levels of H60a transcript in C57BL/6 cells prevent their responsiveness to IFNγ. Future studies will use chromatin immunoprecipitation assays to directly examine whether STAT1 binds the H60a promoter region in response to IFNγ treatment.
Recently, Pedersen et al. (Pedersen et al., 2007) found that IFNs induced a panel of miRNAs that possessed anti-viral properties. We have also published that miRNA-520b is induced by IFNγ and can regulate a human NKG2D ligand (Yadav et al., 2009). In this study, we found that IFNγ acted on the H60a 3′UTR and thus, we suggest that the regulation of H60a by IFNγ could occur via a similar mechanism. At this time, we do not know which IFNγ-inducible miRNA can regulate H60a. A preliminary analysis of the 3′UTR of H60a reveals dozens of potential binding sites for miRNAs (Supplementary Figure S1). Moreover, we found that inhibition of DICER led to significantly increased H60a 3′UTR activity, supporting a role for miRNAs in the regulation of H60a expression. The inhibition of DICER also led to increased levels of H60a protein, but this finding was not significant. Since inhibiting DICER would globally reduce miRNA levels, these results suggest that other miRNAs may participate in regulating H60a protein expression through the regulation of putative trans-acting inhibitory factors.
NKG2D ligands are often found constitutively expressed on primary tumor cells and tumor cell lines, but the level of expression is different depending on the cell line and the tissue type (Cerwenka et al., 2000; Diefenbach et al., 2000; Groh et al., 1999; Groh et al., 2002). Even within a single type of cell line, such as MCA sarcomas of the 129/Sv strain, the basal level of expression of H60a can range from minimal to moderate to very high levels (Bui et al., 2006a). It is not known what determines this basal level of H60a expression, although studies on other NKG2D ligands have implicated control by miRNA (Stern-Ginossar et al., 2008), DNA damage signals (Gasser et al., 2005), and epigenetic mechanisms (Andresen et al., 2007; Kato et al., 2007). Our studies suggest that miRNAs acting through the H60a 3′UTR can contribute to the level of constitutive expression of H60a, and therefore, can regulate how a tumor is recognized by NK cells. We did not examine whether the H60a promoter region can also contribute to the varying levels of H60a expression between cell lines. It will be important to elucidate the factors that control the constitutive expression of H60a on tumor cells, since this aspect of regulation can impact on whether a developing tumor is eradicated by immune cells bearing NKG2D. Our studies on the cis elements that regulate H60a will provide important inroads into this area of investigation.
Supplementary Material
Figure 8. Inhibition of DICER leads to increased H60a 3′UTR activity and surface protein expression.
The H60a-lo 129/Sv cell line f236 was transfected with siRNAs specific for DICER or control siRNA and (A) H60a 3′UTR activity was measured using a luciferase reporter assay or (B) H60a surface protein was measured by flow cytometry. Results are representative of 3 total experiments.
Acknowledgments
We thank Xiaoxia Wang, Deepak Yadav, and Timothy O’Sullivan for critical discussion and reading of the manuscript.
Role of Funding Source This work was supported by grants to J.D.B. from the American Cancer Society (ACS-IRG #70-002), a grant from the Cancer Research Coordinating Committee (6-444951-34384), the V Foundation Scholar Award, the Concern Foundation, and NIH-CA128893. The study sponsors had no role in the study design, collection, analysis, and interpretation of data. They had no role in the writing of the report or the decision to submit the paper for publication.
Abbreviations
- NKG2D
natural killer group 2D
- NK
natural killer
- H60a
histocompatibility antigen 60a
- MULT1
murine ULBP-like transcript
- RAE
retinoic acid early transcript
- MICA/B
MHC class I-like chain A/B
- IFN
interferon
- UTR
untranslated region
- qRT-PCR
quantitative real-time polymerase chain reaction
- miRNA
microRNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andresen L, Jensen H, Pedersen MT, Hansen KA, Skov S. Molecular regulation of MHC class I chain-related protein A expression after HDAC-inhibitor treatment of Jurkat T cells. J Immunol. 2007;179:8235–42. doi: 10.4049/jimmunol.179.12.8235. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Buchau AS, Morizane S, Trowbridge J, Schauber J, Kotol P, Bui JD, Gallo RL. The host defense peptide cathelicidin is required for NK cell-mediated suppression of tumor growth. J Immunol. 184:369–78. doi: 10.4049/jimmunol.0902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui JD, Carayannopoulos LN, Lanier LL, Yokoyama WM, Schreiber RD. IFN-dependent down-regulation of the NKG2D ligand H60 on tumors. J Immunol. 2006a;176:905–13. doi: 10.4049/jimmunol.176.2.905. [DOI] [PubMed] [Google Scholar]
- Bui JD, Uppaluri R, Hsieh CS, Schreiber RD. Comparative analysis of regulatory and effector T cells in progressively growing versus rejecting tumors of similar origins. Cancer Res. 2006b;66:7301–9. doi: 10.1158/0008-5472.CAN-06-0556. [DOI] [PubMed] [Google Scholar]
- Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, Lanier LL. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12:721–7. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol. 2001;1:41–9. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. 2000;1:119–26. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- Diefenbach A, Jensen ER, Jamieson AM, Raulet DH. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413:165–71. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–90. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R, Hobby P, Sutton B, Tigelaar RE, Hayday AC. Regulation of cutaneous malignancy by gammadelta T cells. Science. 2001;294:605–9. doi: 10.1126/science.1063916. [DOI] [PubMed] [Google Scholar]
- Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci U S A. 1999;96:6879–84. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–8. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, Knoblaugh S, Cado D, Greenberg NR, Raulet DH. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–80. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, Tanaka J, Sugita J, Toubai T, Miura Y, Ibata M, Syono Y, Ota S, Kondo T, Asaka M, Imamura M. Regulation of the expression of MHC class I-related chain A, B (MICA, MICB) via chromatin remodeling and its impact on the susceptibility of leukemic cells to the cytotoxicity of NKG2D-expressing cells. Leukemia. 2007;21:2103–8. doi: 10.1038/sj.leu.2404862. [DOI] [PubMed] [Google Scholar]
- Lodoen M, Ogasawara K, Hamerman JA, Arase H, Houchins JP, Mocarski ES, Lanier LL. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med. 2003;197:1245–53. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malarkannan S, Shih PP, Eden PA, Horng T, Zuberi AR, Christianson G, Roopenian D, Shastri N. The molecular and functional characterization of a dominant minor H antigen, H60. J Immunol. 1998;161:3501–9. [PubMed] [Google Scholar]
- Mistry AR, O’Callaghan CA. Regulation of ligands for the activating receptor NKG2D. Immunology. 2007;121:439–47. doi: 10.1111/j.1365-2567.2007.02652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene. 2008;27:5944–58. doi: 10.1038/onc.2008.272. [DOI] [PubMed] [Google Scholar]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–22. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–90. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- Samarakoon A, Chu H, Malarkannan S. Murine NKG2D ligands: “double, double toil and trouble”. Mol Immunol. 2009;46:1011–9. doi: 10.1016/j.molimm.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–11. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. NKG2D function protects the host from tumor initiation. J Exp Med. 2005;202:583–8. doi: 10.1084/jem.20050994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–73. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- Takada A, Yoshida S, Kajikawa M, Miyatake Y, Tomaru U, Sakai M, Chiba H, Maenaka K, Kohda D, Fugo K, Kasahara M. Two novel NKG2D ligands of the mouse H60 family with differential expression patterns and binding affinities to NKG2D. J Immunol. 2008;180:1678–85. doi: 10.4049/jimmunol.180.3.1678. [DOI] [PubMed] [Google Scholar]
- Uppaluri R, Sheehan KC, Wang L, Bui JD, Brotman JJ, Lu B, Gerard C, Hancock WW, Schreiber RD. Prolongation of cardiac and islet allograft survival by a blocking hamster anti-mouse CXCR3 monoclonal antibody. Transplantation. 2008;86:137–47. doi: 10.1097/TP.0b013e31817b8e4b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang MI, Guerra N, Raulet DH. Costimulation of dendritic epidermal gammadelta T cells by a new NKG2D ligand expressed specifically in the skin. J Immunol. 2009;182:4557–64. doi: 10.4049/jimmunol.0802439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav D, Ngolab J, Lim RS, Krishnamurthy S, Bui JD. Cutting edge: down-regulation of MHC class I-related chain A on tumor cells by IFN-gamma-induced microRNA. J Immunol. 2009;182:39–43. doi: 10.4049/jimmunol.182.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama WM. Now you see it, now you don’t! Nat Immunol. 2000;1:95–7. doi: 10.1038/77878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.