Abstract
Among bacterial species demonstrated to have protein O-glycosylation systems, that of Bacteroides fragilis and related species is unique in that extracytoplasmic proteins are glycosylated at serine or threonine residues within the specific three-amino acid motif D(S/T)(A/I/L/M/T/V). This feature allows for computational analysis of the proteome to identify candidate glycoproteins. With the criteria of a signal peptidase I or II cleavage site or a predicted transmembrane-spanning region and the presence of at least one glycosylation motif, we identified 1021 candidate glycoproteins of B. fragilis. In addition to the eight glycoproteins identified previously, we confirmed that another 12 candidate glycoproteins are in fact glycosylated. These included four glycoproteins that are predicted to localize to the inner membrane, a compartment not previously shown to include glycosylated proteins. In addition, we show that four proteins involved in cell division and chromosomal segregation, two of which are encoded by candidate essential genes, are glycosylated. To date, we have not identified any extracytoplasmic proteins containing a glycosylation motif that are not glycosylated. Therefore, based on the list of 1021 candidate glycoproteins, it is likely that hundreds of proteins, comprising more than half of the extracytoplasmic proteins of B. fragilis, are glycosylated. Site-directed mutagenesis of several glycoproteins demonstrated that all are glycosylated at the identified glycosylation motif. By engineering glycosylation motifs into a naturally unglycosylated protein, we are able to bring about site-specific glycosylation at the engineered sites, suggesting that this glycosylation system may have applications for glycoengineering.
Keywords: Bacteria, Cell Division, Glycoprotein, Glycosylation, Membrane Proteins, Bacteroides, Glycosylation Motif
Introduction
Several bacterial species are known to have general protein glycosylation systems where multiple proteins of the organism are modified with glycans. The best studied general bacterial glycosylation system is the N-glycosylation system of Campylobacter jejuni, where more than 65 different extracytoplasmic proteins have been shown to be glycosylated at asparagine residues (1–3) contained within the extended motif (D/E)YNX(S/T) (where X is not equal to P) (4). A similar N-glycosylation system has recently been described in the related epsilon proteobacterial species Helicobacter pullorum (5). Within the last few years, general O-glycosylation systems have been described in Neisseria sp. (6) and Bacteroides sp. (7), and an O-mannosylation system was described previously in several Actinomycetes species (reviewed in Ref. 8). The O-mannosylation system of the Actinomycetes is different from the general glycosylation systems in other bacteria in that mannosylation occurs on the cytoplasmic membrane by the sequential addition of mannose from polyprenol-activated mannose residues to proteins. In contrast, the general glycosylation systems described in Gram-negative bacteria require synthesis of the glycan chain on the lipid carrier undecaprenyl pyrophosphate at the cytoplasmic face of the inner membrane. This assembled glycan chain is then flipped into the periplasm and added en bloc to various extracytoplasmic proteins by the action of oligosaccharyltransferases (Ref. 9; reviewed in Ref. 10).
A common feature of the general glycosylation systems of Gram-negative bacteria is that proteins of extracytoplasmic locations are targeted for glycosylation including those that localize to the outer membrane, periplasm, and outer surface (1, 6, 7). The predicted functions of many of these proteins suggest that they perform important roles for the organism, and abrogation of the N-glycosylation system of C. jejuni reduces the ability of the organism to colonize the mouse intestine and to invade into INT407 cells. However, this glycosylation mutant does not demonstrate an in vitro growth defect (11). In addition, proteins encoded by candidate essential genes have not been demonstrated to be glycosylated in bacteria.
The general O-glycosyation system of Bacteroides fragilis and related species differs from the systems of other bacteria in several regards. The most profound difference is that a defect in protein glycosylation not only abrogates the ability of the organism to competitively colonize the mammalian intestine, but it also has an effect on the in vitro growth of the organism (7). Similar to systems in other Gram-negative bacteria, proteins must be secreted out of the cytoplasm to become glycosylated; however, in B. fragilis, glycosylation occurs at the specific glycosylation motif D(S/T)(A/I/L/M/T/V). Our previous analysis identified eight glycoproteins of B. fragilis whose putative functions suggested their involvement in important cellular processes, but none of these were encoded by candidate essential genes.
The purpose of this study was to obtain a more comprehensive analysis of the number and types of proteins that are glycosylated in B. fragilis to better understand the importance of protein glycosylation to the Bacteroides and why a glycosylation defect impacts the physiology of the organism. Because proteins of B. fragilis are glycosylated at a specific motif, we were able to use a computational approach to predict the number and types of proteins that are glycosylated in this organism and then test the glycosylation status of some of these proteins experimentally. In addition, we began to test the utility of this glycosylation system for glycoengineering applications by determining whether the addition of a glycosylation motifs to a naturally unglycosylated protein leads to its glycosylation.
EXPERIMENTAL PROCEDURES
All of the oligonucleotides used in this study are listed in supplemental Table S1.
Bioinformatics Analyses
The genome of B. fragilis NCTC 9343 (12) comprises both the 5,205,140-bp chromosome and a 36,560-bp resident plasmid and contains 4,231 genes presumed to encode proteins. The Bacteroides thetaiotaomicron VPI-5482 genome (13) encompasses 4,816 genes presumed to encode proteins that are present on a 6,260,361-bp chromosome and a 33,038-bp plasmid. The proteome of each species was obtained from the Kyoto Encyclopedia of Genes and Genomes (14) site, and each was used to create a proteome-specific BLAST (15, 16) database using formatdb (all of the various NCBI utilities used were contained in release 2.2.23 of the Windows version of the BLAST executables and were downloaded from the NCBI ftp site).
Each of the B. fragilis proteins was compared with the B. thetaiotaomicron database using the NCBI blastall program in blastp mode with settings directing the use of locally optimal Smith-Waterman alignments (−s T) and low complexity filtering during the lookup stage only (−F “m S”) (17), and the identity of the B. thetaiotaomicron protein producing the “best hit” (lowest E-value) was retained. The opposite operation was also performed, comparing the B. thetaiotaomicron proteins to the B. fragilis database. These two best hit lists were compared, and orthology was assumed where there was reciprocity as to the identity of the best hit. This B. fragilis-B. thetaiotaomicron reciprocal best hit list was compared with the list of B. thetaiotaomicron candidate essential genes (supplemental Table S2 in Ref. 18) to identify candidate essential B. fragilis genes.
The Linux version 1.0a of the stand alone program LipoP (19) was used to detect members of the B. fragilis 9343 proteome likely to be extracytoplasmic because of the presence of signal peptidase I or II (SpI or SpII)2 cleavage sites. Transmembrane segments were predicted using TMHMM version 2.0c for Linux (20, 21). The proteome was also scanned via a Perl regular expression for sequences containing the glycosylation motif D(S/T)(A/I/L/V/M/T) (7). Custom Perl scripts were also used throughout to facilitate efficient handling of the large datasets.
Each B. fragilis 9343 protein sequence was also compared with the profile hidden Markov models of the Pfam-A and Pfam-B entries of version 24.0 of the Pfam database (22) using version 3.0b3 of the HMMER program (23). Initially, the Pfam database was searched with a maximum expectation value (E-value) cut-off of 1.0; the results returned were then parsed for matches with an E-value equal to or less than 1e-03. These more stringent results were used as the basis for grouping of proteins into functional categories.
Proteins with a match to Pfam families SHNi-TPR (PF10516), TPR_1 (PF00515), TPR_2 (PF07719), TPR_3 (PF07720), or TPR_4 (PF07721) were classified as TPR-containing proteins. Histidine kinase proteins were detected based on the presence of one or more motif defined by the six families of Pfam clan CL0025 (His Kinase A (phospho-acceptor) domain).
The SusC-like proteins of B. fragilis were detected by using the amino acid sequence of the B. thetaiotaomicron VPI-5482 SusC (BT_3702) as a blastp query against the B. fragilis NCTC 9343 database. The identity of B. fragilis proteins where a high scoring segment pair exceeded an alignment length of 850 amino acids was retained (there is a sharp drop-off after this point; the lowest qualifying alignment is for the protein encoded by BF3307, which aligns with the B. thetaiotaomicron SusC protein over a span of 852 amino acids; E-value 1e-87). Protease and peptidase proteins were collected from the B. fragilis proteome on the basis of the genome annotation; all proteins with a description containing “protease” or “peptidase” were included.
Bacterial Strains and Growth Conditions
Escherichia coli strain DH5α containing recombinant plasmids was grown in L broth or on L agar plates containing ampicillin (100 μg/ml). B. fragilis strain NCTC 9343, the isogenic mutant Δgmd-fclΔfkp, or 9343 containing pCMF6-based plasmids were grown anaerobically in basal medium or on brain-heart infusion plates supplemented with hemin (50 μg/ml) and vitamin K1 (0.5 μg/ml), with gentamicin (200 μg/ml) and erythromycin (5 μg/ml) added where appropriate.
Expression of His-tagged Proteins in B. fragilis
The coding regions and ribosome-binding sites of candidate glycoproteins were amplified from the B. fragilis chromosome, digested with BamHI or BglII, and ligated into the BamHI site of pCMF6 (7). The recombinant proteins are modified by the addition of the amino acids GSH10 at the C terminus. The resulting plasmids were transferred from E. coli to wild type B. fragilis or the isogenic mutant Δgmd-fclΔfkp (24) by conjugation.
Purification of His-tagged Proteins from B. fragilis
Cultures (1.5 liters) of B. fragilis harboring plasmids encoding His-tagged proteins were grown to the stationary phase and harvested. His-tagged proteins were purified using 0.5 ml of nickel-nitrilotriacetic acid-agarose resin (Invitrogen). The cells were resuspended in buffers containing an EDTA-free Complete Protease Inhibitor Tablet (Roche Applied Science) and lysed by sonication. Membrane-associated proteins were purified under denaturing conditions, washed with 16 ml of buffer at pH 8.0, 6.0, and 5.3, and eluted with 50 mm EDTA at pH 4.0. Soluble proteins were purified under native conditions with 16-ml washes containing 0 and 20 mm imidazole, a final wash with 16 ml of buffer containing 1 mm EDTA, and eluted with 250 mm imidazole.
Antibodies and Western Blots
Monoclonal antibodies to the His tag were purchased from Genscript and Invitrogen. The anti-glycan antiserum was described previously (7). Western blots were developed with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium phosphatase substrate (KPL) or the WesternBreeze chemiluminescent immunodetection system (Invitrogen).
Site-directed Mutagenesis
Site-directed mutagenesis of protein coding sequences in plasmid pCMF6 was carried out with the QuikChange XL kit (Stratagene) with an elongation time of 12 min. All of the mutations were confirmed by sequencing.
RESULTS
Catalogue of Candidate B. fragilis Glycoproteins
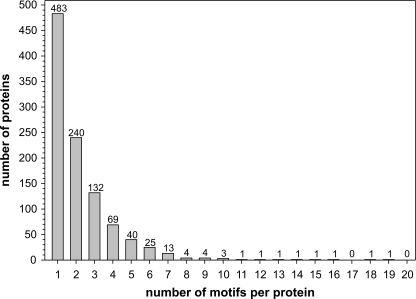
Our previous method of identifying glycoproteins in B. fragilis relied on lectin purification, followed by two-dimensional gel separation and MS/MS identification. This technique detects glycoproteins that are abundantly expressed from in vitro culture. To obtain a complete list of the proteins that may be glycosylated in B. fragilis regardless of expression level, we used a bioinformatic approach. These analyses are based on the fact that proteins must be secreted out of the cytoplasm to be glycosylated and that glycans are added at the three-amino acid glycosylation motif D(S/T)(A/I/L/M/T/V). Using custom Perl scripts, we analyzed the 4231 annotated proteins encoded by the B. fragilis 9343 genome for those containing SpI or SpII cleavage sites. This resulted in a list of 1120 secreted proteins, of which 656 contain one or more glycosylation motif(s) (supplemental Table S2) and were therefore considered candidate glycoproteins. In addition, the TMHMM program was used to identify proteins with membrane-spanning domains. This program identified an additional 762 proteins with transmembrane regions that were not included in the list of proteins containing SpI or SpII cleavage sites. Of these 762 proteins, 365 have at least one glycosylation motif and therefore are candidate glycoproteins (supplemental Table S3). Of the total 1021 candidate glycoproteins, 483 contain a single glycosylation motif, 538 have two or more motifs, and 97 have five or more motifs (Fig. 1 and supplemental Tables S2 and S3).
FIGURE 1.
Number of glycosylation motifs in each of the 1021 candidate glycoproteins. Number of candidate glycoproteins with corresponding numbers of sites matching the glycosylation motif.
Of the eight previously confirmed glycoproteins of B. fragilis, two are predicted protease/peptidases; two have predicted periplasmic chaperone functions; two have TPR domains, one of which meets the criteria set out in the methods section (BF2494); and two are outer surface lipoproteins (Table 1). Both the protease/peptidases and the TPR-containing proteins are highly represented, relative to their total numbers, on the list of candidate glycoproteins. The B. fragilis 9343 genome encodes 44 secreted protease/peptidase type proteins. Of those, 38 have a glycosylation motif. The same is true of secreted proteins containing TPR domains for which 13 of 14 have glycosylation motifs, one previously proven experimentally to be glycosylated. Another highly represented group of proteins on the list are histidine kinases. 37 of the 42 secreted products predicted to be histidine kinases have glycosylation motifs. The histidine kinases are among those candidate glycoproteins containing the highest numbers of glycosylation motifs/protein, collectively they contain 136 glycosylation motifs, and average 3.7 glycosylation motifs/protein. Because these proteins typically have regions that span the inner membrane as well as segments within the periplasm and cytoplasm, we used the transmembrane prediction program TMHMM to determine how many of these glycosylation motifs were likely contained on segments of the protein predicted to be in the periplasm where they would be accessible to the glycosylation machinery. This analysis revealed that 86 of the 136 glycosylation sites are contained on predicted periplasmic regions.
TABLE 1.
Glycoproteins of B. fragilis
| Proteina | No. of amino acids | Motif(s)b | Signalc | Locationd | Sequence features |
|---|---|---|---|---|---|
| Newly confirmed glycoproteins | |||||
| BF0066 | 293 | DSI 128–130; DTI 230–232 | I | Inner membrane | FtsX cell division ABC transporter |
| BF0252* | 246 | DTL 43–45 | I | Inner membrane | FtsQ cell division protein |
| BF0259* | 699 | DTM 83–85; DTA 180–182; DSI 194–196 | I | Inner membrane | FtsI cell division peptidoglycan transpeptidase |
| BF0586 | 285 | DSL 32–34 | II | Outer membrane | SMB-prok_B motif, chromosomal segregation |
| BF1848 | 370 | DSV 39–41; DSL 75–77; DTI 173–175, 179–181, 242–244 | I | (uncertain) | OmpA motif |
| BF2541 | 784 | DSV 31–33; DSI 365–367 | I | Outer membrane | TonB-dependent receptor, β-barrel |
| BF3195 | 394 | DSL 130–2 | I | Periplasm | Aminopeptidase |
| BF3237 | 845 | DSI 42–44, 134–136; DTA 76–78; DTL 404–406 | I | (uncertain) | Peptidase |
| BF3443 | 282 | DSI 19–21; DSI 49–51; DSM 85–87 | II | Outer membrane | Lipoprotein |
| BF3444 | 1016 | DSL 313–315; DST 597–599; DSA 897–899 | I | Outer membrane | SusC-like TonB-dependent receptor |
| BF3556 | 445 | DSI 299–301 | I | (uncertain) | OmpA motif |
| BF4153 | 517 | DSL 149–151; DSV 224–226 | I | Inner membrane | Sensor histidine kinase |
| Previously confirmed glycoproteins | |||||
| BF0447 | 169 | DSL 76–78 | I | Periplasm | Similar to E. coli chaperone Skp |
| BF0522 | 210 | DSV 52–54 | I | Outer membrane, facing surface | |
| BF0935 | 939 | DSI 362–324, 667–669; DTA 834–836 | I | Periplasm | Zinc endopeptidase (M16 family) |
| BF0994 | 712 | DST 229–231; DSA 326–328; DSV 358–360,614–616; DSI 380–382 | II | Periplasm | Proline cis-trans-isomerase (parvulin family) |
| BF2334 | 245 | DSV 58–60 | I | Periplasm | TPR domains (residues 59–92 and 134–167) |
| BF2494 | 403 | DTL 86–88; DTI 177–179; DTA 230–232 | I | Periplasm | TPR domains (residues 181–350) |
| BF3567 | 623 | DSI 139–141, 332–334; DTV 206–208 | I | Outer membrane, facing surface | |
| BF3918 | 677 | DTT 37–39; DSV 111–113; DTV 297–299; DSI 557–559 | I | Outer membrane, facing periplasm | Zinc endopeptidase (M13 family) |
There is also a functional group that is notably under-represented in the list, the SusC orthologues. SusC-like proteins are TonB-dependent outer membrane β-barrel proteins that are involved in the transport of nutrients into the periplasm (25–27). Only 17 of the 56 predicted SpI-containing SusC-like proteins have sites matching the glycosylation motif, and 14 of these have only one motif. The bioinformatic analyses reveal that a glycosylation motif occurs on average 2.8 times/1000 amino acids in secreted proteins. Based on the large size of the SusC-like proteins (averaging 1076 amino acids), these proteins are not only under-represented in the list of candidate glycoproteins but also in the number of motifs per protein; the number of glycosylation sites/1000 amino acids of SusC-like proteins is only 0.4.
A list of 325 candidate essential genes of the related species B. thetaiotaomicron VPI-5482 was generated previously (18). By comparing the genomes of B. thetaiotaomicron VPI-5482 and B. fragilis 9343 using reciprocal best hit analysis, 252 of the 325 candidate essential genes of B. thetaiotaomicron are also present in B. fragilis 9343. Of these 252 putative essential genes of B. fragilis, 68 encode proteins with predicted transmembrane region(s) and/or SpI or SpII cleavage sites, 43 of which have at least one glycosylation motif (supplemental Tables S2 and S3).
Experimental Analysis of Candidate Glycoproteins
We next performed experiments to determine whether certain candidate glycoproteins are in fact glycosylated. We chose to analyze particular and varied types of proteins, including those whose orthologues in B. thetaiotaomicron are encoded by candidate essential genes, those that are predicted to be in the inner membrane because no glycoproteins localizing to this compartment have been identified previously in B. fragilis, additional putative proteases/peptidases, which are highly represented on the list of candidate glycoproteins, and lipoproteins. The glycosylation state of each protein was analyzed as described previously (7). The genes were overexpressed in wild type B. fragilis and Δgmd-fclΔfkp from a vector that creates a fusion protein with a C-terminal His tag. The sizes of the resulting His-tagged proteins from each of these background strains were compared by Western blots using an α-His tag antibody. Δgmd-fclΔfkp is unable to synthesize the fucosylated glycan that is added to glycoproteins (24), and therefore the molecular masses of glycoproteins are smaller for Δgmd-fclΔfkp than they are for wild type bacteria (7).
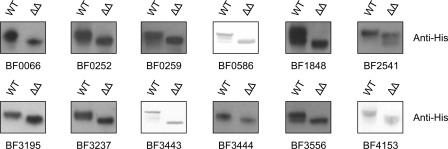
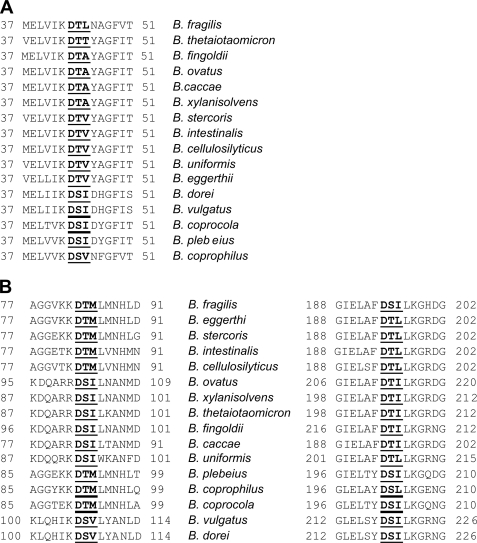
Using this technique, we experimentally confirmed that four proteins with putative functions in cell division and/or chromosomal segregation are glycosylated (Fig. 2 and Table 1). BF0066, BF0252, and BF0259 encode products similar to cell division proteins FtsX, FtsQ, and FtsI of E. coli, respectively, and the product of BF0586 has a putative function in chromosomal segregation. FtsQ and FtsI are essential in E. coli (28), and orthologues of BF0252 (BT3446) and BF0259 (BT3453) are encoded by candidate essential genes in B. thetaiotaomicron (18). All three Fts proteins are predicted to be localized to the inner membrane, and BF0066 has four predicted transmembrane regions. Orthologous proteins in E. coli are similarly inserted in the inner membrane (28–30). These are the first predicted inner membrane proteins shown to be glycosylated in B. fragilis and the first confirmed glycoproteins encoded by candidate essential genes. The glycosylation motif in BF0252 and two of the three motifs in BF0259 are conserved in the orthologous proteins of all 15 other Bacteroides species identified by Blastp searching (the overall similarity of these proteins ranges from 74 to 92%). The precise identity of the amino acids making up the motifs varies considerably in different species (Fig. 3). The conservation of these glycosylation motifs in orthologous proteins suggests that glycosylation at these sites is likely important to these molecules.
FIGURE 2.
Confirmation of twelve new glycoproteins. Whole cell lysates of B. fragilis WT and Δgmd-fclΔfkp (ΔΔ) expressing His-tagged glycoprotein candidates or His-tagged proteins purified from the same strains, separated by SDS-PAGE, blotted, and probed with an anti-His tag antibody. Some blots were developed colorimetrically, whereas others were analyzed using chemiluminescence.
FIGURE 3.
Alignments of the conserved glycosylation motifs of BF0252 (FtsQ) (A) and BF0259 (FtsI) (B) in orthologous proteins of all Bacteroides species in the database. Species are listed in order of the similarity of their orthologue to the B. fragilis protein. The glycosylation sites are in bold and underlined, and the location of the region within the orthologous protein is listed.
Because SusC-like proteins are under-represented in the list of putative glycoproteins, we analyzed the glycosylation state of one of the SusC orthologues that contains glycosylation motifs (BF3444) and one additional protein predicted to be an outer membrane β-barrel protein (BF2541) to determine whether β-barrel outer membrane proteins may be excluded from glycosylation. These analyses revealed that both of these β-barrel outer membrane proteins are glycosylated (Fig. 2). We used three different methods to predict the position of transmembrane β-strand segments for the 17 candidate SusC glycoproteins and then determined whether the glycosylation motifs of these proteins were contained in segments predicted to be external to membrane-spanning regions. Of the 21 glycosylation motifs contained within these SusC orthologues, B2TMR (31) and PRED-TMBB (32) each predicted all sites except one (in BF1816 and in BF4169, respectively) to be outside of a transmembrane segment, whereas TMBETA-NET (33) predicted all 21 motifs to be in nonburied segments.
Six other proteins with predicted inner membrane, periplasmic, and outer membrane localizations were also shown to be glycosylated (Fig. 2 and Table 1). Among these glycoproteins is a sensor histidine kinase and two putative peptidases, both categories of proteins with high relative representation in the list of candidate glycoproteins. Therefore, all 12 of the candidate glycoproteins that we tested, despite differences in extracytoplasmic localizations, in their putative functions, and in their over- or under-representation in the list of candidate glycoproteins, were confirmed to be glycosylated. These data suggest that it is likely that many or most of the 1021 candidate glycoproteins of B. fragilis are in fact glycoproteins.
Analysis of Glycosylation Sites
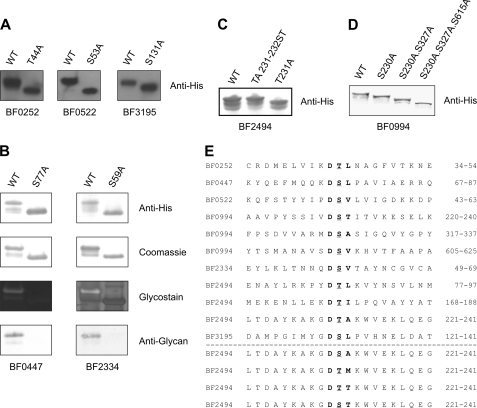
Because the B. fragilis glycosylation motif contains either an Ser or Thr at the second position and an Ala, Ile, Leu, Met, Thr, or Val at the third position, there are 12 different three-amino acid sequences that comprise the glycosylation motif. Previously, we experimentally proved that glycoprotein BF2494 was glycosylated at three different threonine residues contained in the glycosylation sites DTL, DTI, and DTA. Previous alteration of the glycosylation site Thr-231 of BF2494 from DTA to DSA, DTM, or DTT also resulted in glycosylation of that site (7), confirming that each of these three sequences is also a target for glycosylation as predicted. To expand these analyses, we performed additional site-directed mutagenesis to confirm that the site of glycosylation of various proteins was at the expected glycosylation site. Seven of the confirmed glycoproteins of B. fragilis have a single site matching the glycosylation motif (Table 1). We chose five of these proteins (BF0252, BF0447, BF0522, BF2334, and BF3195) for site-directed mutagenesis to confirm that the predicted glycosylation motif was in fact the site of glycosylation. Mutation of each of the putative glycosylation sites of each of these five proteins by replacement of the Ser or Thr at the second position with an Ala, abrogated glycosylation of each of these proteins (Fig. 4, A and B). These analyses increased the number of confirmed glycosylation motifs, adding DSL and DSV to the list of experimentally confirmed sites. In addition, we modified the DTA glycosylation site of BF2494 at amino acid 231 to DST and experimentally confirmed that this site is also used for glycosylation as expected (Fig. 4C). To further support our previous finding that multiple glycosylation motifs of a protein are glycosylated (7), we altered three of the glycosylation sites of BF0994 in series by converting the Ser to Ala and found that all three of these sites are glycosylated (Fig. 4D). Each of the experimentally confirmed glycosylation motifs is shown in Fig. 4E.
FIGURE 4.
Analysis of glycosylation sites. A, whole cell lysates of B. fragilis expressing WT and mutant His-tagged proteins or purified His-tagged proteins, separated by SDS-PAGE, blotted, and probed with antibody to the His tag. B, WT and mutant His-tagged proteins purified from B. fragilis, separated by SDS-PAGE, and stained with Coomassie Blue or Pro-Q Emerald Glycostain or blotted and probed with antibody to the His tag or anti-glycan antiserum. C, whole cell lysates of B. fragilis expressing WT BF2494 and two modifications of the glycosylation motif at amino acid 230–232, separated by SDS-PAGE, blotted, and probed with antibody to the His tag. D, whole cell lysates of B. fragilis expressing WT BF0994 and sequential modifications of the glycosylation motifs at the indicated amino acids, separated by SDS-PAGE, blotted, and probed with antibody to the His tag. E, alignment of the protein sequences surrounding all the confirmed glycosylation sites. Above the line are all the naturally occurring sites confirmed by mutation, and below the line are modifications of the 230–232 glycosylation motif of BF2494 that are confirmed glycosylation sites. Glycosylated residues are underlined, and the glycosylation motifs are in bold type.
Next, we determined the frequency of the 12 different three-amino acid sequences of the glycosylation motif within the 1021 candidate glycoproteins and compared it with the frequency of these motifs in nonsecreted proteins. The overall trend demonstrated that the glycosylation motifs most frequently present in cytoplasmic proteins were also most frequent in the candidate glycoproteins (supplemental Fig. S1). Regardless of the amino acid at the second position, Ile, Leu, and Val were most frequently present at the third position, and motifs with Met at the third position were the rarest in both classes of proteins, likely reflecting the low number of Mets in proteins compared with the other five amino acids at the third position of the motif. Seven of the eight naturally occurring glycosylation sites that we have confirmed end in Ile, Leu, or Val (Fig. 4).
Glycosylation of Engineered Motifs
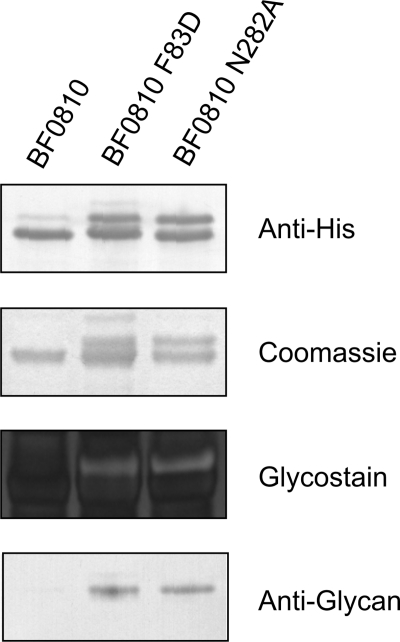
The presence of a defined glycosylation motif, combined with the fact that all secreted proteins with a site matching the glycosylation motif that we have analyzed are glycosylated, suggests that proteins can be targeted for glycosylation by adding a secretion signal and a glycosylation site. Therefore, we set out to determine whether the addition of a glycosylation site to a naturally unglycosylated secreted protein would lead to its glycosylation. BF0810 encodes a 449-amino acid protein that is a putative α-fucosidase with a predicted SpI cleavage site and no predicted transmembrane helices. This protein does not contain a glycosylation motif and is not glycosylated (Fig. 5). Two different sites of BF0810 were separately modified to conform to the glycosylation motif. The alteration F83D near the N terminus of the protein created the glycosylation site DTA from the existing FTA site, and another modification of N282A in the second half of the protein created the glycosylation site DTA from DTN. Analysis of each of these modified proteins by Coomassie staining, glycostaining, and reactivity with anti-glycan serum demonstrated that both of these modifications brought about glycosylation of this protein (Fig. 5).
FIGURE 5.
Analysis of glycosylation status of protein containing engineered glycosylation motifs. Wild type and modified BF0810-His proteins purified from wild type B. fragilis, separated by SDS-PAGE, and stained with Coomassie Blue or Pro-Q Emerald Glycostain or blotted and probed with antibody to the His tag or anti-glycan antiserum.
DISCUSSION
In this paper we build on our initial description of the general O-glycosylation system of B. fragilis by further investigating the scope and nature of protein glycosylation in this organism. We found that more than half of the secreted proteins contain glycosylation motifs. Based on our finding that all of the candidate glycoproteins analyzed were in fact confirmed to be glycosylated, we predict that there are hundreds of proteins glycosylated in this organism. We experimentally showed that an additional 12 proteins are glycosylated, which more than doubles the number of proven glycoproteins to a total of 20. These confirmed B. fragilis glycoproteins now include molecules from all of the extracytoplasmic locations in the cell, with inner membrane glycoproteins being described in this paper for the first time. Also for the first time, we have identified glycoproteins that are likely to be essential to the organism, namely FtsI (BF0259) and FtsQ (BF0252). The glycosylation motif in FtsQ and two of the three motifs in FtsI are conserved in the orthologous proteins of all 15 other Bacteroides species analyzed, suggesting that glycosylation confers an important property to these proteins. Analysis of the conservation of glycosylation motifs in orthologous proteins may be indicative of sites to which the addition of glycans confers an important property such as stabilization, proper localization, or functional properties to the protein. Conversely, it is also likely that glycosylation of some proteins has no effect on the protein and is dispensable.
We demonstrated previously that mutants with defective protein glycosylation were impaired for in vitro growth and could not competitively colonize the mammalian intestine. This study casts light on the significance of the glycosylation system to these bacteria in regard to both the large number of proteins that are likely glycosylated and the types of proteins that are glycosylated. The confirmed glycoproteins have putative roles in crucial cell processes including cell division, chromosomal segregation, signaling processes, chaperone functions, protein-protein interactions, and peptidase/protease reactions.
We observed that the SusC-like β-barrel proteins are under-represented in the list of candidate glycoproteins and have fewer motifs/1000 amino acids than average. When we investigated the glycosylation status of one of the SusC-like proteins with glycosylation motifs (BF3444), we found that it was glycosylated, as was another protein predicted to adopt a β-barrel structure (BF2541). The finding that the few glycosylation motifs in these proteins are predicted to be in nonburied segments suggests that the low frequency of motifs in the β-barrel proteins reflects the fact that significant portions of these proteins are contained in the outer membrane.
In this paper, we confirmed by experimentation that the three-residue motif D(S/T)(A/I/L/M/T/V) is the site of glycosylation of five additional proteins, as suggested by our previous work (7). In N-glycosylation, the NX(S/T) motif forms an Asx turn with the side chain carbonyl of the Asn residue hydrogen bonded to the backbone amide of the Ser or Thr. This conformation is believed to be important both for recognition of the site and for catalysis of the glycosylation reaction, because the hydrogen bond increases the (usually poor) nucleophilicity of the Asn side chain (34). This precedent suggests that residues of the O-glycosylation motif in B. fragilis could be involved in catalysis as well as recognition. The methyl group-containing amino acid required at the third position is unreactive and therefore is likely to play a role only in recognition of the site, whereas the leading Asp residue may play a catalytic role.
We have now investigated 20 proteins that are secreted from the cytoplasm and have one or more glycosylation motifs, and all are proven to be glycosylated. Thus, secretion and the presence of a motif appear to be necessary and may well be sufficient for glycosylation of nonmembrane-spanning protein regions. Consistent with this finding, the introduction of a glycosylation motif in either of two locations was sufficient to allow modification of a secreted protein that is not naturally glycosylated (BF0810). This demonstrates that it may be feasible to use the O-glycosylation system of B. fragilis to precisely target various sites of proteins for O-glycosylation to create engineered glycoproteins.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AI067711.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Fig. S1.
- Sp
- signal peptidase
- TPR
- tetratricopeptide.
REFERENCES
- 1. Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., Jarrell H. C., Cadotte N., St. Michael F., Aberg E., Szymanski C. M. (2002) J. Biol. Chem. 277, 42530–42539 [DOI] [PubMed] [Google Scholar]
- 2. Nothaft H., Szymanski C. M. (2010) Nat. Rev. Microbiol. 8, 765–778 [DOI] [PubMed] [Google Scholar]
- 3. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Hojrup P., Packer N. H., Larsen M. R., Cordwell S. J. (April 1, 2010) Mol. Cell Proteomics 10.1074/mcp.M000031-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kowarik M., Young N. M., Numao S., Schulz B. L., Hug I., Callewaert N., Mills D. C., Watson D. C., Hernandez M., Kelly J. F., Wacker M., Aebi M. (2006) EMBO J. 25, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jervis A. J., Langdon R., Hitchen P., Lawson A. J., Wood A., Fothergill J. L., Morris H. R., Dell A., Wren B., Linton D. (2010) J. Bacteriol. 192, 5228–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vik A., Aas F. E., Anonsen J. H., Bilsborough S., Schneider A., Egge-Jacobsen W., Koomey M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fletcher C. M., Coyne M. J., Villa O. F., Chatzidaki-Livanis M., Comstock L. E. (2009) Cell 137, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Espitia C., Servin-Gonzalez L., Mancilla R. (2010) Mol. Biosyst. 6, 775–781 [DOI] [PubMed] [Google Scholar]
- 9. Castric P. (1995) Microbiology 141, 1247–1254 [DOI] [PubMed] [Google Scholar]
- 10. Hug I., Feldman M. F. (September 24, 2010) Glycobiology 10.1093/glycob/cwq148 [DOI] [PubMed] [Google Scholar]
- 11. Szymanski C. M., Burr D. H., Guerry P. (2002) Infect. Immun. 70, 2242–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cerdeño-Tárraga A. M., Patrick S., Crossman L. C., Blakely G., Abratt V., Lennard N., Poxton I., Duerden B., Harris B., Quail M. A., Barron A., Clark L., Corton C., Doggett J., Holden M. T., Larke N., Line A., Lord A., Norbertczak H., Ormond D., Price C., Rabbinowitsch E., Woodward J., Barrell B., Parkhill J. (2005) Science 307, 1463–1465 [DOI] [PubMed] [Google Scholar]
- 13. Xu J., Bjursell M. K., Himrod J., Deng S., Carmichael L. K., Chiang H. C., Hooper L. V., Gordon J. I. (2003) Science 299, 2074–2076 [DOI] [PubMed] [Google Scholar]
- 14. Kanehisa M., Goto S. (2000) Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 16. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moreno-Hagelsieb G., Latimer K. (2008) Bioinformatics 24, 319–324 [DOI] [PubMed] [Google Scholar]
- 18. Goodman A. L., McNulty N. P., Zhao Y., Leip D., Mitra R. D., Lozupone C. A., Knight R., Gordon J. I. (2009) Cell Host Microbe 6, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Juncker A. S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., Krogh A. (2003) Protein Sci. 12, 1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 21. Sonnhammer E. L., von Heijne G., Krogh A. (1998) Proc Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
- 22. Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) Nucleic Acids Res. 36, D281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eddy S. R. (1998) Bioinformatics 14, 755–763 [DOI] [PubMed] [Google Scholar]
- 24. Coyne M. J., Reinap B., Lee M. M., Comstock L. E. (2005) Science 307, 1778–1781 [DOI] [PubMed] [Google Scholar]
- 25. Reeves A. R., D'Elia J. N., Frias J., Salyers A. A. (1996) J. Bacteriol. 178, 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shipman J. A., Berleman J. E., Salyers A. A. (2000) J. Bacteriol. 182, 5365–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cho K. H., Salyers A. A. (2001) J. Bacteriol 183, 7224–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carson M. J., Barondess J., Beckwith J. (1991) J. Bacteriol. 173, 2187–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gill D. R., Salmond G. P. (1987) Mol. Gen. Genet. 210, 504–508 [DOI] [PubMed] [Google Scholar]
- 30. Bowler L. D., Spratt B. G. (1989) Mol. Microbiol. 3, 1277–1286 [DOI] [PubMed] [Google Scholar]
- 31. Martelli P. L., Fariselli P., Krogh A., Casadio R. (2002) Bioinformatics 18, (Suppl. 1) S46–S53 [DOI] [PubMed] [Google Scholar]
- 32. Bagos P. G., Liakopoulos T. D., Spyropoulos I. C., Hamodrakas S. J. (2004) Nucleic Acids Res. 32, W400–W404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gromiha M. M., Ahmad S., Suwa M. (2004) J. Comput. Chem. 25, 762–767 [DOI] [PubMed] [Google Scholar]
- 34. Imperiali B., Hendrickson T. L. (1995) Bioorg. Med. Chem. 3, 1565–1578 [DOI] [PubMed] [Google Scholar]
- 35. Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., Dao P., Sahinalp S. C., Ester M., Foster L. J., Brinkman F. S. (2010) Bioinformatics 26, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.