Abstract
Glucocorticoids regulate numerous physiological processes and are mainstays in the treatment of inflammation, autoimmune disease, and cancer. The traditional view that glucocorticoids act through a single glucocorticoid receptor (GR) protein has changed in recent years with the discovery of a large cohort of receptor subtypes arising from alternative processing of the GR gene. These isoforms differ in their expression, gene regulatory, and functional profiles. Post-translational modification of these proteins further expands GR diversity. Here, we discuss the origin and molecular properties of the GR isoforms and their contribution to the sensitivity and specificity of the glucocorticoid response.
Keywords: Gene Regulation, Nuclear Receptors, Post-translational Modification, RNA Processing, Transcription Factors, Alternative Splicing, Alternative Translation Initiation, Glucocorticoid Receptor, Glucocorticoids, Protein Isoforms
Introduction
Glucocorticoids are primary stress hormones that function to maintain homeostasis. They are synthesized and released by the adrenal cortex following stress-induced activation of the hypothalamic-pituitary-adrenal axis and affect nearly every organ and tissue in the body. Named for their effects on glucose metabolism, glucocorticoids regulate a plethora of biological processes, including immune function, skeletal growth, reproduction, cognition, behavior, and cell proliferation and survival (1, 2). Because of their powerful anti-inflammatory and immunosuppressive actions, synthetic glucocorticoids are widely prescribed for the treatment of acute and chronic inflammatory diseases, autoimmune diseases, organ transplant rejection, and malignancies of the lymphoid system (3).
The cellular response to glucocorticoids is not uniform, exhibiting profound variability in both magnitude and specificity of action (4–6). Whereas glucocorticoids induce the killing of lymphocytes, they have protective effects on cells of the endometrium, ovarian follicle, liver, and mammary epithelium (7). The sensitivity to glucocorticoids differs not only among individuals but also within tissues of the same individual and even within the same cell during the cell cycle (8, 9). Moreover, tissue-specific glucocorticoid resistance frequently develops in patients on chronic glucocorticoid therapy. The therapeutic benefit of glucocorticoids is also limited by severe side effects such as osteoporosis, abdominal obesity, glaucoma, growth retardation in children, and hypertension (3, 10, 11). Elucidating the molecular mechanisms governing the diversity in the cellular response to glucocorticoids should facilitate the development of new glucocorticoids with improved therapeutic indices.
Both the physiological and pharmacological actions of glucocorticoids are mediated by the glucocorticoid receptor (GR2; NR3C1), a member of the nuclear receptor superfamily of ligand-dependent transcription factors (12). Consistent with the pleiotropic effects of glucocorticoids, the receptor is ubiquitously expressed and necessary for life after birth (13). GR is derived from a single gene, and the prevailing assumption since its cloning in 1985 has been that a single receptor protein is responsible for the diverse actions of glucocorticoids. This simple “one gene-one receptor” paradigm has been challenged by recent studies revealing a large cohort of functionally distinct GR subtypes that arise from alternative processing of the GR gene. In turn, these receptor isoforms are subject to various post-translational modifications that further modulate their activity. Such regulatory mechanisms serve to expand the human proteome to an enormous size, perhaps several orders of magnitude greater than the encoding genome of ∼25,000 genes (14, 15). Accordingly, the sensitivity and specificity of the glucocorticoid response may be determined by the expressed complement and composite actions of the individual GR isoforms. The following minireview focuses on the molecular heterogeneity of GR as a mechanism for generating diversity in glucocorticoid signaling.
GR Signaling Pathway
Like other members of the nuclear receptor superfamily, GR is a modular protein composed of an N-terminal transactivation domain (NTD), a central DNA-binding domain (DBD), and a C-terminal ligand-binding domain (LBD) (Fig. 1) (16). A flexible region of the molecule, termed the hinge region, separates the DBD and LBD. The DBD is the most conserved region across the superfamily and contains two zinc finger motifs that recognize and bind target DNA sequences, termed glucocorticoid-responsive elements (GREs). The NTD contains a strong transcriptional activation function (AF1) that binds various coregulators and components of the basal transcription machinery. The LBD, consisting of 12 α-helices and four β-sheets, forms a hydrophobic pocket for binding glucocorticoids (17). A second activation function (AF2) that interacts with coregulators in a ligand-dependent manner is embedded in the LBD. Two nuclear localization signals, NL1 and NL2, are located at the DBD/hinge region junction and within the LBD, respectively.
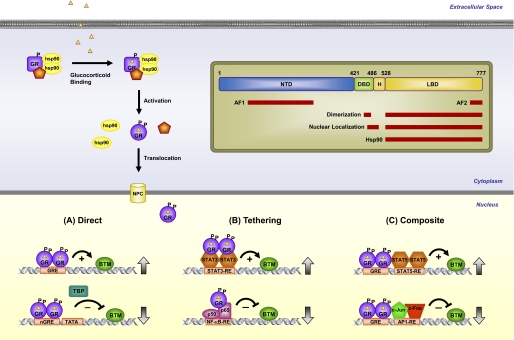
FIGURE 1.
GR signaling pathway. Upon binding glucocorticoids, cytoplasmic GR undergoes a change in conformation (activation), becomes hyperphosphorylated (P), dissociates from accessory proteins, and translocates into the nucleus, where it regulates gene expression. GR enhances or represses transcription of target genes by direct GRE binding (A), by tethering itself to other transcription factors apart from DNA binding (B), or in a composite manner by both direct GRE binding and interactions with transcription factors bound to neighboring sites (C). Inset, GR is composed of an NTD, a DBD, a hinge region (H), and an LBD. Regions involved in transcriptional activation (AF1 and AF2), dimerization, nuclear localization, and chaperone hsp90 binding are indicated. Position numbers are for the human GR. NPC, nuclear pore complex; BTM, basal transcription machinery; TBP, TATA-binding protein; nGRE, negative GRE; RE, response element.
Unliganded GR is found primarily in the cytoplasm of cells as part of a large multiprotein complex that includes various chaperone proteins such as hsp90 (18, 19). Binding of glucocorticoids triggers a conformational change in GR resulting in the dissociation of the heterocomplex, exposure of the nuclear localization signals, and importin-mediated nuclear entry (Fig. 1) (20). The release of Src kinase from the GR heterocomplex and its subsequent phosphorylation of lipocortin-1 are thought to mediate some of the rapid non-genomic effects of glucocorticoids (21). Once in the nucleus, GR dimerizes and binds directly to GREs to stimulate the expression of target genes (Fig. 1A) (22, 23). Alternatively, the receptor associates with less well defined negative GREs to suppress gene activation (24). When bound to the GRE, conformational changes ensue in the receptor that lead to the coordinated recruitment of coregulators and chromatin-remodeling complexes that influence the activity of RNA polymerase II and modulate gene transcription rates (25–27). Both the nature of the bound ligand and the GRE sequence itself can dictate the specific assembly and function of the cofactors through alterations in the receptor structure (28, 29). The receptor interacts only briefly with target genes, rapidly cycling on and off the GRE every few seconds, presumably allowing GR to sample a large number of potential binding sites and interacting proteins (30, 31).
GR can also regulate gene expression by physically associating with other transcription factors. For example, the interaction of GR with the proinflammatory transcription factors AP-1 (activator protein-1) and NF-κB inhibits their activity and accounts for the major anti-inflammatory and immunosuppressive effects of glucocorticoids (32, 33). This repression can be accomplished on some genes by GR tethering itself to these DNA-bound proteins (Fig. 1B), but others require GR to act in a composite manner and bind both a GRE and the transcription factor on an adjacent site (Fig. 1C). In contrast to these inhibitory effects, the physical association of GR with members of the STAT (signal transducer and activator of transcription) family, either apart from or in conjunction with GRE binding, can enhance their transcriptional activity on certain genes (Fig. 1, B and C) (34). Operating through these diverse mechanisms for stimulating or suppressing gene transcription, GR has been shown by microarray analysis to regulate up to 10–20% of the human genome in different cell types (35, 36). Although differences in ligand bioavailability, GR expression levels, and cofactor availability contribute to the tissue-specific effects of glucocorticoids, recent studies have demonstrated that GR heterogeneity may also play an important role in determining the glucocorticoid signaling profile.
GR Isoforms Generated by Alternative Splicing
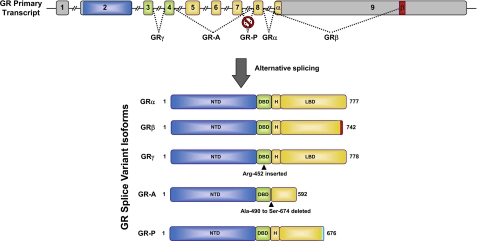
The human GR gene is located on chromosome 5q31–32 and is composed of nine exons. Alternative splicing near the end of the primary transcript generates two receptor isoforms, termed GRα and GRβ, which differ at their extreme C termini (Fig. 2) (37, 38). The classic GRα protein results from the end of exon 8 being joined to the beginning of exon 9. The splice variant GRβ utilizes an alternative splice acceptor site such that the end of exon 8 is joined to downstream sequences in exon 9. The resulting proteins are identical through amino acid 727 but then diverge, with GRα containing an additional 50 amino acids and GRβ an additional non-homologous 15 amino acids. The GRα-specific sequences encode helices 11 and 12 of the LBD, a region crucial not only for glucocorticoid binding but also for coregulator recruitment by AF2. The unique GRβ sequence is predicted to be largely disordered (39), and this structural change confers several distinct properties to the isoform (37, 38). GRβ does not bind glucocorticoids, resides constitutively in the nucleus of cells, and does not directly regulate glucocorticoid-responsive reporter genes. However, when coexpressed with GRα, the splice variant functions as a dominant-negative inhibitor of GRα on genes both positively and negatively regulated by glucocorticoids. Various mechanisms, including competition for GRE binding, competition for transcriptional coregulators, and formation of inactive GRα/GRβ heterodimers, have been proposed to underlie the antagonism.
FIGURE 2.
GR isoforms generated by alternative splicing. The human GR primary transcript is composed of nine exons, with exon 2 encoding most of the NTD, exons 3 and 4 encoding the DBD, and exons 5–9 encoding the hinge region (H) and LBD. The classic GRα protein results from splicing of exon 8 to the beginning of exon 9. GRβ is produced from an alternative splice acceptor site that links the end of exon 8 to downstream sequences in exon 9, encoding a variant with a unique 15-amino acid C terminus (positions 728–742). GRγ is generated by an alternative splice donor site in the intronic sequence separating exons 3 and 4, resulting in a protein with an arginine insertion (Arg-452) between the two zinc fingers of the DBD. GR-A is produced from alternative splicing that joins exon 4 to exon 8, deleting the proximal 185 amino acids of the LBD (Ala-490–Ser-674) encoded by exons 5–7. GR-P is formed by a failure to splice exon 7 to exon 8. The retained intronic sequence introduces a stop codon, resulting in a truncated receptor mutant missing the distal half of the LBD.
The ability of GRβ to inhibit the transcriptional activity of GRα suggests that alterations in the expression level of the splice variant will modulate cellular sensitivity to glucocorticoids. GRβ is widely expressed but generally found at much lower levels than GRα. However, in certain cell types such as neutrophils and specific epithelial cells, GRβ is abundant. Moreover, exposure of cells to proinflammatory cytokines and other immune activators can selectively increase the expression of GRβ, leading to glucocorticoid resistance (40–42). Conversely, agents that increase GRα expression at the expense of GRβ sensitize cells to glucocorticoids (43). Some patients with glucocorticoid-resistant forms of asthma, rheumatoid arthritis, ulcerative colitis, nasal polyposis, systemic lupus erythematous, acute lymphoblastic leukemia, and chronic lymphocytic leukemia present with elevated levels of GRβ (38). The molecular basis for the selective increase in GRβ is poorly understood, but the involvement of the splicing factor SRp30c (serine/arginine-rich protein p30c) has been implicated in several studies (44, 45). Elevated levels of GRβ also result from a naturally occurring polymorphism (A3669G) in the 3′-untranslated region of the GRβ mRNA that disrupts an mRNA destabilization motif (AUUUA) (46, 47). The A3669G allele has been associated with reduced central obesity in women and a more favorable lipid profile in men (48), suggesting that some of the undesirable effects of GRα on fat distribution and lipid metabolism may be antagonized by a rise in GRβ. However, by attenuating the beneficial immunosuppressive and anti-inflammatory actions of GRα, the increase in GRβ may also underlie the elevated risk of A3669G carriers for pathologies with known inflammatory components such as autoimmune disease, myocardial infarction, and coronary artery disease (49).
A broader role for GRβ in cell signaling and physiology has emerged in recent years. Global gene expression analyses have shown that GRβ can directly induce and repress a large number of genes not controlled by GRα (50, 51). The ability of GRβ to constitutively induce histone deacetylation may account for its repression of certain genes (52, 53). GRβ was also found to bind the glucocorticoid antagonist mifepristone (RU486), and this binding silenced its activity on many of the regulated genes (51). Collectively, these data suggest that GRβ has an intrinsic gene regulatory function that may contribute to alterations in glucocorticoid signaling independent of GRα antagonism. Another provocative finding has been the discovery of GRβ in several animal species (54, 55). This result was unanticipated because the alternative splice acceptor site utilized in humans for generating GRβ is only partially conserved across species and is absent from rodents (55, 56). GRβ isoforms in zebrafish (zGRβ) and mouse (mGRβ) arise from a distinct mechanism that employs alternative splice donor sites in the intron separating exons 8 and 9. The resulting GRβ isoforms are strikingly similar in structure and functionality to human GRβ. The point of divergence from GRα is at the end of exon 8, with mGRβ and zGRβ containing an additional 15- and 50-amino acid C terminus, respectively. In addition, mGRβ and zGRβ exhibit ubiquitous expression, nuclear localization, inability to bind glucocorticoid agonists, and antagonism of GRα. Genetic manipulation of GRβ in these animal models may shed new light on the biological importance of this intriguing splice variant.
Several additional GR isoforms arise from alternative splicing and impact glucocorticoid signaling (Fig. 2). GRγ contains an insertion of a single arginine residue between the two zinc fingers of the DBD and originates from the use of an alternative splice donor site in the intron separating exons 3 and 4 (57). This widely expressed isoform binds glucocorticoids and DNA with a capacity similar to GRα, but it is compromised in its ability to stimulate glucocorticoid-responsive reporters and exhibits a transcriptional profile distinct from GRα on a subset of commonly regulated genes (28, 57). GRγ expression is associated with glucocorticoid resistance in small cell lung carcinoma cells, corticotroph adenomas, and childhood acute lymphoblastic leukemia (57–59). Two non-hormone-binding GR splice variants missing large regions of the LBD were initially discovered in glucocorticoid-resistant multiple myeloma cells (60). GR-A, derived from alternative splicing linking the end of exon 4 to the beginning of exon 8, is missing the N-terminal half of the LBD encoded by exons 5–7. GR-P is missing exons 8 and 9, which encode the C-terminal half of the LBD due to a failure to splice at the exon 7/8 boundary. Although little is known about GR-A, GR-P is detected in many tissues and appears to be the predominant receptor variant in several glucocorticoid-insensitive cancer cells (61–63). Depending on the cell type, GR-P has been shown to either repress or stimulate the transcriptional activity of GRα on glucocorticoid-responsive reporter genes (61).
GR Isoforms Generated by Alternative Translation Initiation
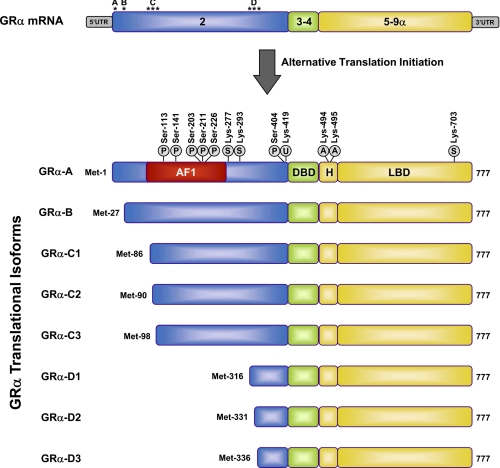
Although the generation of distinct GR isoforms by alternative splicing has been recognized for many years, only recently was it demonstrated that an additional cohort of receptor proteins is produced by alternative translation initiation from a single GR mRNA species (64, 65). Sequence alignment of human, monkey, rat, and mouse GRs revealed eight conserved AUG start codons derived from exon 2, and these were shown to produce eight GRα isoforms with progressively shorter NTDs (GRα-A, GRα-B, GRα-C1, GRα-C2, GRα-C3, GRα-D1, GRα-D2, and GRα-D3) (Fig. 3). Both ribosomal leaky scanning and ribosomal shunting mechanisms are involved in the generation of the GRα subtypes with truncated N termini (64). GRα-A is the classic full-length 777-amino acid human receptor that is generated from the first initiator codon. Each of the GR splice variants (GRβ, GRγ, GR-A, and GR-P) would also be expected to give rise to a similar complement of translational isoforms.
FIGURE 3.
GRα isoforms generated by alternative translation initiation and sites of post-translational modification. Initiation of translation from eight different AUG start codons in a single GRα mRNA generates receptor isoforms with progressively shorter NTDs. Approximate locations of the AUG start codons in the exon 2 sequences of the GRα mRNA are designated by asterisks. The initiator methionines, AF1 region (amino acids 77–262), and sites of post-translational modifications (phosphorylation (P), sumoylation (S), ubiquitination (U), and acetylation (A)) are indicated for the human GRα isoforms. H, hinge region.
The GRα translational isoforms, distinguished only by the length of their N termini, possess both common and unique properties. No significant differences have been observed in their affinity for glucocorticoids or their capacity to interact with GREs following ligand activation (36). However, the subcellular distribution of the isoforms differs, with GRα-D residing constitutively in the nucleus of cells (64). This result suggests that sequences in the NTD may play a previously unappreciated role in nuclear translocation, nuclear export, and/or cytoplasmic retention of the receptor. The nuclear localized GRα-D isoforms also exhibit constitutive binding to certain GRE-containing promoters (36). The most striking difference among the translational isoforms is, however, in their gene regulatory profiles (36, 64). The GRα-C isoforms are the most active, and the GRα-D subtypes are the most deficient in their ability to enhance transcription of glucocorticoid-responsive reporter and endogenous genes. These isoform-selective transcriptional effects appear to be due to differences among the subtypes in the recruitment of coregulators and RNA polymerase II to target gene promoters. In global gene expression assays performed on human osteosarcoma cells selectively expressing the individual isoforms, the GRα subtypes were found to regulate both common and unique sets of genes (36). Remarkably, the majority of glucocorticoid-responsive genes were selectively regulated by different GRα isoforms, with <10% being commonly regulated by all the subtypes. In addition, the GRα-D isoform, which is missing the entire AF1 region and almost 80% of the NTD, still regulated ∼1800 genes in response to hormone. These isoform-unique gene regulatory profiles produced functional differences in glucocorticoid-induced apoptosis. Cells expressing GRα-C were the most sensitive to the cell-killing effects of glucocorticoids, whereas cells expressing GRα-D were the most resistant.
With each GRα translational isoform possessing a unique genetic fingerprint, the cellular response to glucocorticoids may be determined by the expressed complement of receptor subtypes. The receptor isoforms show a widespread tissue distribution, but their relative levels differ both within and between tissues (64). In rodents, for example, the GRα-C isoforms are found at low levels in the liver but at high levels in the pancreas, lung, and colon. The GRα-D subtypes are prevalent in the spleen and bladder but are expressed at low levels in heart and pancreas. Within the same tissue, GRα-B is more abundant than GRα-A in the thymus and colon but is less abundant in other tissues. In human cell lines examined so far, GRα-A and GRα-B are the predominant isoforms, with the exception of trabecular meshwork cells that preferentially express the GRα-C and GRα-D proteins (66). Recent studies have also demonstrated signal- and time-dependent changes in the cellular complement of GRα translational isoforms. Treatment of differentiated murine skeletal muscle cells with a selective estrogen-related receptor β/γ agonist induced a selective increase in the GRα-D isoforms (67). Moreover, the relative levels of the GRα subtypes expressed in the human brain were found to change during development and the aging process (68). The molecular mechanisms governing the expressed complement of translational isoforms are poorly understood. Polymorphisms in the GR gene, as well as heterogeneity in the 5′-untranslated region of the GRα mRNA, have been reported to influence the efficiency of alternative start codon usage (69, 70). Additionally, changes in the relative levels of the GRα subtypes may be achieved by post-translational modifications that differentially affect receptor half-life.
Other nuclear receptors closely related to GR also have the potential for generating multiple translational isoforms from internal AUG start codons. The mineralocorticoid receptor contains 10 methionine residues in its NTD that are conserved across human, rat, and mouse; the androgen receptor (AR) contains 7; the progesterone receptor has 2; and estrogen receptor (ER) α and ERβ have 6 and 3, respectively (64). Several of these internal AUG codons have been reported to function as translational start sites, including Met-15, which gives rise to the B isoform of the mineralocorticoid receptor (71); Met-174, which produces ERα-46 (72); and Met-188, which gives rise to a shorter isoform of AR, termed AR-A (73). These findings suggest that alternative translation initiation may be a common mechanism by which steroid receptors mediate diverse signaling responses.
Post-translational Modification of GR Isoforms
Each individual GR isoform is subject to various post-translational modifications that further modulate receptor activity and expand the functional pool of receptor proteins available for glucocorticoid signaling (Fig. 3). Phosphorylation was the first identified covalent modification of GR and has been the focus of most studies (74–76). At least 6 serine residues (Ser-113, Ser-141, Ser-203, Ser-211, Ser-226, and Ser-404) are phosphorylated on human GRα, and these sites are conserved in mouse and rat. The receptor displays a basal level of phosphorylation and becomes hyperphosphorylated upon binding glucocorticoids, with the extent of phosphorylation dependent on the nature of the bound ligand (77, 78). Interdependence of several of the phosphorylation sites suggests that the phosphorylation state of an individual receptor isoform may depend on prior phosphorylation/dephosphorylation events. The major kinases that phosphorylate GRα include MAPKs, cyclin-dependent kinases, and GSK-3 (glycogen synthase kinase-3).
Phosphorylation of GRα changes its transcriptional activity, often in a gene-selective manner. Early studies showed that phosphorylation-deficient GRα mutants were compromised in their ability to activate reporter genes in a promoter-dependent fashion (79). Subsequently, it was reported that phosphorylation of Ser-211 correlated with the transcriptionally active form of GRα, whereas phosphorylation of Ser-226 impaired its signaling capability (78, 80, 81). Phosphorylation of Ser-211 appears to be necessary for glucocorticoid-induced apoptosis of lymphoid cells, suggesting that a deficiency in this phosphorylation event may be a mechanism by which lymphocytes become resistant to glucocorticoids (82, 83). Phosphorylation of Ser-211 also provides GRα with the means to cross-talk with other signaling pathways. Estrogen treatment of several breast cancer cell lines promotes the dephosphorylation of Ser-211 via enhanced expression of protein phosphatase PP5 and leads to suppressed GRα activity on several target genes involved in growth inhibition (84). Glucocorticoid-dependent phosphorylation of Ser-404 also has major consequences on GRα signaling, impairing both activation and repression of target genes (85). Cells expressing a GRα mutant incapable of Ser-404 phosphorylation show a redirection of the global transcriptional response to hormone that includes activation of distinct signaling pathways. Differences in cofactor recruitment have been implicated in these transcriptional effects, consistent with the sites of phosphorylation being located within or nearby the AF1 domain (Fig. 3). Phosphorylation of Ser-211 enhances the interaction of GRα with the coactivator MED14 (81), whereas phosphorylation of Ser-404 diminishes its interaction with the coactivator p300/CBP (cAMP-responsive element-binding protein-binding protein) and the p65 subunit of NF-κB (85).
Several other properties of GRα are affected by receptor phosphorylation leading to alterations in glucocorticoid signaling. For example, phosphorylation of GRα influences the expression level of the receptor protein by modulating its half-life. The tumor suppressor gene TSG101 interacts preferentially with non-phosphorylated GRα to protect the ligand-free receptor from proteasome-dependent degradation (86). Conversely, phosphorylation of the receptor in response to glucocorticoids appears to promote GRα decay, as phosphorylation-deficient mutants display a marked ligand-dependent stabilization (79). Phosphorylation also modulates the cellular trafficking of the receptor. GRα phosphorylated on Ser-203 is preferentially retained in the cytoplasm of cells and, accordingly, is poorly recruited to glucocorticoid-responsive target genes (78, 80). Similarly, the diminished signaling of GRα phosphorylated on Ser-226 or Ser-404 may be due in part to its enhanced nucleocytoplasmic transport (85, 87).
GRα also serves as a substrate for a variety of other post-translational modifications (Fig. 3). The receptor is ubiquitinated at a conserved lysine residue located in a PEST degradation motif at the end of the NTD, and this modification targets the receptor for degradation by the proteasome (88, 89). Mutation of this lysine residue blocks ligand-dependent down-regulation of GRα and enhances its transcriptional activity on reporter genes (89). In addition, an E3 ubiquitin ligase has been identified for GRα, and alterations in the expression of this enzyme modulate receptor levels and cellular responsiveness to glucocorticoids (90). GRα is also a substrate for sumoylation, in which SUMO (small ubiquitin-related modifier) peptides are covalently attached to the receptor at specific lysine residues (Lys-277, Lys-293, and Lys-703). Sumoylation of GRα dramatically promotes its degradation and inhibits the transcriptional activity of the receptor in a promoter-dependent fashion through the recruitment of corepressors (91–97). Furthermore, recent reports have demonstrated that GRα is acetylated at Lys-494 and Lys-495 in response to glucocorticoids, and this modification impairs its antagonism of NF-κB (98). Clearly, multiple aspects of GRα function can be regulated by various post-translational modifications, providing cells with additional receptor heterogeneity for controlling the glucocorticoid response and integrating GRα action with other signaling pathways. To what extent the various splicing and translational isoforms of GR are subject to and regulated by these modifications remains to be investigated.
Summary and Perspective
The traditional view that glucocorticoids exert their diverse effects through one receptor protein has changed dramatically over the last 2 decades with the discovery of multiple GR isoforms arising from the single GR gene. GR subtypes with unique expression and gene regulatory profiles are generated by alternative splicing of the nascent transcript, alternative translation initiation of the mature mRNA, and post-translational modifications of the receptor protein. The capacity of a cell to generate dozens of GR isoforms that control specific sets of genes and/or differentially regulate common sets provides enormous potential for signaling diversity. Further contributing to the tissue- and cell-specific effects of glucocorticoids is the potential for these isoforms to heterodimerize with each other and cross-talk with other signaling molecules. A critical goal of future studies will be to use genetic approaches to assess the contribution an individual GR isoform makes to the actions of glucocorticoids in the whole animal. Additionally, it will be important to determine whether changes in the cellular complement of GR isoforms underlie pathologies characterized by glucocorticoid resistance and/or the severe side effects that accompany glucocorticoid treatment. Finally, future studies should look to identify the operative factors that govern the production of GR splice variants and translational isoforms. A greater understanding of the role that GR heterogeneity plays in the cellular response to glucocorticoids should aid in the development of safer and more effective glucocorticoid therapies.
Supplementary Material
This work was supported, in whole or in part, by the NIEHS Intramural Research Program of the National Institutes of Health. This is the fourth article in the Thematic Minireview Series on Nuclear Receptors in Biology and Diseases. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- GR
- glucocorticoid receptor
- NTD
- N-terminal transactivation domain
- DBD
- DNA-binding domain
- LBD
- ligand-binding domain
- GRE
- glucocorticoid-responsive element
- zGRβ
- zebrafish GRβ
- mGRβ
- mouse GRβ
- AR
- androgen receptor
- ER
- estrogen receptor.
REFERENCES
- 1. Barnes P. J. (1998) Clin. Sci. 94, 557–572 [DOI] [PubMed] [Google Scholar]
- 2. Sapolsky R. M., Romero L. M., Munck A. U. (2000) Endocr. Rev. 21, 55–89 [DOI] [PubMed] [Google Scholar]
- 3. Rhen T., Cidlowski J. A. (2005) N. Engl. J. Med. 353, 1711–1723 [DOI] [PubMed] [Google Scholar]
- 4. Chrousos G. P., Kino T. (2005) Sci. STKE 2005, pe48. [DOI] [PubMed] [Google Scholar]
- 5. Kino T., De Martino M. U., Charmandari E., Mirani M., Chrousos G. P. (2003) J. Steroid Biochem. Mol. Biol. 85, 457–467 [DOI] [PubMed] [Google Scholar]
- 6. Lamberts S. W., Huizenga A. T., de Lange P., de Jong F. H., Koper J. W. (1996) Steroids 61, 157–160 [DOI] [PubMed] [Google Scholar]
- 7. Viegas L. R., Hoijman E., Beato M., Pecci A. (2008) J. Steroid Biochem. Mol. Biol. 109, 273–278 [DOI] [PubMed] [Google Scholar]
- 8. Gorovits R., Ben-Dror I., Fox L. E., Westphal H. M., Vardimon L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4786–4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsu S. C., DeFranco D. B. (1995) J. Biol. Chem. 270, 3359–3364 [DOI] [PubMed] [Google Scholar]
- 10. Miner J. N., Hong M. H., Negro-Vilar A. (2005) Expert Opin. Investig. Drugs 14, 1527–1545 [DOI] [PubMed] [Google Scholar]
- 11. Schäcke H., Döcke W. D., Asadullah K. (2002) Pharmacol. Ther. 96, 23–43 [DOI] [PubMed] [Google Scholar]
- 12. Evans R. M. (1988) Science 240, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cole T. J., Blendy J. A., Monaghan A. P., Krieglstein K., Schmid W., Aguzzi A., Fantuzzi G., Hummler E., Unsicker K., Schütz G. (1995) Genes. Dev. 9, 1608–1621 [DOI] [PubMed] [Google Scholar]
- 14. Nilsen T. W., Graveley B. R. (2010) Nature 463, 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Walsh C. T., Garneau-Tsodikova S., Gatto G. J., Jr. (2005) Angew. Chem. Int. Ed. Engl. 44, 7342–7372 [DOI] [PubMed] [Google Scholar]
- 16. Kumar R., Thompson E. B. (2005) J. Steroid Biochem. Mol. Biol. 94, 383–394 [DOI] [PubMed] [Google Scholar]
- 17. Bledsoe R. K., Montana V. G., Stanley T. B., Delves C. J., Apolito C. J., McKee D. D., Consler T. G., Parks D. J., Stewart E. L., Willson T. M., Lambert M. H., Moore J. T., Pearce K. H., Xu H. E. (2002) Cell 110, 93–105 [DOI] [PubMed] [Google Scholar]
- 18. Grad I., Picard D. (2007) Mol. Cell. Endocrinol. 275, 2–12 [DOI] [PubMed] [Google Scholar]
- 19. Pratt W. B., Toft D. O. (1997) Endocr. Rev. 18, 306–360 [DOI] [PubMed] [Google Scholar]
- 20. Freedman N. D., Yamamoto K. R. (2004) Mol. Biol. Cell 15, 2276–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song I. H., Buttgereit F. (2006) Mol. Cell. Endocrinol. 246, 142–146 [DOI] [PubMed] [Google Scholar]
- 22. Beato M. (1989) Cell 56, 335–344 [DOI] [PubMed] [Google Scholar]
- 23. Freedman L. P. (1992) Endocr. Rev. 13, 129–145 [DOI] [PubMed] [Google Scholar]
- 24. Dostert A., Heinzel T. (2004) Curr. Pharm. Des. 10, 2807–2816 [DOI] [PubMed] [Google Scholar]
- 25. Lonard D. M., O'Malley B. W. (2005) Trends Biochem. Sci. 30, 126–132 [DOI] [PubMed] [Google Scholar]
- 26. Jenkins B. D., Pullen C. B., Darimont B. D. (2001) Trends Endocrinol. Metab. 12, 122–126 [DOI] [PubMed] [Google Scholar]
- 27. Rosenfeld M. G., Glass C. K. (2001) J. Biol. Chem. 276, 36865–36868 [DOI] [PubMed] [Google Scholar]
- 28. Meijsing S. H., Pufall M. A., So A. Y., Bates D. L., Chen L., Yamamoto K. R. (2009) Science 324, 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ronacher K., Hadley K., Avenant C., Stubsrud E., Simons S. S., Jr., Louw A., Hapgood J. P. (2009) Mol. Cell. Endocrinol. 299, 219–231 [DOI] [PubMed] [Google Scholar]
- 30. McNally J. G., Müller W. G., Walker D., Wolford R., Hager G. L. (2000) Science 287, 1262–1265 [DOI] [PubMed] [Google Scholar]
- 31. Stavreva D. A., Müller W. G., Hager G. L., Smith C. L., McNally J. G. (2004) Mol. Cell. Biol. 24, 2682–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Necela B. M., Cidlowski J. A. (2004) Proc. Am. Thorac. Soc. 1, 239–246 [DOI] [PubMed] [Google Scholar]
- 33. Newton R., Holden N. S. (2007) Mol. Pharmacol. 72, 799–809 [DOI] [PubMed] [Google Scholar]
- 34. Rogatsky I., Ivashkiv L. B. (2006) Tissue Antigens 68, 1–12 [DOI] [PubMed] [Google Scholar]
- 35. Galon J., Franchimont D., Hiroi N., Frey G., Boettner A., Ehrhart-Bornstein M., O'Shea J. J., Chrousos G. P., Bornstein S. R. (2002) FASEB J. 16, 61–71 [DOI] [PubMed] [Google Scholar]
- 36. Lu N. Z., Collins J. B., Grissom S. F., Cidlowski J. A. (2007) Mol. Cell. Biol. 27, 7143–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kino T., Su Y. A., Chrousos G. P. (2009) Cell. Mol. Life Sci. 66, 3435–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lewis-Tuffin L. J., Cidlowski J. A. (2006) Ann. N.Y. Acad. Sci. 1069, 1–9 [DOI] [PubMed] [Google Scholar]
- 39. Yudt M. R., Jewell C. M., Bienstock R. J., Cidlowski J. A. (2003) Mol. Cell. Biol. 23, 4319–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hauk P. J., Hamid Q. A., Chrousos G. P., Leung D. Y. (2000) J. Allergy Clin. Immunol. 105, 782–787 [DOI] [PubMed] [Google Scholar]
- 41. Tliba O., Cidlowski J. A., Amrani Y. (2006) Mol. Pharmacol. 69, 588–596 [DOI] [PubMed] [Google Scholar]
- 42. Webster J. C., Oakley R. H., Jewell C. M., Cidlowski J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6865–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goecke I. A., Alvarez C., Henríquez J., Salas K., Molina M. L., Ferreira A., Gatica H. (2007) Mol. Immunol. 44, 2115–2123 [DOI] [PubMed] [Google Scholar]
- 44. Xu Q., Leung D. Y., Kisich K. O. (2003) J. Biol. Chem. 278, 27112–27118 [DOI] [PubMed] [Google Scholar]
- 45. Zhu J., Gong J. Y., Goodman O. B., Jr., Cartegni L., Nanus D. M., Shen R. (2007) Biochim. Biophys. Acta 1773, 1087–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Derijk R. H., Schaaf M. J., Turner G., Datson N. A., Vreugdenhil E., Cidlowski J., de Kloet E. R., Emery P., Sternberg E. M., Detera-Wadleigh S. D. (2001) J. Rheumatol. 28, 2383–2388 [PubMed] [Google Scholar]
- 47. Schaaf M. J., Cidlowski J. A. (2002) Steroids 67, 627–636 [DOI] [PubMed] [Google Scholar]
- 48. Syed A. A., Irving J. A., Redfern C. P., Hall A. G., Unwin N. C., White M., Bhopal R. S., Weaver J. U. (2006) Obesity 14, 759–764 [DOI] [PubMed] [Google Scholar]
- 49. van den Akker E. L., Koper J. W., van Rossum E. F., Dekker M. J., Russcher H., de Jong F. H., Uitterlinden A. G., Hofman A., Pols H. A., Witteman J. C., Lamberts S. W. (2008) Arch. Intern. Med. 168, 33–39 [DOI] [PubMed] [Google Scholar]
- 50. Kino T., Manoli I., Kelkar S., Wang Y., Su Y. A., Chrousos G. P. (2009) Biochem. Biophys. Res. Commun. 381, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lewis-Tuffin L. J., Jewell C. M., Bienstock R. J., Collins J. B., Cidlowski J. A. (2007) Mol. Cell. Biol. 27, 2266–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kelly A., Bowen H., Jee Y. K., Mahfiche N., Soh C., Lee T., Hawrylowicz C., Lavender P. (2008) J. Allergy Clin. Immunol. 121, 203–208.e1 [DOI] [PubMed] [Google Scholar]
- 53. Kim S. H., Kim D. H., Lavender P., Seo J. H., Kim Y. S., Park J. S., Kwak S. J., Jee Y. K. (2009) Exp. Mol. Med. 41, 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hinds T. D., Jr., Ramakrishnan S., Cash H. A., Stechschulte L. A., Heinrich G., Najjar S. M., Sanchez E. R. (2010) Mol. Endocrinol. 24, 1715–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schaaf M. J., Champagne D., van Laanen I. H., van Wijk D. C., Meijer A. H., Meijer O. C., Spaink H. P., Richardson M. K. (2008) Endocrinology 149, 1591–1599 [DOI] [PubMed] [Google Scholar]
- 56. Otto C., Reichardt H. M., Schütz G. (1997) J. Biol. Chem. 272, 26665–26668 [DOI] [PubMed] [Google Scholar]
- 57. Ray D. W., Davis J. R., White A., Clark A. J. (1996) Cancer Res. 56, 3276–3280 [PubMed] [Google Scholar]
- 58. Beger C., Gerdes K., Lauten M., Tissing W. J., Fernandez-Munoz I., Schrappe M., Welte K. (2003) Br. J. Haematol. 122, 245–252 [DOI] [PubMed] [Google Scholar]
- 59. Rivers C., Levy A., Hancock J., Lightman S., Norman M. (1999) J. Clin. Endocrinol. Metab. 84, 4283–4286 [DOI] [PubMed] [Google Scholar]
- 60. Moalli P. A., Pillay S., Krett N. L., Rosen S. T. (1993) Cancer Res. 53, 3877–3879 [PubMed] [Google Scholar]
- 61. de Lange P., Segeren C. M., Koper J. W., Wiemer E., Sonneveld P., Brinkmann A. O., White A., Brogan I. J., de Jong F. H., Lamberts S. W. (2001) Cancer Res. 61, 3937–3941 [PubMed] [Google Scholar]
- 62. Gaitan D., DeBold C. R., Turney M. K., Zhou P., Orth D. N., Kovacs W. J. (1995) Mol. Endocrinol. 9, 1193–1201 [DOI] [PubMed] [Google Scholar]
- 63. Krett N. L., Pillay S., Moalli P. A., Greipp P. R., Rosen S. T. (1995) Cancer Res. 55, 2727–2729 [PubMed] [Google Scholar]
- 64. Lu N. Z., Cidlowski J. A. (2005) Mol. Cell 18, 331–342 [DOI] [PubMed] [Google Scholar]
- 65. Yudt M. R., Cidlowski J. A. (2001) Mol. Endocrinol. 15, 1093–1103 [DOI] [PubMed] [Google Scholar]
- 66. Nehmé A., Lobenhofer E. K., Stamer W. D., Edelman J. L. (2009) BMC Med. Genomics 2, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang S. C., Myers S., Dooms C., Capon R., Muscat G. E. (2009) Mol. Cell. Endocrinol. 315, 146–152 [DOI] [PubMed] [Google Scholar]
- 68. Sinclair D., Webster M. J., Wong J., Weickert C. S. (2010) Mol. Psychiatry doi:10.1038/mp.2010.28, 1–12 [DOI] [PubMed] [Google Scholar]
- 69. Pedersen K. B., Geng C. D., Vedeckis W. V. (2004) Biochemistry 43, 10851–10858 [DOI] [PubMed] [Google Scholar]
- 70. Russcher H., van Rossum E. F., de Jong F. H., Brinkmann A. O., Lamberts S. W., Koper J. W. (2005) Mol. Endocrinol. 19, 1687–1696 [DOI] [PubMed] [Google Scholar]
- 71. Pascual-Le Tallec L., Lombès M. (2005) Mol. Endocrinol. 19, 2211–2221 [DOI] [PubMed] [Google Scholar]
- 72. Barraille P., Chinestra P., Bayard F., Faye J. C. (1999) Biochem. Biophys. Res. Commun. 257, 84–88 [DOI] [PubMed] [Google Scholar]
- 73. Wilson C. M., McPhaul M. J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1234–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Beck I. M., Vanden Berghe W., Vermeulen L., Yamamoto K. R., Haegeman G., De Bosscher K. (2009) Endocr. Rev. 30, 830–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Galliher-Beckley A. J., Cidlowski J. A. (2009) IUBMB Life 61, 979–986 [DOI] [PubMed] [Google Scholar]
- 76. Kumar R., Calhoun W. J. (2008) Biologics 2, 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Avenant C., Ronacher K., Stubsrud E., Louw A., Hapgood J. P. (2010) Mol. Cell. Endocrinol. 327, 72–88 [DOI] [PubMed] [Google Scholar]
- 78. Wang Z., Frederick J., Garabedian M. J. (2002) J. Biol. Chem. 277, 26573–26580 [DOI] [PubMed] [Google Scholar]
- 79. Webster J. C., Jewell C. M., Bodwell J. E., Munck A., Sar M., Cidlowski J. A. (1997) J. Biol. Chem. 272, 9287–9293 [DOI] [PubMed] [Google Scholar]
- 80. Blind R. D., Garabedian M. J. (2008) J. Steroid Biochem. Mol. Biol. 109, 150–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen W., Dang T., Blind R. D., Wang Z., Cavasotto C. N., Hittelman A. B., Rogatsky I., Logan S. K., Garabedian M. J. (2008) Mol. Endocrinol. 22, 1754–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Miller A. L., Garza A. S., Johnson B. H., Thompson E. B. (2007) Cancer Cell Int. 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Miller A. L., Webb M. S., Copik A. J., Wang Y., Johnson B. H., Kumar R., Thompson E. B. (2005) Mol. Endocrinol. 19, 1569–1583 [DOI] [PubMed] [Google Scholar]
- 84. Zhang Y., Leung D. Y., Nordeen S. K., Goleva E. (2009) J. Biol. Chem. 284, 24542–24552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Galliher-Beckley A. J., Williams J. G., Collins J. B., Cidlowski J. A. (2008) Mol. Cell. Biol. 28, 7309–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ismaili N., Blind R., Garabedian M. J. (2005) J. Biol. Chem. 280, 11120–11126 [DOI] [PubMed] [Google Scholar]
- 87. Itoh M., Adachi M., Yasui H., Takekawa M., Tanaka H., Imai K. (2002) Mol. Endocrinol. 16, 2382–2392 [DOI] [PubMed] [Google Scholar]
- 88. Deroo B. J., Rentsch C., Sampath S., Young J., DeFranco D. B., Archer T. K. (2002) Mol. Cell. Biol. 22, 4113–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wallace A. D., Cidlowski J. A. (2001) J. Biol. Chem. 276, 42714–42721 [DOI] [PubMed] [Google Scholar]
- 90. Wang X., DeFranco D. B. (2005) Mol. Endocrinol. 19, 1474–1482 [DOI] [PubMed] [Google Scholar]
- 91. Davies L., Karthikeyan N., Lynch J. T., Sial E. A., Gkourtsa A., Demonacos C., Krstic-Demonacos M. (2008) Mol. Endocrinol. 22, 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Holmstrom S., Van Antwerp M. E., Iñiguez-Lluhi J. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15758–15763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Holmstrom S. R., Chupreta S., So A. Y., Iniguez-Lluhi J. A. (2008) Mol. Endocrinol. 22, 2061–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Iñiguez-Lluhí J. A., Pearce D. (2000) Mol. Cell. Biol. 20, 6040–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Le Drean Y., Mincheneau N., Le Goff P., Michel D. (2002) Endocrinology 143, 3482–3489 [DOI] [PubMed] [Google Scholar]
- 96. Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M. (2006) Mol. Cell 24, 341–354 [DOI] [PubMed] [Google Scholar]
- 97. Tian S., Poukka H., Palvimo J. J., Jänne O. A. (2002) Biochem. J. 367, 907–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ito K., Yamamura S., Essilfie-Quaye S., Cosio B., Ito M., Barnes P. J., Adcock I. M. (2006) J. Exp. Med. 203, 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.