Abstract
Most hemoglobins serve for the transport or storage of O2. Although hemoglobins are widespread in “entomostracan” Crustacea, malacostracans harbor the copper-containing hemocyanin in their hemolymph. Usually, only one type of respiratory protein occurs within a single species. Here, we report the identification of a hemoglobin of the shore crab Carcinus maenas (Malacostraca, Brachyura). In contrast to the dodecameric hemocyanin of this species, C. maenas hemoglobin does not reside in the hemolymph but is restricted to the gills. Immunofluorescence studies and cell fractioning showed that C. maenas hemoglobin resides in the membrane of the chief cells of the gill. To the best of our knowledge, this is the first time that a membrane-bound hemoglobin has been identified in eukaryotes. Bioinformatic evaluation suggests that C. maenas hemoglobin is anchored in the membrane by N-myristoylation. Recombinant C. maenas hemoglobin has a hexacoordinate binding scheme at the Fe2+ and an oxygen affinity of P50 = 0.5 Torr. A rapid autoxidation rate precludes a function as oxygen carrier. We rather speculate that, analogous to prokaryotic membrane-globins, C. maenas hemoglobin carries out enzymatic functions to protect the lipids in cell membrane from reactive oxygen species. Sequence comparisons and phylogenetic studies suggested that the ancestral arthropod hemoglobin was most likely an N-myristoylated protein that did not have an O2 supply function. True respiratory hemoglobins of arthropods, however, evolved independently in chironomid midges and branchiopod crustaceans.
Keywords: Blood, Hemoglobin, Membrane Proteins, Oxidative Stress, Oxygen Binding, Oxygen Transport, Reactive Oxygen Species (ROS), Gills, Hemocyanin, Myristoylation
Introduction
In the animal kingdom, oxygen transport and storage is facilitated by three distinct types of metalloproteins that are classified according to their active sites: hemoglobin (Hb), hemerythrin, and hemocyanin (Hc).2 Hc is a copper-containing respiratory protein in the hemolymph of many arthropod and mollusk species (1, 2). Although the active sites in molluscan and arthropodan Hcs are similar, both proteins are, if at all, only distantly related and emerged independently in evolution (3, 4).
Hbs are small proteins of ∼140 to 150 amino acids that contain an heme iron prosthetic group (iron protoporphyrin IX), by which they reversibly bind gaseous ligands such as O2, CO, and NO. Hbs are the most widespread respiratory proteins and have been found in animals, bacteria, fungi, protists, and plants (5). For a long time, it has been assumed that any Hb has a respiratory function either in transporting O2 in the body fluid or in intracellular O2 supply. Only recently, it has become evident that certain Hbs may carry out various other functions, e.g., maize globin regenerates NAD+ under anaerobic conditions (6). Some prokaryotic globins are involved in O2 sensing (7). Escherichia coli Hb decomposes NO (8), and a similar function has been demonstrated for mammalian myoglobin (9). Myoglobin may also be involved in decomposition of reactive oxygen species (ROS) (10).
In the arthropod subphylum Crustacea, both Hbs and hemocyanins have been identified. In several “entomostracan” Crustacea, including Branchiopoda, Ostracoda, Copepoda, and Cirripedia, an extracellular Hb mediates O2 transport in the hemolymph (11–14). Depending on the species, crustacean Hbs occur as dimers or large multisubunit proteins with masses ranging from 220 to 800 kDa (12, 14, 15). By contrast, Malacostraca and Remipedia possess hemocyanins (1, 16–18). Arthropod hemocyanins are hexamers or oligohexamers of six similar or identical subunits, each of which is capable to bind one O2 molecule. Both subunit compositions and quaternary structures are species-specific and are highly variable even among related malacostracan species (18), e.g., the hemocyanin of the brachyuran crab Carcinus maenas is a 2 × 6-mer that is composed of four distinct subunit types (18, 19).
In most animals, only a single type of respiratory protein occurs. However, there are two species in which a co-existence of Hb and hemocyanin has been reported. Lieb et al. (20) found in the hemolymph of the planorbid snail Biomphalaria glabrata trace amounts of hemocyanin in addition to Hb. The amphipod crustacean Cyamus scammoni has both Hb and hemocyanin, which appear to have distinct roles in O2 transport (21, 22). Here, we report the identification of a Hb in the shore crab C. maenas, which does not co-occur with the hemocyanin in the hemolymph but surprisingly is located in the cell membranes of gill cells. We also provide evidence against a respiratory function of this Hb but suggest a role in protection of membrane lipids from ROS.
EXPERIMENTAL PROCEDURES
Experimental Animals
C. maenas were obtained from the Meeresbiologische Forschungsanstalt Helgoland. Hemolymph was withdrawn from living crabs by the aid of a syringe and diluted with an equal volume of 100 mm Tris/HCl, pH 7.8, 10 mm MgCl2, 10 mm CaCl2. After centrifugation at 10,000 × g for 5 min, the supernatant was stored at −20 °C until use. The animals were dissected, and selected tissues were removed and immediately used for the experiments or kept frozen at −80 °C.
Cloning and Sequencing of CmaHb cDNA and Gene
Total RNA was extracted from whole animals according to the guanidinium-isothiocyaniate method (23). First-strand cDNA synthesis and subsequent PCR reactions were carried out by using SuperScript III reverse transcriptase and Platinum TaqDNA Polymerase (Invitrogen) according to the manufacturer's instructions. Specific oligonucleotide primers for nested PCR were designed using the tentative coding sequence of C. maenas Hb (CmaHb) as inferred from the expressed sequence tag (EST) databases of C. maenas. The primers 5′-GCTACTCTCAAGACCCACAC-3′ and 5′-GTAGTAGGACTGAAGGACTG-3′ were used in the first PCR, whereas the second PCR was performed with the two oligonucleotide primers 5′-ATGGGAGCCGTGCTGAGTGTG-3′ and 5′-TCATGCATCATAAGAAGT-3′. Genomic DNA of C. maenas was isolated using the Qiagen DNeasy kit. Several oligonucleotide primers were designed according to the CmaHb cDNA sequence (supplemental Table 1). Various combinations of the specific forward primers with the reverse primers were employed, and several partial gene fragments were amplified using the Expand Long Template PCR System (Roche Applied Science). The PCR products were cloned into the pGEM-T Easy vector (Promega), and the sequences were determined by a commercial sequencing service (GENterprise GmbH). The final cDNA sequence of C. maenas Hb has been submitted to EMBL/GenBankTM under the accession no. FN995203.
Sequence Analyses and Phylogenetic Studies
The BLAST algorithm (24) was employed to search the EST database available at GenBankTM. DNA translation and analyses of primary structures were performed with the programs available at the ExPASy Molecular Biology Server. Exon-intron structure of the CmaHb gene was predicted by the online mRNA-to-genomic alignment program Spidey. Subcellular localization and posttranslational modifications were predicted by PSORT II (25) and Myristoylater. Globin structure was analyzed using the program SeqView, which is based on a multiple alignment of 859 metazoan globin amino acid sequences (26).
Hb amino acid sequences from selected arthropod species (see supplemental Table 1 and supplemental Fig. 1) were aligned by hand with GeneDoc (version 2.6; 27). We added Hb sequences of the ticks (Chelicerata) Ixodes scapularis and Rhipicephalus appendiculatus, the beetle Dascillus cervinus (Coleoptera), and the silkworm Bombyx mori (Lepidoptera) that had been derived from the EST databases. The appropriate model of amino acid sequence evolution was selected by ProtTest (28) using the Akaike Information Criterion. MrBayes (version 3.1.2) was employed for Bayesian phylogenetic analyses (29) assuming the WAG model with Γ distribution of rates (four Γ categories). The analyses were run for 2,000,000 generations with random starting trees. The final average S.D. of split frequencies were <0.02. Trees were sampled every 100th generation, and posterior probabilities were estimated on the last 15,000 trees (burnin = 5000).
Recombinant C. maenas Hemoglobin
For recombinant expression in E. coli, the CmaHb cDNA was cloned into the expression vector pET3a using PCR-generated NdeI and BamHI restriction sites. The E. coli strain BL21(DE3)pLysS was transformed with the construct, and cultures were grown at 26 °C in low salt Luria-Bertani medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl, pH 7.4) supplemented with 100 μg/ml ampicillin, 34 μg/ml chloramphenicol, and 1 mm δ-aminolevulinic acid. The culture was induced at A600 = 0.5 by the addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.4 mm, and expression was continued overnight. Cells were harvested by centrifugation at 4000 × g for 30 min and resuspended in 50 mm Tris/HCl, pH 8.0, 1 mm DTT, 1 mm MgCl2, 8 μg/ml DNase, 4 μg/ml RNase supplemented with Pefabloc SC protease inhibitor (Roth) and Complete protease inhibitor mixture (Roche Applied Science). Bacteria were disrupted by sonication (5 × 1 min), and the sample was incubated for 2 h at 37 °C to digest the DNA and RNA, followed by a centrifugation for 1 h at 10,000 × g and 4 °C to remove the cell debris. The supernatant was fractionated by ammonium sulfate precipitation and the Hb-containing reddish 60% ammonium sulfate pellet was dissolved in 20 mm Tris/HCl, pH 8.5. After desalting the sample with an Amicon Ultra filter (Millipore), it was loaded on a DEAE ion exchange column and eluted with a gradient of 0 to 1 m NaCl in 20 mm Tris/HCl, pH 8.5. Further purification was achieved by a size exclusion chromatography using a High LoadTM 16/60 SuperdexTM 75 prep grade column (GE Healthcare). Final CmaHb fractions were analyzed by SDS-PAGE, pooled, and stored at −20 °C. Protein concentrations were determined according to Bradford (30).
Spectra and Ligand Binding Measurements
Absorption spectra were measured on a Varian Cary400 spectrophometer. Samples in 50 mm Tris-HCl, 150 mm NaCl buffer at pH 8.5 were reduced with sodium dithionite under an atmosphere of nitrogen to obtain the deoxy spectrum (31). CO was then added to obtain the CO-bound form. Attempts to obtain the oxy form led rather to an oxidized state. However, samples in 10 mm DTT gave a spectrum similar to oxyHb; the spectra shifted toward the oxy form with increasing oxygen partial pressure, and could finally be converted to the CO form, confirming a ferrous state. DTT could also be used to convert the oxidized form to the ferrous state. To measure the autoxidation rate, the oxy form was prepared in the presence of 10 mm DTT. The DTT was then stripped, and the absorption spectra were measure versus time. Kinetic measurements of ligand binding were made with a flash photolysis system, as described previously (31, 32). Kinetics at different CO concentrations and under a mixed CO and O2 atmosphere were used to study the competitive ligand binding.
mRNA Expression Analyses
RNA extraction of selected tissues (gills, heart, hepatopancreas, ovary, and muscles) was performed using the Qiagen RNeasy mini kit. 1.5 μg of total RNA per tissue were converted into cDNA with an oligo(dT) primer by Superscript II reverse transcriptase (Invitrogen). About 10% of the total cDNA was used for standard PCR employing the CmaHb-specific oligonucleotide primers 5′-ATGGGAGCCGTGCTGAGTGTG-3′ and 5′-TCATGCATCATAAGAAGT-3′.
Antibody Preparation
To generate polyclonal antibodies against CmaHb, the two peptides FKGFAGKSIEELKNC (amino acids 83–96) and AMVDNLEDVSVLVEL (amino acids 113–127) were synthesized, coupled to keyhole limpet hemocyanin, and used for the immunization of rabbits (Eurogentec). Specific antibodies were affinity purified from the rabbit serum using a HiTrap NHS-activated HP column (GE Healthcare) according to the manufacturer's instructions.
SDS-PAGE and Western Blotting
Selected tissues from C. maenas were homogenized in 10 mm Tris/HCl, pH 7.4, 10 mm NaCl, 5 mm MgCl2, 1 mm DTT, 0.1% Nonidet P40, 1 mm Pefabloc SC protease inhibitor (Roth) and Complete protease inhibitor mixture (Roche Applied Science). After precipitation for 10 min at 10,000 × g and 4 °C, the protein concentrations of the supernatants were determined (30). Subcellular fractionation of gill tissue was carried out by using the Qproteome Cell Compartment kit (Qiagen) according to the manufacturer's instructions.
For SDS-PAGE, protein extracts were heat-denatured in sample buffer (65 mm Tris-HCl, pH 6.8, 1% SDS, 5% β-mercaptoethanol, 10% glycerol) at 95 °C for 5 min and loaded onto a 15% polyacrylamide gel. Semi-dry electroblotting of proteins from the polyacrylamide gels onto nitrocellulose membranes (Hartenstein) was carried out for 2 h at 0.8 mA/cm2. Nonspecific binding sites were blocked for 1 h with 2% nonfat dry milk in TBS (10 mm Tris/HCl, pH 7.4; 140 mm NaCl). Immunodetection was performed for 2 h at room temperature with affinity-purified polyclonal anti-CmaHb antibodies diluted 1:100 in 2% milk/TBS. The nitrocellulose filters were washed three times with TBS for 15 min and incubated for 1 h with the goat anti-rabbit antibody coupled with alkaline phosphatase (Dianova) diluted 1:10,000 in TBS. After the final washing step, detection was carried out with nitro-blue-tetrazolium-chloride and 5-bromo-4-chloro-3-indolyl-phosphate as substrates.
Immunofluorescence Studies
Dissected gills from C. maenas were fixed for 48 h in 4% paraformaldehyde in PBS (8 mm Na2HPO4, 1.5 mm KH2PO4, 140 mm NaCl, 2.7 mm KCl). Cryosections (6 μm) were prepared and placed onto glass slides. Nonspecific binding sites were blocked for 15 min in 1% bovine serum albumin, 0.3% Triton X-100 in PBS. The sections were incubated with affinity-purified anti-CmaHb antibodies (1:10) in the blocking solution overnight at 4 °C. In control experiments, the affinity-purified anti-CmaHb antibody was only used on cryosections after incubation overnight with 1–8 μg of recombinant CmaHb. After washing the sections 3 × 15 min in PBS, incubation with the secondary antibody coupled with Cy3 (Dianova), diluted 1:500 in blocking solution, was carried out for 2 h at room temperature in the dark. The sections were washed 3 × 15 min with PBS, and cell nuclei were stained with Hoechst dye (Invitrogen). Cryosections were mounted in Mowiol 4–88 (Calbiochem) and examined with an Olympus BX51 epifluorescence microscope.
RESULTS
Identification of a Hemoglobin in C. maenas
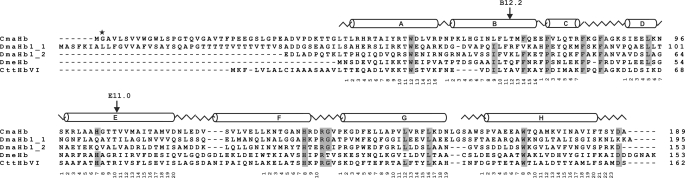
Systematic BLAST searches for Hb-like sequences were carried out using the databases of ESTs. Seven Hb-coding cDNAs (DN635027, DN551432, DN551331, DN161664, DN161580, DN1611354, and DN161266) were identified in the ESTs from C. maenas. The tentative cDNA sequence of CmaHb, which was inferred from the assembled ESTs, covered the complete coding region of 567 bp. Specific oligonucleotide primers were designed according to this sequence and used to amplify the cDNA from total RNA. RT-PCR experiments resulted in a single product, which was cloned and sequenced (supplemental Fig. 2). The nucleotide sequence was found to be identical to that deduced from the ESTs. Using genomic DNA as template, we identified two introns located in positions B12.2 (i.e. between the second and third bp of codon 12 of globin helix B) and E11.0 (i.e. between the third bp of codon 10 and the first bp of codon 11 of globin helix E). The intron in B12.2 measures 690 bp, and the intron in E11.0 covers 471 bp.
The CmaHb cDNA was translated into a polypeptide and compared with other arthropod Hb sequences. The alignment shows that the globin fold, which covers ∼140–150 amino acids of the standard α-helices A through H, is conserved (Fig. 1). The CmaHb protein harbors the highly conserved key residues important for oxygen binding, such as the Trp at A12, the Phe at CD1, as well as the characteristic distal and proximal histidines in the positions E7 and F8 that are involved in heme binding. The full-length protein consists of 189 amino acids with a predicted molecular mass of 20.7 kDa. It thus exceeds the length of a typical globin, which is due to an N-terminal extension of ∼40 amino acids. Computer prediction for subcellular localization using PSORT II program (25) suggested that the CmaHb protein does not contain any signal peptide for extracellular secretion. An N-terminal myristoylation site, which is typical for membrane-bound proteins, was predicted at the Gly at amino acid position 2 with high confidence (93%).
FIGURE 1.
Comparison of the C. maenas hemoglobin with other arthropod globins. The secondary structure of the D. melanogaster hemoglobin (DmeHb) is superimposed in the upper row. The α-helices are designated A through H, and conserved amino acids are shaded. The globin consensus numbering is given below the sequences. The two intron positions B12.2 and E11.0 in the CmaHb gene are indicated by black arrows, the N-terminal glycine predicted for protein N-myristoylation is marked by an asterisk. DmaHb_1, D. magna hemoglobin domain 1, DmaHb_2, D. magna hemoglobin domain 2; CttHbVI, Chironomus thummi thummi hemoglobin VI.
Hemoglobin mRNA and Protein in C. maenas Tissues
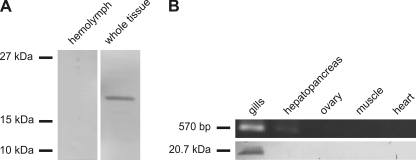
In Western blotting experiments, a polyclonal antibody against CmaHb-specific peptides specifically stained a protein in the extract from whole crab tissue, whereas no signal was observed in the hemolymph (Fig. 2A). The detected protein band had an apparent molecular mass of ∼20 kDa, which is in good agreement with the expected mass of CmaHb. The tissue distribution of CmaHb was further analyzed by RT-PCR and Western blotting. Total RNA and proteins were extracted from five selected crab tissues. RT-PCR revealed the presence of CmaHb mRNA in the gills, whereas no amplification product was observed in the muscle, heart, ovary, and hepatopancreas (Fig. 2B, upper row). Likewise, CmaHb protein was only detected by Western blotting in extracts from the gills but not in other tissues (Fig. 2B, lower row). We therefore conclude that CmaHb is a gill-specific protein.
FIGURE 2.
C. maenas hemoglobin is a gill-specific protein. About 70 μg hemolymph proteins and protein extracts from whole tissue were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and stained with purified polyclonal anti-CmaHb antibodies. A, CmaHb was detected in the fraction containing proteins from whole tissue. B, RNA and protein were isolated from the indicated crab tissues. CmaHb mRNA was determined by RT-PCR (B, upper row). Western blotting experiments were performed with ∼70 μg total proteins of tissue extracts (B, lower row). CmaHb protein and mRNA are restricted to the gills.
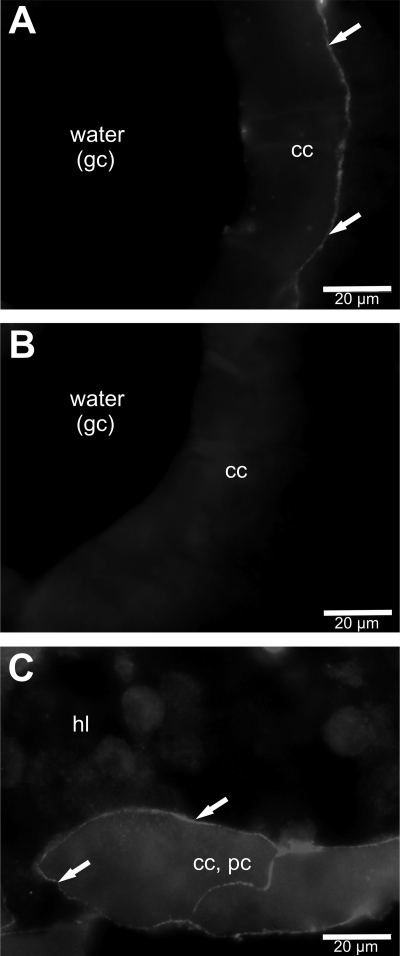
The cellular and subcellular localization of CmaHb in the phyllobranchiate gills of crab were studied by indirect immunofluorescence experiments. The affinity purified anti-CmaHb antibody was applied to 6-μm thick cryosections prepared from paraformaldehyde-fixed gills. Although we observed strong autofluorescence that resulted in high background, unambiguous CmaHb immune reactivity was observed. The images showed that the gill consists of paired serial lamellae along a central stem, which joins the incurrent (afferent) and excurrent (efferent) hemolymph channels. CmaHb was present in the tissue areas that are exposed to the water in the gill chamber. Closer examination showed that CmaHb was located in the membrane of the chief cells of the central stem (Fig. 3A) and the lamellae (Fig. 3C). The specificity of the CmaHb antibody was verified by preabsorption tests with its recombinant antigen, resulting in background fluorescence but no specific staining (Fig. 3B).
FIGURE 3.
Localization of C. maenas hemoglobin in gills. An affinity purified anti-CmaHb antibody was applied on cryosections of gills. CmaHb immune reactivity was found at the membrane of chief cells in the lateral area of the central stem (A) and at the membrane of cells of the lamellae (C). In preabsorption tests no specific immune staining but only background fluorescence was observed (B). cc, chief cells; gc, gill chamber; hl, hemolymph; pc, pillar cells.
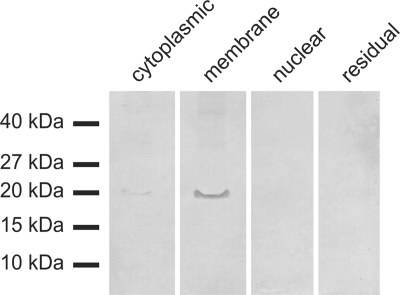
The subcellular distribution of CmaHb was further analyzed by fractioning the proteins from C. maenas gill tissue by a discontinuous centrifugation approach. The four fractions contained (i) cytosolic proteins, (ii) proteins from the membrane and from the lumen of organelles (e.g. ER and mitochondria), (iii) nuclear proteins, and (iv) the residual, mainly cytoskeletal, proteins. The samples were analyzed by Western blotting applying the anti-CmaHb antibody. In good agreement with the results of the immunofluorescence studies, the CmaHb protein was mainly detected in the fraction that includes the membrane proteins, whereas only a weak signal was observed in the cytosolic proteins (Fig. 4).
FIGURE 4.
Subcellular localization of hemoglobin by Western blot analysis. About 70 μg protein extract isolated from four different cellular compartments were applied on each lane and used for Western blotting. CmaHb protein was mainly detected in the fraction that includes the membrane proteins. An additional weak signal was observed in the cytosolic fraction, whereas no CmaHb protein was detectable in the protein samples containing nuclear proteins and the residual ones.
Biochemical Characterization of C. maenas Hemoglobin
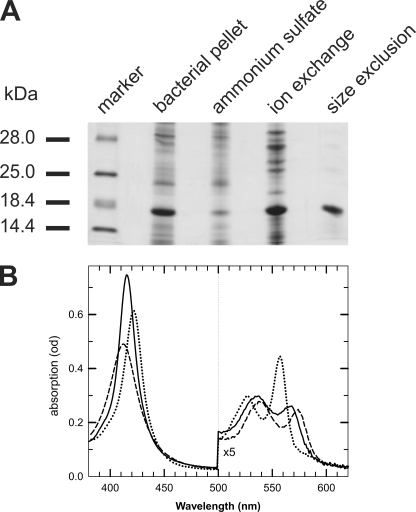
CmaHb was recombinantly expressed in E. coli BL21(DE3)pLysS employing the pET system. We could not obtain detectable amounts of soluble full-length CmaHb protein, which precipitated as insoluble inclusion bodies. Likewise, expression of a truncated version of CmaHb that excludes 41 amino acids of the N-terminal extension (CmaHbΔN1–41) resulted in the formation of inclusion bodies. Expression of a truncated form that lacks amino acids 1–21 (CmaHbΔN1–21) gave rise to soluble protein. Purification of CmaHbΔN1–21 from the bacterial supernatant was carried out by a three-step protocol, which includes ammonium sulfate precipitation followed by ion exchange and size exclusion chromatography (Fig. 5A).
FIGURE 5.
Recombinant expression and characterization of C. maenas hemoglobin. A, CmaHbΔN1–20 was recombinantly expressed in E. coli and purified using a three-step protocol consisting of ammonium sulfate precipitation followed by ion exchange and size exclusion chromatography. B, absorbance spectra of hexacoordinate CmaHb. Deoxygenated ferrous form (dotted line), CO form (solid line), and in the presence of oxygen (dashed line).
After reduction with an excess of sodium dithionite, absorbance spectra display large amplitudes of the α-band (560 nm) and the Soret band (426 nm) for the ferrous deoxy form with unique spectra for the His-Fe-His species in the absence of external ligands (Fig. 5B). These spectra resemble those of neuroglobin (Ngb) and indicate that CmaHb is a hexacoordinate globin (33). A pure oxy form was difficult to obtain, but samples reduced with DTT provided a state of ∼90% of the oxy form.
The ligand binding kinetics displayed high association rates of CO and oxygen that are comparable with the state of Ngb with the disulfide bond and confirmed a weaker binding of the internal His ligand (Table 1). For the pentacoordinated Hb and Mb systems, where the sixth coordination site is open in the deoxy state, the O2 on- and off-rates are sufficient to determine the oxygen affinity (K). For systems such as Ngb or CmaHb, which are hexacoordinate in the deoxy state, one needs to take into account KHis, the affinity of the His residue, which is in competition for binding to the iron atom. Here, the observed oxygen affinity is Kobs = K/(1 + KHis).
TABLE 1.
Ligand binding parameters for CmaHb compared with wild type human Ngb with the disulfide bond (31)
Note that Ngb and DTT (to reduce the cysteines) or the mutant Ngb without cysteines has a 10-fold lower oxygen affinity due mainly to a lower His dissociation rate. On/off-rates were measured at 25 °C in 50 mm Tris/HCl, 150 NaCl, 5 mm DTT, pH 8.5.
| CmaHb | Human Ngb | |
|---|---|---|
| kon(CO) (μm/s) | 40 | 40 |
| kon(O2) (μm/s) | 100 | 140 |
| koff(O2) (s) | 5.8 | 0.8 |
| K(O2) (nm) | 55 | 5.7 |
| kon(His) (s) | ≈100 | 2000 |
| koff(His) (s) | 5 | 7 |
| KHis | 20 | 280 |
| P50(O2) (Torr) | 0.5a | 0.9 |
a This value is also measured at equilibrium.
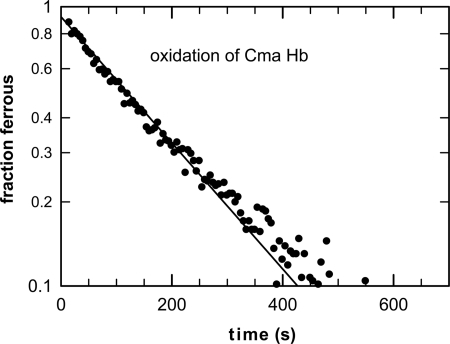
From the competitive ligand binding studies, the association and dissociation rates of O2 and the internal ligand could be extracted. At 25 °C, O2 binds to CmaHb with an affinity P50 = 0.5 Torr. After stripping the DTT, CmaHb was found unstable and rapidly autoxidized within minutes (t1/2 = ∼3 min at 25 °C) to the ferric Fe3+ form that is unable to bind O2 (Fig. 6). This rate is even faster than of those measured for mouse and human Ngb at 37 °C, pH 7.0 (t1/2 = ∼5 min and ∼11 min, respectively) (33) and much faster than a typical autoxidaton reaction for a similar O2 affinity pentacoordinate species such as Mb, which typically takes a few days.
FIGURE 6.
Autoxidation of hexacoordinate C. maenas hemoglobin: transition of the ferrous oxy to ferric hexacoordinated form.
Phylogenetic Position of C. maenas Hemoglobin
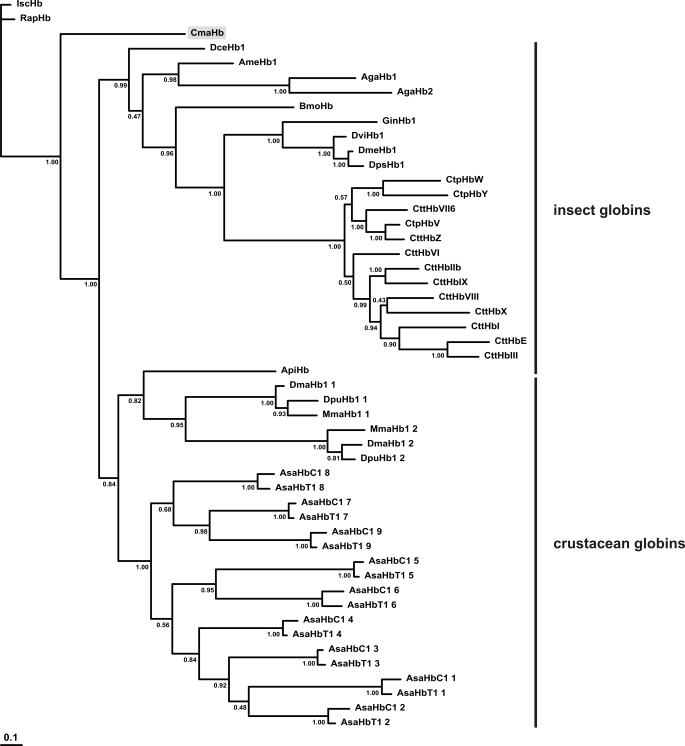
BLAST searches did not identify any protein that is obviously orthologous with CmaHb (data not shown). The amino acid sequence of CmaHb was included in a multiple sequence alignment consisting of selected insect and crustacean Hbs (supplemental Fig. 1). Phylogenetic analysis was carried out by a Bayesian method. The chelicerate Hbs were employed as outgroup. Branchiopod and insect globins were found to form two independent monophyletic clades. C. maenas Hb was in sister group position to all other crustacean and insect globins (Fig. 7).
FIGURE 7.
Phylogeny of the arthropod globins. The numbers at the nodes are Bayesian posterior probabilities. The bar represents 0.1 PAM distance. The hemoglobin of C. maenas is shaded in gray.
DISCUSSION
Two Types of Respiratory Proteins in C. maenas
Besides C. maenas, the amphipod C. scammoni is the only known arthropod species in which Hb and Hc co-exist (21, 22, 34). In C. scammoni, both types of respiratory proteins circulate freely dissolved in the body fluid. Here, Hb might be the principal O2 carrier, whereas the contribution of Hc might be minimal (22). It has also been speculated that C. scammoni Hc may rather function as phenoloxidase (like in some other arthropods) because no such specific enzyme could be identified in this species. The situation is quite different in C. maenas. Although Hb occurs in this brachyuran crab along with a 2 × 6 Hc (18, 19), a respiratory function of CmaHb is, however, unlikely.
Rapid Autoxidation of C. maenas Hemoglobin Precludes a Respiratory Function
Spectroscopic and ligand-binding studies indicate that the heme iron atom is hexacoordinated in CmaHb. However, the ferrous spectrum for CmaHb was not as distinct (compared with other globins) as that of Ngb, indicating that there was not a complete hexacoordination (33).
The O2 affinity of CmaHb (P50 = 0.5 Torr) is similar to that of Ngb and other hexacoordinate globins (33). Relative to Ngb with the disulfide bond, the O2 affinity of the active site is higher, but the affinity of His to Fe2+ is lower. Thus, there is a compensation between the two ligand affinities resulting in an overall O2 affinity in the range commonly found for globins. Note that the time to equilibrate with oxygen depends on both the oxygen and histidine dissociation rates; although CmaHb could equilibrate with oxygen faster than Ngb, it is still much slower than classical pentacoordinated Hbs (31, 33).
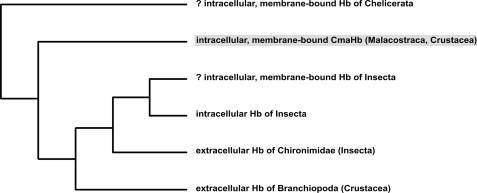
Although the kinetic data demonstrate reversible O2 binding of CmaHb, the very rapid autoxidation precludes a function as respiratory protein (Fig. 6). Autoxidation causes the formation of ferric (Fe3+) Hb that is not able to bind O2. A role of CmaHb in O2 supply is also difficult to reconcile with its prominent expression in gills; the excellent O2 supply to the gill cells from the surrounding water renders a specific O2 transport or storage protein in this tissue unnecessary. Based on phylogenetic considerations (Figs. 7 and 9), it may be speculated that a CmaHb-like gill globin represents the original version of crustacean Hbs and that respiratory arthropod Hbs represent derived forms (see below).
FIGURE 9.
Simplified phylogenetic tree of arthropod Hbs. N-Myristoylated Hbs represent the ancient branch of arthropod globins. CmaHb is shaded in gray, computer predicted membrane binding is indicated by a question mark. See text for further explanations.
Structural Basis of C. maenas Hemoglobin Kinetics and Oxidation
CmaHb harbors the highly conserved key residues Trp(A12), Phe(CD1), His(E7), and His(F8), consistent with the typical globin fold. However, the polar Thr(F4) is unusual. In most globins, apolar amino acids at positions F4, FG5, and G5 form a hydrophobic cavity that prevents solvent access to the proximal site of the heme (35). Inspection of 859 metazoan globin sequences (26) revealed that in 89.8% of the globins F4 is occupied by Leu, and additional 8.4% harbor another hydrophobic amino acid. To date, only seven other globins are known with ThrF4, which all have been identified in nematode genomes (SwissProt accession nos. P27613, P51535, P51536, Q26332, Q27302, Q27430, and Q8WS56). Nothing is known about the functions of these globins. However, it may be speculated that the replacement of Leu by Thr at position F4 facilitates the access of water to the active center of CmaHb, thereby explaining its rapid autoxidation. Liong et al. (35) also noted that Gly(F4) and Ser(F4) mutants of myoglobin loose the heme more readily than the Leu(F4) wild type. However, we did not observe significant heme loss, usually associated with protein precipitation, during purification or in kinetic experiments. Moreover, hexacoordination tends to stabilize the heme.
C. maenas Gill Hemoglobin Is Associated with Cell Membrane
The phyllobranchiate gills of Carcinus consist of paired serial lamellae connected medially to a central stem, which joins the incurrent (afferent) and excurrent (efferent) hemolymph channels (36). CmaHb is localized in the lamellae as well as in the central stem (Fig. 3). Interestingly, this protein is restricted to tissue areas adjacent to the water in the gill chamber. Within this epithelium, chief cells are the predominant cells that likely play a role in respiration.
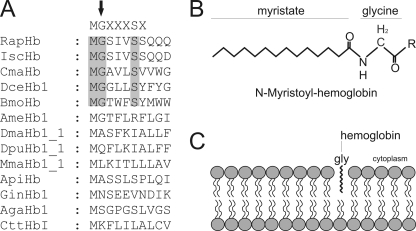
Immunofluorescence studies suggest that CmaHb localizes in the membrane of the chief cells, a result that is confirmed by cell fractioning (Fig. 4). CmaHb harbors an N-terminal extension that is not part of the conserved eight globin helices (Fig. 1). Computer predictions indicated the presence of a myristoylation site at Gly2. N-Myristoylation is a covalent attachment of myristate, a 14-carbon saturated fatty acid, to the N-terminal glycine of eukaryotic proteins (Fig. 8B) (37, 38). N-Myristoylation is an irreversible protein modification, which mostly occurs co-translationally following removal of the initiator Met residue (39, 40). It promotes weak and reversible protein-membrane and protein-protein interactions (Fig. 8C) (41, 42).
FIGURE 8.
Membrane-association of globins promoted by N-myristoylation. Identification of N-myristoylation signal sequences within the arthropod globins by computer predictions. N-Myristoylation sites are shaded gray. Consensus sequence for N-myristoylation is superimposed in the upper row. The N-terminal glycine required for the attachment of the fatty acid myristate is marked by an arrow. RapHb, R. appendiculatus hemoglobin; IscHb, I. scapularis hemoglobin; DceHb1, D. cervinus hemoglobin; BmoHb, B. mori hemoglobin; AmeHb1, A. mellifera hemoglobin; DmaHb1_1, D. magna hemoglobin domain 1; DpuHb1, Daphnia pulex hemoglobin domain 1; MmaHb1_1, M. macrocopa hemoglobin domain 1; ApiHb, Acyrthosiphon pisum hemoglobin; GinHb1, Gasterophilus intestinalis hemoglobin; AgaHb1, A. gambiae hemoglobin; CttHbI, Chironomous thummi thummi hemoglobin. A, myristoylated hemoglobin. After removal of the initiator Met residue, the 14-carbon saturated fatty acid myristate is covalently attached to the N-terminal glycine of eukaryotic proteins (B). Shown is the membrane association of globins by N-myristoylation (C).
A Hb in the cell membrane is unprecedented among eukaryotes. However, an association of Hb with membrane lipids has already been found in prokaryotes (43–47). For example, Vitreoscilla Hb is concentrated adjacent to the cytosolic side of the cell membrane (45). It has been demonstrated that the active site of Vitreoscilla Hb interact with membrane lipids (46). Likewise, E. coli flavohemoglobin is able to bind to lipid extracts of E. coli membranes (43, 47). These bacterial Hbs may actually provide hints to the function of CmaHb.
C. maenas Hemoglobin May Protect Membrane Lipids from ROS Stress
Most Hbs are considered being involved in transport or storage of O2, a function that is highly unlikely for CmaHb. Within the past 12 years, various roles for Hb have been suggested (48). None of these actually fit to the properties of CmaHb. For example, a role as O2 sensor, as previously demonstrated for various Hbs from Archaea and bacteria (7, 49) is incompatible with the observed autoxidation rate. However, this feature along with the localization in the membrane of gill cells allows drawing conclusions for CmaHb function. It is tempting to assume that CmaHb has a similar role like the bacterial membrane-bound Hbs. Several hypotheses of the function of Hb in bacterial membranes have been put forward. These proteins have been suggested to facilitate oxygen diffusion to membrane-located oxidases, to protect these oxidases from reactive oxygen species (ROS) or reactive nitrogen species, or to preserve the integrity of the membrane lipids by peroxide-reducing activities as a response to ROS/reactive nitrogen species stress (47).
Although we could readily exclude the first two functions because of the absence of a respiratory chain in the cell membrane of eukaryotes, a role of CmaHb in protection of membrane lipids is conceivable. CmaHb resides in cells adjacent to the water in the gill chamber, which causes relatively high oxygen partial pressures and thus promotes the generation of ROS. These ROS can cause damage of lipids (50). The exact mechanism of ROS protection remains uncertain, although it may be speculated that autoxidation of CmaHb is an essential step. Interestingly, CmaHb may also be reduced by DTT, suggesting a function in a cycling redox reaction in the presence of a reducing system.
Implication for Evolution of Crustacean Hemoglobins
CmaHb does not group with the extracellular, respiratory Hbs from branchiopods (Artemia salina, Daphnia magna, Daphnia pulex, and Moina macrocopa), rendering crustacean Hbs paraphyletic (Figs. 7 and 8). Database searches identified partial ESTs from the malacostracans Homarus americanus (accompanying nos. EX471677) and Litopenaeus vannamei (FE137590; FE081411), which resemble CmaHb, and suggest that similar Hbs exist in other crustaceans. CmaHb is a membrane-associated Hb and computer prediction suggests that association of CmaHb with the cell membrane is promoted by N-myristoylation at Gly (Fig. 1). Interestingly, further sequence analyses showed the presence of N-myristoylation sites in various other arthropod Hbs (Fig. 8A). Such N-terminal modification sites were identified in chelicerate Hbs, represented in this study by the Hbs of the ticks I. scapularis and R. appendiculatus, in the Hbs from the beetle D. cervinus and the silkworm B. mori, and can be derived from the partial Hb EST of H. americanus. This observation implies that these Hbs are also associated with the membrane and may carry out a similar function like CmaHb.
The Hbs from most insects do not harbor N-myristoylation sites and are either cytoplasmic or hemolymph proteins. Cytoplasmic Hbs have been identified in several holometabolous insects (51–56). The Hbs of the mosquito Anopheles gambiae, the honeybee Apis mellifera, and the fruit fly Drosophila melanogaster globin 1 are mainly expressed in the tracheal system (52, 54, 55). Their exact functions are still unknown, but O2-dependent regulation suggests a role other than O2 supply, such as O2-buffer or in ROS protection (57). Moreover, the tracheal-based expression of insect Hbs can be analogous to the gill-based expression of CmaHb. It may be speculated that these proteins have similar roles in the O2 metabolism of the organism, which is possibly related to ROS protection.
The phylogenetic tree suggests that N-myristoylation and membrane association is an ancestral feature of arthropod Hbs, whereas cytoplasmic and hemolymph-based Hbs represent derived forms (Fig. 9). Thus, a respiratory function was not the original role of arthropod Hb but later evolved in some taxa. It is evident that hemolymph-based O2-transporting Hbs emerged independently within Chironimidae (Insecta) and Branchiopoda (Crustacea) from intracellular ancestors. Most likely, such “true” Hbs evolved in response to the hypoxic lifestyle of chironomid midges and branchiopods (58, 59).
Supplementary Material
Acknowledgments
We thank Ralph Pirow and Rüdiger Paul for help with the oxygen binding studies, Roy Weber for advice with the enzymatic reduction system, and Katja Reimann for help with protein purification.
This work has been supported in part by a grant from the German Research Foundation (Deutsche Forschungsgemeinschaft Bu956/9) and INSERM.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) FN995203.
- Hc
- hemocyanin
- CmaHb
- C. maenas hemoglobin
- EST
- expressed sequence tag
- Ngb
- neuroglobin
- ROS
- reactive oxygen species.
REFERENCES
- 1. Markl J., Decker H. (1992) Adv. Comp. Environ. Physiol. 13, 325–376 [Google Scholar]
- 2. van Holde K. E., Miller K. I. (1995) Adv. Protein Chem. 47, 1–81 [DOI] [PubMed] [Google Scholar]
- 3. Burmester T. (2001) Mol. Biol. Evol. 18, 184–195 [DOI] [PubMed] [Google Scholar]
- 4. van Holde K. E., Miller K. I., Decker H. (2001) J. Biol. Chem. 276, 15563–15566 [DOI] [PubMed] [Google Scholar]
- 5. Hardison R. (1998) J. Exp. Biol. 201, 1099–1117 [DOI] [PubMed] [Google Scholar]
- 6. Sowa A. W., Duff S. M., Guy P. A., Hill R. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10317–10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hou S., Freitas T., Larsen R. W., Piatibratov M., Sivozhelezov V., Yamamoto A., Meleshkevitch E. A., Zimmer M., Ordal G. W., Alam M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9353–9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gardner P. R., Gardner A. M., Martin L. A., Salzman A. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10378–10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flögel U., Merx M. W., Godecke A., Decking U. K., Schrader J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flögel U., Gödecke A., Klotz L. O., Schrader J. (2004) FASEB J. 18, 1156–1158 [DOI] [PubMed] [Google Scholar]
- 11. Fox H. M. (1957) Nature 179, 148. [DOI] [PubMed] [Google Scholar]
- 12. Weber R. E., Vinogradov S. N. (2001) Physiol. Rev. 81, 569–628 [DOI] [PubMed] [Google Scholar]
- 13. Mangum C. P. (1983) The Biology of Crustacea (Mantel L. H. ed.) pp. 373–429, Academic Press, New York [Google Scholar]
- 14. Terwilliger N. B. (1998) J. Exp. Biol. 201, 1085–1098 [DOI] [PubMed] [Google Scholar]
- 15. Ilan E., Daniel E. (1979) Comp. Biochem. Physiol. B 63, 303–308 [Google Scholar]
- 16. Burmester T. (2002) J. Comp. Physiol. B 172, 95–107 [DOI] [PubMed] [Google Scholar]
- 17. Ertas B., von Reumont B. M., Wägele J. W., Misof B., Burmester T. (2009) Mol. Biol. Evol. 26, 2711–2718 [DOI] [PubMed] [Google Scholar]
- 18. Markl J. (1986) Biol. Bull. 171, 90–115 [Google Scholar]
- 19. Markl J., Hofer A., Bauer G., Markl A., Kempter B., Brenzinger M., Linzen B. (1979) J. Comp. Physiol. B 133, 167–175 [Google Scholar]
- 20. Lieb B., Dimitrova K., Kang H. S., Braun S., Gebauer W., Martin A., Hanelt B., Saenz S. A., Adema C. M., Markl J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12011–12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Terwilliger N. B. (1991) Structure and Function of Invertebrate Oxygen Carriers (Vingoradov S. N., Kapp O. H. eds.) pp. 59–63, Springer Verlag, New York [Google Scholar]
- 22. Terwilliger N. B. (2008) Protein Reviews: Dioxygen Binding and Sensing Proteins (Bolognesi M., di Prisco G., Verde C. eds.) pp. 203–209, Springer-Verlag Italy [Google Scholar]
- 23. Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 24. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakai K., Horton P. (1999) Trends Biochem. Sci. 24, 34–36 [DOI] [PubMed] [Google Scholar]
- 26. Marden M. C., Dewilde S., Kiger L., Hamdane D., Uzan J., Burmester T., Hankeln T., Moens L., Baudin-Creuza V., Celier C., Wajcman H. (2007) Gene 398, 35–41 [DOI] [PubMed] [Google Scholar]
- 27. Nicholas K. B., Nicholas H. B., Jr., Deerfield D. W., 2nd (1997) EMBNEWS 4, 14 [Google Scholar]
- 28. Abascal F., Zardoya R., Posada D. (2005) Bioinformatics 21, 2104–2105 [DOI] [PubMed] [Google Scholar]
- 29. Huelsenbeck J. P., Ronquist F. (2001) Bioinformatics 17, 754–755 [DOI] [PubMed] [Google Scholar]
- 30. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 31. Hamdane D., Kiger L., Dewilde S., Green B. N., Pesce A., Uzan J., Burmester T., Hankeln T., Bolognesi M., Moens L., Marden M. C. (2003) J. Biol. Chem. 278, 51713–51721 [DOI] [PubMed] [Google Scholar]
- 32. Uzan J., Dewilde S., Burmester T., Hankeln T., Moens L., Hamdane D., Marden M. C., Kiger L. (2004) Biophys. J. 87, 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dewilde S., Kiger L., Burmester T., Hankeln T., Baudin-Creuza V., Aerts T., Marden M. C., Caubergs R., Moens L. (2001) J. Biol. Chem. 276, 38949–38955 [DOI] [PubMed] [Google Scholar]
- 34. Terwilliger N. B., Ryan M. C. (2006) Biol. Bull. 210, 38–50 [DOI] [PubMed] [Google Scholar]
- 35. Liong E. C., Dou Y., Scott E. E., Olson J. S., Phillips G. N., Jr. (2001) J. Biol. Chem. 276, 9093–9100 [DOI] [PubMed] [Google Scholar]
- 36. Goodman S. H., Carvey M. J. (1990) Cell Tissue Res. 260, 495–505 [Google Scholar]
- 37. Farazi T. A., Waksman G., Gordon J. I. (2001) J. Biol. Chem. 276, 39501–39504 [DOI] [PubMed] [Google Scholar]
- 38. Resh M. D. (1999) Biochim. Biophys. Acta 1451, 1–16 [DOI] [PubMed] [Google Scholar]
- 39. Towler D. A., Adams S. P., Eubanks S. R., Towery D. S., Jackson-Machelski E., Glaser L., Gordon J. I. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 2708–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zha J., Weiler S., Oh K. J., Wei M. C., Korsmeyer S. J. (2000) Science 290, 1761–1765 [DOI] [PubMed] [Google Scholar]
- 41. Murray D., Ben-Tal N., Honig B., McLaughlin S. (1997) Structure 5, 985–989 [DOI] [PubMed] [Google Scholar]
- 42. Peitzsch R. M., McLaughlin S. (1993) Biochemistry 32, 10436–10443 [DOI] [PubMed] [Google Scholar]
- 43. Bonamore A., Farina A., Gattoni M., Schininà M. E., Bellelli A., Boffi A. (2003) Biochemistry 42, 5792–5801 [DOI] [PubMed] [Google Scholar]
- 44. Liu C., He Y., Chang Z. (2004) Biochem. Biophys. Res. Commun. 316, 1163–1172 [DOI] [PubMed] [Google Scholar]
- 45. Ramandeep, Hwang K. W., Raje M., Kim K. J., Stark B. C., Dikshit K. L., Webster D. A. (2001) J. Biol. Chem. 276, 24781–24789 [DOI] [PubMed] [Google Scholar]
- 46. Rinaldi A. C., Bonamore A., Macone A., Boffi A., Bozzi A., Di Giulio A. (2006) Biochemistry 45, 4069–4076 [DOI] [PubMed] [Google Scholar]
- 47. Di Giulio A., Bonamore A. (2008) Methods Enzymol. 436, 239–253 [DOI] [PubMed] [Google Scholar]
- 48. Hankeln T., Ebner B., Fuchs C., Gerlach F., Haberkamp M., Laufs T. L., Roesner A., Schmidt M., Weich B., Wystub S., Saaler-Reinhardt S., Reuss S., Bolognesi M., De Sanctis D., Marden M. C., Kiger L., Moens L., Dewilde S., Nevo E., Avivi A., Weber R. E., Fago A., Burmester T. (2005) J. Inorg. Biochem. 99, 110–119 [DOI] [PubMed] [Google Scholar]
- 49. Freitas T. A., Hou S., Alam M. (2003) FEBS Lett. 552, 99–104 [DOI] [PubMed] [Google Scholar]
- 50. Muller F. L., Lustgarten M. S., Jang Y., Richardson A., Van Remmen H. (2007) Free Radic. Biol. Med. 43, 477–503 [DOI] [PubMed] [Google Scholar]
- 51. Burmester T., Hankeln T. (1999) Mol. Biol. Evol. 16, 1809–1811 [DOI] [PubMed] [Google Scholar]
- 52. Burmester T., Klawitter S., Hankeln T. (2007) Insect Mol. Biol. 16, 133–142 [DOI] [PubMed] [Google Scholar]
- 53. Burmester T., Storf J., Hasenjäger A., Klawitter S., Hankeln T. (2006) FEBS J. 273, 468–480 [DOI] [PubMed] [Google Scholar]
- 54. Hankeln T., Jaenicke V., Kiger L., Dewilde S., Ungerechts G., Schmidt M., Urban J., Marden M. C., Moens L., Burmester T. (2002) J. Biol. Chem. 277, 29012–29017 [DOI] [PubMed] [Google Scholar]
- 55. Hankeln T., Klawitter S., Krämer M., Burmester T. (2006) J. Insect. Physiol. 52, 701–710 [DOI] [PubMed] [Google Scholar]
- 56. Burmester T., Hankeln T. (2007) J. Insect Physiol. 53, 285–294 [DOI] [PubMed] [Google Scholar]
- 57. Gleixner E., Abriss D., Adryan B., Kraemer M., Gerlach F., Schuh R., Burmester T., Hankeln T. (2008) FEBS J. 275, 5108–5116 [DOI] [PubMed] [Google Scholar]
- 58. Bergtrom G. (1977) Insect Biochem. 7, 313–316 [Google Scholar]
- 59. Osmulski P. A., Leyko W. (1986) Comp. Biochem. Physiol. B 85, 701–722 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.