Abstract
Regulation of apoptosis during infection has been observed for several viral pathogens. Programmed cell death and regulation of apoptosis in response to a viral infection are important factors for host or virus survival. It is not known whether Crimean-Congo hemorrhagic fever virus (CCHFV) infection regulates the apoptosis process in vitro. This study for the first time suggests that CCHFV induces apoptosis, which may be dependent on caspase-3 activation. This study also shows that the coding sequence of the S segment of CCHFV contains a proteolytic cleavage site, DEVD, which is conserved in all CCHFV strains. By using different recombinant expression systems and site-directed mutagenesis, we demonstrated that this motif is subject to caspase cleavage. We also demonstrate that CCHFV nucleocapsid protein (NP) is cleaved into a 30-kDa fragment at the same time as caspase activity is induced during infection. Using caspase inhibitors and cells lacking caspase-3, we clearly demonstrate that the cleavage of NP is caspase-3-dependent. We also show that the inhibition of apoptosis induced progeny viral titers of ∼80–90%. Thus, caspase-3-dependent cleavage of NP may represent a host defense mechanism against lytic CCHFV infection. Taken together, these data suggest that the most abundant protein of CCHFV, which has several essential functions such as protection of viral RNA and participation in various processes in the replication cycle, can be subjected to cleavage by host cell caspases.
Keywords: Apoptosis, Caspase, Negative-strand RNA Viruses, Protein Assembly, Viral Replication, CCHFV, Crimean-Congo Hemorrhagic Fever Virus, Nucleocapsid Protein
Introduction
Programmed cell death and regulation of apoptosis in response to a viral infection are important factors for host or virus survival. Protease caspases (cysteine aspartate-specific proteases) play an important role in apoptosis. Caspase activation leads to a proteolytic cascade, where procaspase-8, -9, -10 is activated. These initiator caspases activate caspase-3, -6, and -7, which organize the death of the cell. Caspase-3 is activated as a mediator in the effector phase of programmed cell death. The activated caspases cleave the substrates at specific sites, making them very specific members of the proteases (1). Poly(ADP-ribose) polymerase (PARP)2 is one substrate responsible for the destruction of cellular structures (2, 3). It has been demonstrated that PARP can be cleaved by activated caspase-3 and -7.
To date, several studies have demonstrated the regulation of apoptosis in the replication cycle and spread of viruses. Epstein-Barr virus encodes proteins for inhibition of different steps of the apoptotic pathway in order to maintain virus persistence (4). In contrast to Epstein-Barr virus, vesicular stomatitis virus (VSV) induces cell death. Induced apoptosis during VSV infection is triggered at an early stage of infection and does not depend on viral protein synthesis (5). Intravascular apoptosis and destruction of immune cells have also been observed during infection in fatal cases of Ebola virus. However, the mechanism behind this event is not fully known.
The family Bunyaviridae is one of the largest virus groups, comprising over 350 arthropod- and rodent-borne viruses. All viruses within the Bunyaviridae contain a three-single-stranded RNA genome of negative polarity. The large (L) segment encodes the RNA-dependent RNA polymerase, the medium (M) segment encodes the two envelope glycoproteins (G1 and G2), and the small (S) segment encodes the nucleocapsid protein (6). Several members of this family have been demonstrated to regulate apoptosis. Findings in vivo and in vitro demonstrate that induction of apoptosis affects cellular function (e.g. during La Crosse virus infection) (7). Rift Valley fever virus is another member of the Bunyaviridae that stimulates induction of apoptosis during infection (8). Very recently, it has been shown that Oropuche virus, yet another member of the Bunyaviridae, causes cytopathic effects and induction of programmed cell death by the intrinsic pathway and that induced apoptosis during infection requires viral protein synthesis and is triggered by, but not necessary for, viral replication (9). For members of the Hantavirus genus in the family Bunyaviridae, there are conflicting results, with some researchers having observed apoptosis during infection, (10, 11) and other researchers arguing that hantaviruses are poor inducers of apoptosis in cultured cells (12). However, a direct link has now been demonstrated between Hantaan virus (HTNV) nucleocapsid protein (N) and the modulation of apoptosis through NF-κB (13).
Crimean-Congo hemorrhagic fever virus (CCHFV) is a member of the Nairovirus genus of the family Bunyaviridae. The mortality rate is around 30% in humans, and among other clinical findings, severe dysfunction of the coagulation system is one of the most common symptoms of hemorrhagic fevers. Damage to endothelial cells and vascular leakage seen in these patients may either be a direct result of the virus infection or an immune response-mediated effect (14). Better understanding of virus-host cell interaction is necessary to understand the pathogenesis and the effect of reactions mediated by the immune response during infection with bunyaviruses. This study examined whether CCHFV nucleocapsid protein has a specific cleavage site for caspase-3 and whether it is cleaved when caspase activity is induced during infection.
EXPERIMENTAL PROCEDURES
Cells, Antibodies, and Viruses
Vero (African green monkey kidney epithelial), BHK (baby hamster kidney), and A549 (human alveolar epithelial cell line) cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics (10 units/ml penicillin and 10 μg/ml streptomycin). SW13 cells (human adrenal cortex adenocarcinoma cells) were maintained in Leibovitz's medium (L15), and the MCF-7 (caspase-3-deficient) human breast cancer cell line was grown in RPMI 1640. Stable caspase-3-transfected MCF-7 cells (kindly provided by Prof. Reiner U. Janicke) were grown in RPMI 1640, containing G418. Human umbilical vein endothelial cells (HUVEC) (Lonza, Walkersville, MD) were grown according to the manufacturer's instructions. All growth media contained fetal bovine serum and antibiotics as described above. Antibodies used in this study included a rabbit polyclonal anti-CCHFV nucleocapsid antibody (15) and a mouse monoclonal anti-CCHFV nucleocapsid protein antibody (mAb CCHFV NP). A rabbit polyclonal anti-calnexin antibody was used to detect equal amounts of loaded sample. Anti-rabbit PARP antibody 9542, mouse monoclonal anti-caspase-3 antibody 9668 (Cell Signaling Technology, Beverly, MA), anti-FLAG polyclonal antibody (Sigma), anti-c-Myc monoclonal antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and VSV-G B2709 were diluted and used according to the manufacturer's instructions.
Secondary antibodies (goat anti-mouse, goat anti-rabbit (2 mg/ml), highly cross-adsorbed (Alexa Fluor, Molecular Probes), and horseradish peroxidase (HRP)-conjugated (Bio-Rad) were used according to the manufacturer's instructions. Nigerian CCHFV strain Ibar10200, originally isolated in Nigeria, was used in the experiments (15), and all handling of live virus took place in the BSL-4 facility of the Swedish Institute for Infectious Disease Control (Solna, Sweden).
Fluorescence Focus Units
The CCHFV strain was serially 10-fold diluted and then titrated on Vero-E6 cells in 96-well plates. After 24 h postinfection, cells were fixed with 80% acetone and stained by immunofluorescence assay. The fluorescent foci in each well were counted, and the titer was determined using a rabbit polyclonal anti-CCHFV nucleocapsid antibody, diluted in PBS containing 0.2% BSA and 0.1% Triton X-100 followed by FITC-conjugated anti-rabbit antibody as described previously (16).
Transient Transfection
Expression plasmids were constructed using the pXJ40myc or pXJ3′ vector.3 For easy detection, the sequences coding for 12 residues of the human c-Myc were linked to the NP genes at either the N or C terminus. Transient transfection of Vero cells was performed using Fugene (Roche Applied Science), according to the manufacturer's protocol. Approximately 24 h after transfection, the cells were harvested and subjected to Western blot analysis.
SDS-PAGE and Western Blot Analysis
Samples were resuspended in reducing sample buffer (10 mm Tris-HCl, 0.5% SDS, 10% glycerol, 2% β-mercaptoethanol, bromphenol blue), boiled for 5 min, and separated by SDS-PAGE using precast gels from Invitrogen. When cell culture medium containing detached cells was collected separately, the cell culture medium was centrifuged at 16,000 rpm for 10 min and resuspended and handled as described above. Electrophoresis was carried out at 200 V, and proteins were transferred to nitrocellulose membranes using a transfer buffer containing 25 mm Tris, 192 mm glycine, and 20% methanol at 100 V for 1 h. Membranes were blocked in 5% nonfat dried milk overnight at +4 °C. After washing in PBS containing 0.01% Tween (PBST), the membranes were incubated with a mouse monoclonal anti-CCHFV nucleocapsid antibody for 1 h at room temperature or rabbit polyclonal anti-PARP antibody. Membranes were then washed with PBST before the addition of IgG HRP-conjugated antibody. After incubation at room temperature for 1 h, the membranes were washed in PBST. Proteins were detected with ECL Plus Western blotting detection reagents (Amersham Biosciences) according to the manufacturer's instructions. All antibodies were diluted in blocking buffer as described above.
Generation of Recombinant Semliki Forest Virus (SFV)
Recombinant SFV NP was produced as described previously (15, 17). In brief, recombinant SFV plasmids (containing sequences for the nucleocapsid protein) were purified using the QIAprep Spin Miniprep kit (Qiagen) and linearized by NruI digestion. Linearized plasmids were used as templates for in vitro RNA transcription using SP6 polymerase. In vitro transcripts made from pSFV-CCHFV NP were electroporated into BHK cells together with equal amounts of mRNA transcripts from SpeI-linearized pSFV helper. Electroporated cells were then diluted in DMEM, supplemented with 10% FBS and antibiotics, seeded in 6-well plates, and incubated at 37 °C. Recombinant virus particles were collected after 24 h.
PCR methods were used to generate substitution mutants of CCHFV NP, where the aspartic acid residue at either position 266 or 269 in the primary sequence was changed to alanine. Next, mutated sequences were ligated into the SpeI site of SFV expression plasmids. The recombinant mutants and full-length NP were then transfected with helper plasmid (containing structural proteins) into electrocompetent TOP 10 cells (Invitrogen). Sequence analysis was carried out to confirm amino acid substitutions. Generation of recombinant virus expressing deletions within the nucleocapsid protein (SFV mutants) was carried out as described above.
Virus Infection
A549, HUVEC, SW13, and MCF-7 cells were seeded in 6-well plates until they reached 80% confluence. Cells were then infected with CCHFV (MOI = 1). Attached cells and cell culture medium containing detached cells were harvested separately 24, 48, and 72 h postinfection and analyzed for NP, PARP, and caspase-3 by Western blot.
BHK cells grown in 6-well plates were infected with recombinant SFV-expressing native or mutants of NP (MOI = 0.5). Cells were harvested 6, 12, 24, and 48 h postinfection and analyzed with rabbit polyclonal anti-PARP antibody and mAb anti-CCHFV NP antibody by Western blot.
TUNEL Assay
SW13 cells were infected with CCHFV (MOI = 1 and MOI = 10) and VSV (1 MOI) for 48 and 72 h postinfection. Infected cells were washed in PBS, followed by 1 h of incubation in 4% paraformaldehyde (pH 7.4) at room temperature. Cells were then incubated with permeabilization solution containing 0.1% Triton X-100 and 0.1% sodium citrate for 2 min on ice, washed, and air-dried. Labeling of DNA strand breaks was performed with the TMR-red cell death detection kit (Roche Applied Science) according to the manufacturer's instructions. Cells were then stained with rabbit polyclonal anti-CCHFV NP and mouse anti-mouse VSV-G.
Annexin V Assay
Cells were infected with CCHFV (MOI = 1) for 48 and 72 h postinfection. Cells were treated and labeled according to the manufacturer's instructions (Annexin V-Cy3TM apoptosis detection kit, Sigma).
Caspase Inhibition Assays
SW13 cells were seeded in 6-well plate. Cells were infected with CCHFV (MOI = 1) and after 1 h were treated with caspase-3-inhibitor (Z-DEVD-fmk) or a negative caspase inhibitor (Z-FA-fmk) or left untreated. Both inhibitors were dissolved in DMSO at a concentration of 20 μm and were obtained from BD Biosciences. The inhibitors remained in the medium throughout the experiment. Infected cells were collected 24, 48, and 72 h postinfection and analyzed by Western blot. The antibodies used in the assay (PARP, mAb CCHFV NP antibody, and polyclonal CCHFV NP antibody A) and control cells were treated with staurosporine (Cell Signaling), dissolved in 1 μm DMSO solution.
Statistical Analyses
Student's t test was used for calculation of significant differences between different cell lines of mock- or drug-treated cells. p < 0.05 was considered significant.
RESULTS
CCHFV NP Has a Specific Cleavage Motif for Caspase-3
Examination of the primary structure of CCHFV NP demonstrated a potential motif for caspase cleavage (DEVD) at positions 266–269 of the nucleocapsid protein (Fig. 1A). Comparison with available sequences of the S segment of CCHFV in GenBankTM revealed that this motif was conserved in all available strains (data not shown).
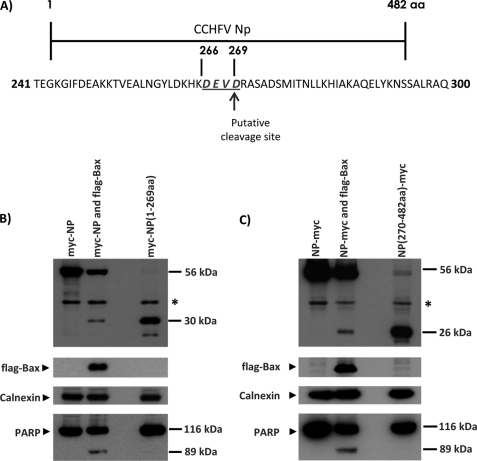
FIGURE 1.
CCHFV NP is cleaved during Bax-induced apoptosis. A, map of the entire NP. The amino acid sequence of residues 240–300 of NP is shown. The DEVD motif is indicated, and the putative cleavage site between residues 269 and 270 is indicated. B, Vero cells were transiently transfected with plasmids encoding N-terminal c-Myc-tagged full-length NP or residues 1–269 of NP. To induce apoptosis, co-transfection with Bax plasmid was performed. The cells were harvested 24 h after transfection, and the expressions of the transfected proteins were analyzed by Western blot analysis with anti-Myc and anti-FLAG antibodies. Expressions of endogenous calnexin and PARP were also analyzed by Western blot analysis. C, a similar experiment was performed with plasmids encoding C-terminal c-Myc-tagged full-length NP or residues 270 to 482 of NP. *, an unknown endogenous protein detected by the anti-Myc antibody. aa, amino acids.
To investigate whether this motif in NP is cleaved during apoptosis, we constructed several different plasmids expressing the full-length NP with the c-Myc tag at either the N or C terminus. We also constructed plasmid coding either residues 1–269 of NP (Myc-NP(1–269)) or residues 270–482 of NP (NP(270–482)-Myc). To determine if this motif in NP is cleaved during apoptosis, cells were transfected with the plasmid coding for full-length NP and plasmid coding for Bax, a potent inducer of apoptosis, or plasmid coding for the fragment of NP. After 24 h, the cells were harvested and analyzed by antibodies against c-Myc. As seen in Fig. 1B, we clearly demonstrated that Myc-NP was cleaved in cells expressing Bax, and the size of the cleavage product is ∼30 kDa. This cleavage product of Myc-NP migrated at the same rate as Myc-NP(1–269), suggesting that cleavage occurred at the DEVD motif in NP. As expected, endogenous PARP was cleaved in the cells expressing Myc-NP and Bax. Consistently, NP-Myc was also cleaved in the presence of Bax, and the cleavage product migrated at the same rate as NP(270–482)-Myc (Fig. 1C).
To confirm these results, recombinant SFV, known to induce apoptosis via the intrinsic pathway (18), was produced in order to express NP. BHK cells were infected with recombinant SFV (MOI = 1), and cells were harvested 6, 12, 24, and 48 h postinfection. As shown in Fig. 2B, in contrast to mock-infected cells, the cleavage form of PARP was present at 24 h and more pronounced in cells collected at 48 h postinfection, which indicates the activation of caspase-3. In addition to the expected full-length protein of 56 kDa, several fragments of NP were detected, including a smaller 30-kDa fragment of NP at 48 h postinfection (Fig. 2A). By comparison with the results in Fig. 1, it appears that this 30-kDa fragment corresponds to the cleavage product containing residues 1–269 of NP. Calnexin was used as a loading control.
FIGURE 2.
The DEVD motif, in CCHFV NP, is a subject for caspase-3 cleavage. A, BHK cells were infected with recombinant SFV-expressing CCHFV NP. Lysates from mock-infected or SFV NP-infected cells were harvested at different times postinfection and analyzed for CCHFV NP by Western blot. The arrows show full-length NP (56 kDa) and the cleavage product (30 kDa). B, cell lysates were further analyzed for PARP by Western blot (the 116 kDa band represents uncleaved PARP, and the 85 kDa band represents cleaved PARP). C, BHK cells were infected with recombinant SFV expressing native CCHFV NP or the mutated form (at positions 266 and 269) of NP. Lysates from infected cells were harvested at different period of times postinfection and analyzed for CCHFV NP by Western blot.
In order to confirm that the DEVD at positions 266–269 of NP is subjected to caspase cleavage, we created two mutants of CCHFV NP, lacking this predicted cleavage site of NP. BHK cells were infected with rSFV-expressing mutants and the native form of NP, and after 24, 48, and 72 h postinfection, cells were harvested, and the expression pattern of NP was analyzed. As shown in Fig. 2C, we were unable to detect the 30-kDa fragment of NP in cells infected by mutants. However, we found cleavage of PARP already at 24 h postinfection (data not shown). We also observed a 50-kDa fragment in cells infected with recombinant SFV NP, and the expression was more pronounced in cells infected with recombinant mutants compared with cells infected with the native form of NP.
Apoptosis and Cleavage of NP in Late Stages of CCHFV Infection
To determine if cleavage of NP occurs during CCHFV infection, SW13, HUVEC, and A549 cells were infected with CCHFV (MOI = 1). Supernatants were collected at 24, 48, and 72 h postinfection and analyzed. Due to safety concerns in the handling of BSL4 virus, the culture media containing detached cells and the attached cells were harvested separately for Western blot analysis. It is expected that the culture medium will contain cells in late apoptotic stages because they are not strongly adhered to the culture plates, whereas the cells will contain non-apoptotic cells or cells in early apoptotic stages (see also Fig. 5). As shown in Fig. 3, we were able to demonstrate the appearance of a cleaved 30-kDa NP product in detached cells at 48 h postinfection. However, this fragment was more strongly manifested at 72 h postinfection. Careful examination of Fig. 3 reveals a nonspecific band in mock-infected HUVEC and A549 cells. Because the appearance of the cleaved 30-kDa NP product was most pronounced in the SW13 cells, further analysis was performed with this cell line.
FIGURE 5.
Caspase-3 is activated at late time post CCHFV infection. SW13 cells were infected with CCHFV (MOI = 1). At 72 h postinfection, the culture media containing detached cells (A) and the attached cells (B) were harvested separately. The lysates were analyzed for NP and caspase-3, and calnexin was used as a loading control.
FIGURE 3.
A smaller, 30-kDa fragment of NP appears at a late time postinfection. A549, HUVEC, and SW13 cells were infected with CCHFV at MOI = 1. The culture medium containing detached cells was harvested at 24, 48, and 72 h postinfection and analyzed for the expression pattern of NP by Western blot. Several lanes from separate gels have been merged in this figure.
To determine if CCHFV infection induces apoptosis, we performed a TUNEL assay (intrachromosomal DNA strand breaks). SW13 cells were mock-infected or infected with CCHFV (MOI = 1). At 48 and 72 h postinfection, the detached were collected and analyzed by a TUNEL assay (Fig. 4A). These results demonstrated that CCHFV infection induced significantly greater amounts of TUNEL-positive cells compared with mock-infected cells. However, to confirm these results, we analyzed the expression of Annexin V in infected SW13 cells. Briefly, SW13 cells were mock-infected or infected with CCHFV (MOI = 1). At 48 or 72 h postinfection, the detached were collected and analyzed by an Annexin V assay (Fig. 4B). These results demonstrated that CCHFV infection induced significantly greater amounts of Annexin-positive cells compared with mock-infected cells.
FIGURE 4.
Programmed cell death is induced during CCHFV infection. A, SW13 cells were mock-infected or infected with CCHFV (MOI = 1 or 10). Mock-infected and CCHFV-infected cells were fixed for 1 h with 4% paraformaldehyde (pH 7.4) at room temperature. Cells were then incubated with permeabilization solution containing 0.1% Triton X-100 and 0.1% sodium citrate. Labeling of DNA strand breaks was performed with the TMR-red cell death detection kit. Values given are the average of three independent experiments. Error bars, S.D. B, SW13 cells were mock-infected or infected with CCHFV (MOI = 1). Cells were stained for Annexin V as described under “Experimental Procedures.” Values given are the average of three independent experiments. Error bars, S.D. Significance is illustrated with p < 0.001 (**) and p < 0.0001 (***). hpi, hours postinfection.
Caspase-3 Cleavage of CCHFV Nucleocapsid Protein
To study cleavage of NP in more detail, SW13 cells were mock-infected or infected by CCHFV (MOI = 1). Culture media containing detached cells and attached cells were harvested separately and analyzed for detection of caspase-3. As shown in Fig. 5, we were able to demonstrate the presence of activated caspase-3 in infected cells at the same time course as observed cleavage of NP. We also observed more activated caspase-3 in the detached cells, which is consistent with the apoptotic cells lifting off of the dish (Fig. 5).
In order to confirm that cleavage of NP is caspase-3-dependent, SW13 cells were mock-infected or infected with CCHFV (MOI = 1), and after 1 h, cells were mock-treated or treated with Z-FA-fmk or Z-DEVD-fmk or only with DMSO at different concentrations. The culture media containing detached cells and the attached cells were harvested separately at 48 and 72 h postinfection. The expression patterns of NP and of PARP were analyzed by Western blot.
We were able to clearly demonstrate that caspase-3 inhibitor, Z-DEVD-fmk, completely inhibited cleavage of nucleocapsid protein compared with the controls (Fig. 6A). In addition, the presence of caspase inhibitors reduced cleavage of PARP (Fig. 6B).
FIGURE 6.
Inactivation of caspase-3 inhibits cleavage of NP. A and B, SW13 cells were infected with CCHFV (MOI = 1). After 1 h, cells were mock-treated or treated with Z-FA-fmk or Z-DEVD-fmk. The culture media containing detached cells and the attached cells were collected separately at 72 h postinfection and analyzed for NP (A) and PARP (B) by Western blot. C, mock-infected MCF-7, stable MCF-7 caspase cells, and SW13 cell lysate were analyzed for the presence of caspase-3 by Western blot. D, MCF-7 and stable MCF-7-caspase-3 cells infected with CCHFV (MOI = 1). The culture media containing detached cells and the attached cells were collected separately at 48 and 72 h postinfection and analyzed for PARP and NP by Western blot. The arrows show full-length CCHFV NP (56 kDa), cleaved NP (30 kDa), uncleaved PARP (116 kDa), and cleaved PARP (89 kDa). hr.p.i., hours postinfection. E, MCF-7 and MCF-7-caspase cells were mock-infected or infected with CCHFV (MOI = 1). Mock-infected and CCHFV-infected cells were fixed for 1 h with 4% paraformaldehyde (pH 7.4) at room temperature. Cells were then incubated with permeabilization solution containing 0.1% Triton X-100 and 0.1% sodium citrate. Labeling of DNA strand breaks was performed with the TMR-red cell death detection kit. Values given are the average of three independent experiments. Error bars, S.D. Significance is illustrated with p < 0.05 (*).
To confirm these results, we compared cleavage of NP in MCF-7 cells, which lack caspase-3, and also stable MCF-7 cells, which had been transfected with caspase-3 plasmid and constitutively expressed caspase-3. These cells were infected by CCHFV (MOI = 1). The culture media containing detached cells and the attached cells were harvested separately at 24, 48, and 72 h postinfection and prepared for Western blot analysis. We showed that MCF-7 cells in contrast to the stable MCF-7-caspase-3 cells did not express caspase-3 (Fig. 6C).
As shown in Fig. 6D, in contrast to MCF-7 cell lines, we were able to detect the 30-kDa fragment of NP in stable MCF-7 casapse-3 cells. However, we were able to detect cleavage of PARP (Fig. 6D) in both cell lines. These results confirm previous data that demonstrate that cleavage of PARP can be performed by caspase-7. To determine if CCHFV infection induces more apoptosis in MCF-7-caspase-3 cells compared with MCF-7 cells lacking caspase-3, we performed a TUNEL assay. Cells were mock-infected or infected with CCHFV (MOI = 1). At 72 h postinfection, the detached cells were collected and analyzed by a TUNEL assay (Fig. 6E). These results demonstrated that CCHFV infection induced significantly greater amounts of TUNEL-positive cells compared with MCF cells lacking caspase-3.
Caspase-3-induced Cleavage of NP Inhibits the Yield of Progeny Virus
To study whether caspase-3-induced apoptosis has an inhibitory effect on yield of progeny virus, SW13 cells were infected with CCHFV (MOI = 1). These infected cells were then mock-treated or treated with Z-DEVD-fmk or Z-FA-fmk. Supernatants were collected at 24 and 48 h postinfection, and the amount of infectious virus particles was determined by focus-forming units. The virus titer increased by almost 1 log in cells treated with Z-DEVD-fmk at 48 h postinfection compared with mock-treated cells or cells treated with the control (Fig. 7). To confirm these results, The MCF-7 and stable MCF-7-caspase-3 cells were infected with CCHFV (MOI = 1). The supernatants were collected at 24 and 48 h postinfection, and the amount of infectious virus particles was determined by focus-forming units. As shown in Fig. 7B, we found that MCF-7 cells expressing caspase-3 produced a significantly smaller amount of the virus.
FIGURE 7.
Inhibition of or absence of caspase activation increases the yield of progeny virus. SW13-, MCF-7-, and MCF-7-expressing caspase-3 cells were infected with CCHFV (MOI = 1). SW13 cells were mock-treated or treated with Z-DEVD-fmk or Z-FA-fmk. Supernatants of SW13 cells (A) and MCF-7 cells (B) were harvested at 24 and 48 h postinfection (hpi) and analyzed for yield of progeny virus by focus-forming units (FFU). Values given are the average of three independent experiments. Error bars, S.D. Significance is illustrated with p < 0.05 (*), p < 0.001 (**), and p < 0.0001 (***).
DISCUSSION
The primary pathophysiological events in CCHF patients appear to be leakage of erythrocytes and plasma through the vasculature into tissues (19). Endothelial damage can contribute to coagulopathy by deregulated stimulation of platelet aggregation, which in turn activates the intrinsic coagulation cascade, ultimately leading to clotting factor deficiency and hemorrhages. For CCHFV, vascular leakage may be caused by destruction of endothelial cells, but it is unclear whether these events are a direct consequence of infection or whether virus-induced host factors cause the endothelial dysfunction (14). It has been suggested that the hemorrhages and coagulation disturbances may be indirect effects, possibly caused by high levels of proinflammatory cytokines due to massive destruction of blood cells. In fact, for other hemorrhagic fever viruses, such as Ebola and Dengue virus, there is a correlation between the strength of the proinflammatory response, vascular leakage, and disease severity (20–24).
In this study, we were able to clearly demonstrate that CCHFV NP has a conserved cleavage site for caspase-3. By using transient transfection and the SFV expression systems, we found that this motif is subjected to caspase cleavage during an apoptosis event. By mutation of a single residue, we were able to clearly demonstrate that the 30-kDa form of NP disappeared, but at the same time we found other cleavage products that were more pronounced in cells infected with the mutated forms of NP. These data suggest that the conserved motif DEVD is the major subject for caspase-3 cleavage, but there are other sites or motifs available for cleavage during apoptosis, especially when the DEVD site is disrupted.
The DEVD motif is conserved in all known strains of CCHFV, and because NP is an important structural protein and necessary for viral replication cycle, it remains to be determined whether cleavage of NP during apoptosis has a specific function for viral replication or pathogenesis. The fact that the caspase cleavage site has not been eliminated during evolution suggests either that this motif must be important for the replication cycle of this particular virus or that CCHFV has evolved a strategy to inhibit the activation events for apoptosis and thereby escape cleavage at early stages of infection.
It has previously been demonstrated for some viruses that structural proteins are subjected to cleavage by activated caspases during an apoptosis process (25). During transmissible gastroenteritis coronavirus infection, caspase activation causes cleavage of the viral nucleocapsid protein (26). During infection with influenza, virus cleavage of the nucleocapsid protein has been observed during the process of apoptosis in late stages of the infection, and the primary structure of NP of human influenza viruses (A and B) has been shown to have proteolytic sites, which engage involvement of caspases (27).
Using different human cell lines, we demonstrated that NP underwent cleavage at the late postinfection stages. By performing detailed analysis with SW13 cells, numerous hallmarks of apoptosis were clearly observed in CCHFV-infected cells, especially at 72 h postinfection. Importantly, through the use of caspase-3 inhibitors, we were able to clearly demonstrate that inhibition of apoptosis inhibits cleavage of CCHFV NP. Using MCF-7 cells, deficient in caspase-3, we further confirmed that cleavage of NP to a 30-kDa fragment is a caspase-3-dependent process. However, cleavage of PARP was observed late during infection even in this cell type. It has previously been suggested that cleavage of PARP can be performed by caspase-7 in the absence of activated caspase-3, and CCHFV infection may stimulate several caspases directly or indirectly (28). Comparison of the yield of progeny virus between mock-treated cells and cells treated with caspase inhibitors revealed that inhibition of apoptosis increased the yield of progeny virus by at least 80–90%. These data suggest that cleavage of CCHFV NP affects the replication cycle of CCHFV. They also indicate that induced caspase activity is not beneficial for the virus replication cycle and may be induced by the host cell immune response. A similar observation was recently reported for the Kaposi sarcoma-associated herpesvirus ORF57 protein (29). In this case, there is a caspase-7 cleavage site in ORF57, and inhibition of cleavage led to increased expression of viral lytic genes and production of cell-free virus particles.
This study did not analyze whether apoptosis induction is a direct effect of the CCHFV replication cycle or a consequence of indirect processes induced by CCHFV infection. Type I interferons (IFN-α/β) are produced in infected cells as a host defense mechanism against infection. Type I interferons may act as initiators of cell death together with double-stranded RNA or as modulators of a cascade of immune reactions upon viral infection (30). IFNs induce a large number of proteins, such as PKR (RNA-activated protein kinase). The involvement of PKR in cell stress and cell death has been demonstrated in a number of studies. Induced programmed cell death by PKR mainly involves the FADD/caspase-8 pathway, but activation of the Apaf-1/caspase-9 pathway has also been observed (31). To date, there is no evidence of induction of PKR during early CCHFV infection. In fact, we have previously demonstrated that CCHFV delays induction and secretion of IFNs and induces translocation of IRF-3 (interferon regulatory factor) and activation of interferon-stimulated genes late during infection (32). Some RNA viruses activate IRF-3, which induces a set of interferon-stimulated genes in order to express proteins to establish an antiviral state, which in the end can lead to programmed cell death (33). Previous studies have demonstrated that Bunyamwera virus induces apoptotic cell death by activating IRF-3 (34). We recently demonstrated that CCHFV has evolved a new strategy to escape recognition of RIG-I and thereby escape the activation of innate immune responses in early replication (35).
Overall, in this study, we suggest that induced apoptosis by CCHFV late postinfection may be dependent on caspase-3 activation. We also demonstrated that cleavage of nucleocapsid protein at the DEVD consensus site during apoptosis; these observations may be associated with limiting virus replication.
The reason why caspases become activated and programmed cell death is induced late during CCHFV infection in vitro and mechanism involved is unclear. In future studies, we will examine whether cleavage of NP plays a role in the viral pathogenesis of CCHF and attempt to characterize the mechanism behind the regulation of apoptosis in CCHFV-infected cells.
Acknowledgment
MCF-7 cells expressing caspase-3 were kindly provided by Prof. Reiner U. Janicke.
This work was supported in part by Swedish Medical Research Council Grant K2010-57X-0349-01-3 (to A. M.).
E. Manser and T. Leung, personal communication.
- PARP
- poly(ADP-ribose) polymerase
- VSV
- vesicular stomatitis virus
- CCHFV
- Crimean-Congo hemorrhagic fever virus
- HUVEC
- human umbilical vein endothelial cell(s)
- NP
- nucleocapsid protein
- SFV
- Semliki Forest virus
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone.
REFERENCES
- 1. Zimmermann K. C., Bonzon C., Green D. R. (2001) Pharmacol. Ther. 92, 57–70 [DOI] [PubMed] [Google Scholar]
- 2. Thornberry N. A. (1998) Chem. Biol. 5, R97–R103 [DOI] [PubMed] [Google Scholar]
- 3. Thornberry N. A., Lazebnik Y. (1998) Science 281, 1312–1316 [DOI] [PubMed] [Google Scholar]
- 4. Tarodi B., Subramanian T., Chinnadurai G. (1994) Virology 201, 404–407 [DOI] [PubMed] [Google Scholar]
- 5. Gadaleta P., Vacotto M., Coulombié F. (2002) Virus Res. 86, 87–92 [DOI] [PubMed] [Google Scholar]
- 6. Elliott R. M. (1990) J. Gen. Virol. 71, 501–522 [DOI] [PubMed] [Google Scholar]
- 7. Pekosz A., Phillips J., Pleasure D., Merry D., Gonzalez-Scarano F. (1996) J. Virol. 70, 5329–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Won S., Ikegami T., Peters C. J., Makino S. (2007) J. Virol. 81, 13335–13345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Acrani G. O., Gomes R., Proença-Módena J. L., da Silva A. F., Carminati P. O., Silva M. L., Santos R. I., Arruda E. (2010) Virus Res. 149, 56–63 [DOI] [PubMed] [Google Scholar]
- 10. Akhmatova N. K., Yusupova R. S., Khaiboullina S. F., Sibiryak S. V. (2003) Russ. J. Immunol. 8, 37–46 [PubMed] [Google Scholar]
- 11. Kang J. I., Park S. H., Lee P. W., Ahn B. Y. (1999) Virology 264, 99–105 [DOI] [PubMed] [Google Scholar]
- 12. Hardestam J., Klingström J., Mattsson K., Lundkvist A. (2005) J. Med. Virol. 76, 234–240 [DOI] [PubMed] [Google Scholar]
- 13. Ontiveros S. J., Li Q., Jonsson C. B. (2010) Virology 401, 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schnittler H. J., Feldmann H. (2003) Thromb. Haemost. 89, 967–972 [PubMed] [Google Scholar]
- 15. Andersson I., Simon M., Lundkvist A., Nilsson M., Holmström A., Elgh F., Mirazimi A. (2004) J. Med. Virol. 72, 83–93 [DOI] [PubMed] [Google Scholar]
- 16. Andersson I., Bladh L., Mousavi-Jazi M., Magnusson K. E., Lundkvist A., Haller O., Mirazimi A. (2004) J. Virol. 78, 4323–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liljeström P., Garoff H. (1991) Biotechnology 9, 1356–1361 [DOI] [PubMed] [Google Scholar]
- 18. Barry G., Fragkoudis R., Ferguson M. C., Lulla A., Merits A., Kohl A., Fazakerley J. K. (2010) J. Virol. 84, 7369–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ergönül O. (2006) Lancet Infect. Dis. 6, 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bray M., Geisbert T. W. (2005) Int. J. Biochem. Cell Biol. 37, 1560–1566 [DOI] [PubMed] [Google Scholar]
- 21. Bray M., Pilch R. (2006) Expert Rev. Anti Infect. Ther. 4, 917–921 [DOI] [PubMed] [Google Scholar]
- 22. Chen S. T., Lin Y. L., Huang M. T., Wu M. F., Cheng S. C., Lei H. Y., Lee C. K., Chiou T. W., Wong C. H., Hsieh S. L. (2008) Nature 453, 672–676 [DOI] [PubMed] [Google Scholar]
- 23. Chen H. C., Hofman F. M., Kung J. T., Lin Y. D., Wu-Hsieh B. A. (2007) J. Virol. 81, 5518–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lei H. Y., Yeh T. M., Liu H. S., Lin Y. S., Chen S. H., Liu C. C. (2001) J. Biomed. Sci. 8, 377–388 [DOI] [PubMed] [Google Scholar]
- 25. Best S. M. (2008) Annu. Rev. Microbiol. 62, 171–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eléouët J. F., Slee E. A., Saurini F., Castagné N., Poncet D., Garrido C., Solary E., Martin S. J. (2000) J. Virol. 74, 3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhirnov O. P., Konakova T. E., Garten W., Klenk H. (1999) J. Virol. 73, 10158–10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jänicke R. U. (2009) Breast Cancer Res. Treat. 117, 219–221 [DOI] [PubMed] [Google Scholar]
- 29. Majerciak V., Kruhlak M., Dagur P. K., McCoy J. P., Jr., Zheng Z. M. (2010) J. Biol. Chem. 285, 11297–11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Samuel C. E. (2001) Clin. Microbiol. Rev. 14, 778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. García M. A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C., Esteban M. (2006) Microbiol. Mol. Biol. Rev. 70, 1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andersson I., Karlberg H., Mousavi-Jazi M., Martínez-Sobrido L., Weber F., Mirazimi A. (2008) J. Med. Virol. 80, 1397–1404 [DOI] [PubMed] [Google Scholar]
- 33. Clemens M. J. (2005) Semin. Cell Dev. Biol. 16, 13–20 [DOI] [PubMed] [Google Scholar]
- 34. Kohl A., Clayton R. F., Weber F., Bridgen A., Randall R. E., Elliott R. M. (2003) J. Virol. 77, 7999–8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Habjan M., Andersson I., Klingström J., Schümann M., Martin A., Zimmermann P., Wagner V., Pichlmair A., Schneider U., Mühlberger E., Mirazimi A., Weber F. (2008) PLoS ONE 3, e2032. [DOI] [PMC free article] [PubMed] [Google Scholar]