Abstract
Retinoic acid (RA) regulates clustered Hox gene expression during embryogenesis and is required to establish the anterior-posterior body plan. Using mutant embryonic stem cell lines deficient in the RA receptor γ (RARγ) or Hoxa1 3′-RA-responsive element, we studied the kinetics of transcriptional and epigenomic patterning responses to RA. RARγ is essential for RA-induced Hox transcriptional activation, and deletion of its binding site in the Hoxa1 enhancer attenuates transcriptional and epigenomic activation of both Hoxa and Hoxb gene clusters. The kinetics of epigenomic reorganization demonstrate that complete erasure of the polycomb repressive mark H3K27me3 is not necessary to initiate Hox transcription. RARγ is not required to establish the bivalent character of Hox clusters, but RA/RARγ signaling is necessary to erase H3K27me3 from activated Hox genes during embryonic stem cell differentiation. Highly coordinated, long range epigenetic Hox cluster reorganization is closely linked to transcriptional activation and is triggered by RARγ located at the Hoxa1 3′-RA-responsive element.
Keywords: Chromatin, Differentiation, Embryonic Stem Cell, Epigenetics, Histones, Transcription, Vitamin A, Homeodomain, Polycomb, Retinoic Acid Receptor (RAR)
Introduction
During development, the differentiation of embryonic cells is modulated by extrinsic patterning signals of immense complexity. Remarkably, all-trans-retinoic acid (RA)3 induces sequential activation of the clustered Hox (homeobox) genes in an anteroposterior order that resembles their positions in the chromosomal cluster, and this patterning activity is replicated when cultured embryonic stem cells (ESCs) are exposed to RA (1, 2). In both mice and humans, the Hox genes are organized into four clusters (Hoxa to Hoxd), with each cluster containing 9–11 genes (3, 4). In the developing CNS, individual Hox genes within these chromosomal clusters are expressed in a precise spatiotemporal pattern along the anteroposterior axis (5, 6). Hox gene expression is tightly regulated during the differentiation of adult stem cells, and dysregulated Hox gene expression is found in many human malignancies (7–12). The mechanisms by which the sequential activation of Hox genes is achieved are not fully understood.
Several interdependent signaling pathways (e.g. RA, Wnt pathway, sonic hedgehog, and fibroblast growth factors) exert their patterning effects by regulating Hox gene expression in the developing embryo (13, 14). Both local and distant DNA cis-regulatory elements have been implicated in the co-linear transcriptional control of Hox gene expression during development (4, 15). RA plays an essential role in Hox gene regulation by binding to specific retinoic acid receptors (RARs: NR1B1 (RARα), NR1B2 (RARβ), or NR1B3 (RARγ)) at RA-responsive DNA elements (RAREs) located within the Hox clusters (2, 16–33). Elegant genetic analyses have revealed critical cross-regulation of Hoxa1 and Hoxb1 during rhombomere segmentation (18, 34–39). Although these auto- and para-regulatory controls govern the duration and degree of Hoxb1 expression and are known to be subordinate to upstream RA signaling, the extent to which this cross-regulation dictates the epigenomic and transcriptional reorganization of the entire Hoxa and Hoxb clusters is less well appreciated.
Specific epigenetic modifications are associated with, and tend to predict, the transcriptional state of the chromatin. Several modifications located in close proximity to one another form the basis of a “histone code” that is recognized by proteins interacting in either a synergistic or antagonistic manner to control access to the underlying genetic information encoded by the DNA (40–42). Tri-methylation of histone H3 lysine 4 (H3K4me3) is associated with the body and promoters of genes poised for transcription, and the relative enrichment of this mark generally correlates with expression level (43, 44). Acetylation at H3 lysines 9 and 14 (acH3) tends to co-localize with H3K4me3 and is associated with accessible euchromatin (44–46). In contrast, genes marked for chromatin condensation and expression silencing are often associated with tri-methylation of H3 lysine 27 and lysine 9 (H3K27me3 and H3K9me3) and/or CpG methylation (47–51). In pluripotent ESCs, some genes are marked by both the H3K4me3 modification and the repressive H3K27me3 chromatin mark. Such “bivalent” domains are believed to underlie ESC plasticity and mark genes for either silencing or activation as differentiation proceeds (52–55).
Master epigenetic regulatory proteins of the homeobox, trithorax, jumonji, and polycomb group families orchestrate the transcriptional activation and remodeling of Hox gene chromatin. Although Hox genes are silenced in ESCs, their transcription is rapidly induced early in development, and patterned tissue-specific Hox gene subsets are expressed throughout life. Tight regulation of Hox gene expression is also required for ESC differentiation in vitro (56, 57). Polycomb group proteins control the transition from stem cells to differentiated cells (58–61). Polycomb group-repressive complexes (PRC1 and PRC2) mark Hox genes for silencing in ESCs (59, 62). Ezh2 is a component of the PRC2 complex that places the H3K27me3 mark recognized by PRC1 factors, which in turn bring Ring1 to mono-ubiquitinylate histone H2A lysine 119 (H2Aub) (63). The DNA elements that PRCs bind have not been characterized but may include CpG islands (64, 65). Noncoding RNAs within the Hox clusters interact with PRCs to regulate Hox gene expression (66, 67) reinforcing the tight interdependence of expression and epigenetic remodeling. These repressive PRC marks must be removed by the histone lysine demethylase activity of Jumonji C (JmjC) domain-containing enzymes for physiological Hox gene induction and normal development (58, 68–73). Global (cluster) control regions coordinate the recruitment of histone acetyltransferases, JmjC domain-containing demethylases, and H3K4 methylases of the trithorax family, at least partially through their interactions with JmjC proteins such as UTX (74).
In this study, we utilized mutant ESCs to probe the spatiotemporal sequence of RA-induced transcriptional and epigenomic reorganization of the clustered Hox genes. These mutant ESC lines allowed us to answer the following questions concerning the functions of RARγ and the Hoxa1 3′-RARE in RA signaling. Are RARs required to establish the basal epigenomic configuration of the clustered Hox genes? Which RAR is most important for transcriptional activation of the clustered Hox genes in ESCs? To what extent is the co-linear activation of these genes dependent upon RA-induced epigenomic reorganization of the locus and how extensive is this reorganization? Is the removal of the H3K27me3 mark necessary for transcriptional activation? How are the epigenetic changes coordinated over time across the Hoxa and Hoxb chromosomal clusters?
EXPERIMENTAL PROCEDURES
Derivation and Culture of the ESC Lines
We isolated the RARγ−/−, RARβ+/− ESC line (RARγ-KO) by crossing RARγ/RARβ heterozygous male and female mice (75, 76) and establishing ESC lines in culture from isolated blastocysts (77). RARβ−/− ESCs were generated similarly from mating RARβ−/− mice. WT ESCs were established by this method at the same time. RARE-KO ESCs were cloned following transfection of WT cells with a vector targeting the Hoxa1 3′-RARE enhancer (Fig. 1). Two rounds of selection were performed to knock out both enhancer alleles. All genotypes were verified by Southern blotting. WT, RARE-KO, and RARγ-KO ESCs were cultured as described previously (78). All experiments were performed in the presence of leukemia inhibitory factor (Millipore, Billerica, MA). All-trans retinoic acid (RA) was from Sigma. Additional methods are provided in the supplemental “Experimental Procedures”.
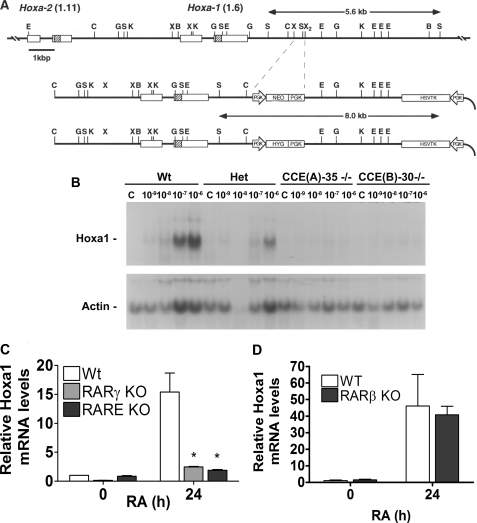
FIGURE 1.
Hoxa1 is transcriptionally activated by RA in WT and RARβ/KO but not in RARγ/KO and RARE/KO cells. A, schematic of the Hoxa1 3′-RARE targeting construct. B, Northern blot showing dose response of Hoxa1 induction 48 h after different doses of RA in WT, heterozygous RARE/KO, and two independently derived lines with RARE/KO in both alleles. C, relative Hoxa1 expression prior to and 24 h after 1 μm RA in WT, RARE/KO, and RARγ/KO ESCs. Quantitative RT-PCR was used to quantify transcripts normalized to a 36B4 reference control (mean ± S.E.). Asterisk denotes p < 0.05 for the relative expression of Hoxa1 mRNA in WT compared with the RARE/KO and RARγ/KO ESCs. D, relative Hoxa1 expression prior to and 24 h after 1 μm RA in WT and RARβ/KO ESCs. Quantitative RT-PCR was used to quantify transcripts normalized to a 36B4 reference control (mean ± S.E.).
ChIP, ChIP-Chip, and Gene Expression Profiling
ESCs were plated in gelatin-coated tissue culture dishes ∼48 h prior to harvesting. At various times after RA addition (0, 1, 4, 8, and 24 h), cells were cross-linked (1% formaldehyde, 10 min), quenched (200 mm glycine, 5 min), washed with ice-cold phosphate-buffered saline (PBS), and harvested by scraping. At least three biological replicate ChIP experiments were performed, as described previously (31, 32). ChIP DNA was amplified by ligation-mediated PCR and hybridized to custom-designed Agilent tiling oligonucleotide arrays per the manufacturer's instructions (Agilent). For expression analysis, cells were plated as described and then harvested at various times after RA treatment. Total RNA was extracted using TRIzol, and RNA was labeled and hybridized to mouse-ref8 expression arrays (Illumina), per manufacturer's instructions. Data were analyzed using custom-designed R language routines utilizing Bioconductor packages. Detailed methods are provided in the supplemental Experimental Procedures”.
ChIP Antibodies, RNA Polymerase II
Anti-phospho-Ser-5 carboxyl-terminal domain of RNA polymerase II (H14) was purchased from Covance Research Products (catalog no. MMS-134R, Richmond, CA). acH3 antibody was purchased from Upstate (catalog no. 06-599, Billerica, MA); H3K4me3 antibody was purchased from Upstate (catalog no. MC315) or Abcam (catalog no. ab8580, Cambridge, MA); and H3K27 antibody was purchased from Upstate (catalog no. 07-449). Anti-RARγ serum was generated in rabbit against a peptide corresponding to 15 amino acids at the carboxyl terminus of RARγ. Polyclonal anti-RARγ IgG was purified from the crude serum through use of a DEAE-Affi-Gel blue column (Bio-Rad). Ash2l antibody was a kind gift from Dr. Jeffrey Dilworth.
Custom Array Design
Long oligonucleotide microarrays were designed using the Agilent eArray platform. Probes were tiled through each of the murine Hox clusters and through biological control regions such as Cyp26a1 and Gapdh. Additional probes used for quality control and normalization were also included on the arrays (Agilent). A total of 44,000 probes were tiled at 100–300-bp intervals through these loci with the median gap of 43 bp between the probes. Detailed methods are provided in the supplemental “Experimental Procedures”.
RESULTS
RA Activation of Hoxa1 Requires RARγ Bound to a 3′-Enhancer
We generated ESC lines in which either RARγ (RARγ-KO) or its cognate binding site downstream of Hoxa1 (RARE-KO) (Fig. 1A) are deleted. In WT ESCs, RA triggers a 15-fold increase in Hoxa1 transcripts within 24 h (Fig. 1, A and B). This response is severely blunted in both the RARγ-KO and RARE-KO mutants (Fig. 1, B and C). These results demonstrate that RARγ is essential for RA-mediated activation of Hoxa1 transcription and that RARγ acts through the Hoxa1 3′-RARE enhancer. Moreover, these results demonstrate that neither RARα nor RARβ can functionally compensate for RARγ in ESCs at these early times, as we have previously reported in F9 embryonal carcinoma cells (31, 32). We also show that Hoxa1 transcript levels in RARβ KO ES cells are similar to those in WT ES cells (Fig. 1D), demonstrating that RARβ is not essential for the RA-induced transcriptional activation of Hoxa1 in ES cells at these early time points. We next examined the epigenetic changes that accompanied Hoxa1 transcriptional activation.
RA Triggers Broad Epigenomic Reorganization of the Hoxa Cluster
We studied the spatiotemporal epigenetic reorganization of the entire ∼120-kb Hoxa chromosomal cluster (24) by examining three epigenetic marks (acH3, H3K4me3, and H3K27me3) prior to and 1, 8, and 24 h after RA addition (Hoxa cluster genes Hoxa1 to Hoxa13 plus Evx1, Fig. 2D). We also assessed the transcriptional activity of the Hoxa cluster genes using expression microarrays (Fig. 2E).
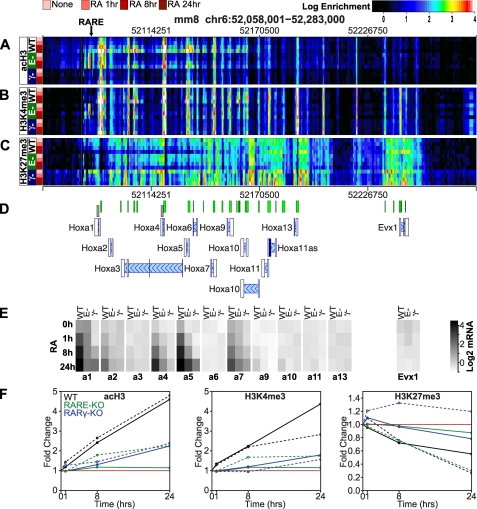
FIGURE 2.
Epigenetic landscape of the Hoxa gene cluster with RA treatment. The Hoxa gene cluster is shown with the genomic coordinates of the locus indicated at the top of the panel and the location of the Hoxa1 3′-RARE indicated by an arrow. ChIP-chip data for acH3 (A), H3K4me3 (B), and H3K27me3 (C) is presented as heatmaps with rows representing ChIP-chip data sets (replicate means), columns indicating genomic loci, and color representing log2-transformed ChIP enrichment. ChIP-chip was performed for three ESC lines as follows: WT, RARE-KO (E−), and RARγ-KO (γ−). ESCs were cultured ±1 μm RA for 1, 8, and 24 h, as indicated. The color scale of the log2 enrichment is indicated at the top of the figure. D, gene locations (blue tones) and CpG islands (green tones) are indicated schematically. The direction of transcription for each gene is indicated by light blue hash marks and proceeds from right to left. E, log2-transformed average (triplicate) mRNA microarray expression signal is represented for each gene in the locus as a grayscale level. The expression scale is indicated in the right-hand portion of the panel. At least three biological replicates were performed for each experiment. F, quantitative analyses of the Hoxa1 and Hoxa4 regions shown in D as gray boxes. The -fold change in enrichment in acH3 (1st subpanel), H3K4me3 (2nd subpanel), and H3K27me3 (3rd subpanel) is shown for the Hoxa1 locus (solid lines) and the Hoxa4 locus (dashed lines) and each of the ESC lines as follows: WT (black), RARE-KO (green), and RARγ-KO (blue).
In self-renewing WT ESCs, the clustered Hoxa genes reside within a large bivalent epigenomic domain broadly coated by the polycomb (PRC) repressive mark, H3K27me3, and punctated with foci of the oppositional H3K4me3 modification (Fig. 2, WT). These foci, representing regions poised for transcription, are coupled with acetylation of H3K9/K14 (acH3) and are highly correlated with the CpG islands and gene transcription start sites within the cluster (Fig. 2).
Immediately following RA exposure, the bivalent character of the proximal Hoxa genes (Hoxa1 to Hoxa7) is rapidly remodeled (within 1 h), and transcription is induced in WT ESCs. The regions with H3K4me3 and acH3 marks increase in magnitude and spread laterally, with the regions with H3 acetylation extending more broadly than those with H3K4me3. These changes are clearly evident at 1 h, the earliest time point shown (Fig. 2, A and B, WT). RA induces transcription of the proximal Hoxa cluster genes, with the relative exclusion of Hoxa3 and Hoxa6 (Fig. 2E, WT), and this transcriptional activation is associated with the increased placement of the acH3 and H3K4me3 chromatin marks surrounding the transcription start site of the activated genes. In contrast, removal of the PRC repressive mark, H3K27me3, occurs several hours later (Fig. 2C, WT). Only a minor local reduction of H3K27me3 is seen surrounding the Hoxa1 3′-RARE early after RA exposure (1 h). Further removal of this mark occurs more broadly at 8 h, but full erasure of H3K27 does not occur until the 24-h time point. These findings strongly suggest that placement of activating marks and induction of transcription are not directly coupled to removal of H3K27me3, at least within the context of a bivalent chromatin domain.
Hoxa1 3′-RARE Is a Key Organizer of Hoxa Cluster Activation by RARγ at Early Times after RA Addition
We examined the same epigenetic marks (acH3, H3K4me3, and H3K27me3) and gene expression in the RARγ-KO and RARE-KO ESC lines to assess how deletion of RARγ or of the 3′-Hoxa1 RARE affects activation of the Hoxa cluster. We found that in the absence of RA, neither deletion of RARγ nor of its cognate binding site downstream of Hoxa1 altered the transcriptional or qualitative epigenetic configuration of the Hoxa cluster (Fig. 2, None/0 h). We observed a modest increase in activation marks (acH3 and H3K4me3) in the RARE-KO ESCs surrounding the site of the deleted RARE where an active PGK promoter was inserted. These results indicate that RARγ and other factors capable of binding or being recruited to the Hoxa1 3′-RARE are dispensable for the establishment of a bivalent Hoxa chromatin domain in undifferentiated ESCs.
In contrast, RA must signal through RARγ to induce transcription of the proximal Hoxa genes and to trigger reorganization of the epigenomic configuration of the cluster (Fig. 2, γ−). In the absence of RARγ, all of the normal transcriptional responses to RA within the Hoxa cluster are severely blunted or absent. Aside from the small and delayed induction of Hoxa1, Hoxa5, and Hoxa7, the proximal Hoxa genes remain unaltered by RA exposure, and the entire locus remains in a bivalent configuration throughout the first 24 h of RA exposure. These results demonstrate that RARγ is essential for Hoxa cluster transcriptional activation within the first 24 h of RA exposure.
We next asked whether RARγ acts exclusively through the 3′-Hoxa1 RARE to activate the entire Hoxa cluster. Compared with WT ESCs (Fig. 2, WT), RA-mediated Hox gene induction was severely attenuated in RARE-KO ESCs, and the deposition of activating histone modifications (acH3 and H3K4me3) was constrained (Fig. 2, E−). Yet these deficits were far less comprehensive than those seen in the RARγ-KO cells (e.g. Hoxa4, -a5, and -a7), suggesting that aspects of RA signaling are mediated either directly or indirectly through other genomic elements or target genes. Remarkably, the spatiotemporal pattern of H3K27me3 removal from the Hoxa locus in RARE-KO ESCs was virtually identical to the WT, suggesting that activation or recruitment of the JmjC domain-containing enzymes is uncoupled from Hoxa locus-specific recruitment of RARγ. These results provide direct biochemical evidence that RA-mediated activation of the Hoxa cluster is triggered by RARγ bound to the 3′-Hoxa1 RARE but that normal epigenetic remodeling requires indirect signaling. To highlight the magnitude of epigenetic changes seen in the heat map (Fig. 2, A–C), we show the quantitative analysis of the acH3, H3K4me3, and H3K27me3 marks at the indicated regions in the Hoxa1 and Hoxa4 genes (Fig. 2F).
RA-induced Activation of the Hoxb Cluster Requires RARγ Activity Mediated through the Hoxa1 3′-Enhancer
Coordinate activation of the Hoxa and Hoxb clusters occurs during central nervous system development, and we observed synchronous activation of these clusters in ESCs (Figs. 2D and 3D). Both Hoxa1 and Hoxb1 have essential 3′-RAREs required for RA-induced transcriptional activation (19–21, 29), yet in hindbrain development, activation of Hoxb1 is subordinate to Hoxa1 because of a Hoxa1 responsive enhancer upstream of the Hoxb1 promoter (34, 36, 38). We explored how genetic lesions affecting the Hoxa locus alter the transcriptional and epigenomic responses of the Hoxb cluster to RA.
FIGURE 3.
Epigenetic landscape of the Hoxb gene cluster with RA treatment. The Hoxb cluster is shown with the genomic coordinates of the locus indicated at the top of the panel, and the location of the Hoxb1 3′-RARE is indicated by an arrow. ChIP-chip data for acH3 (A), H3K4me3 (B), and H3K27me3 (C) is presented as heatmaps with rows representing ChIP-chip data sets (replicate means), columns indicating genomic loci, and color representing log2-transformed ChIP enrichment. ChIP-chip was performed for the following three ESC lines: WT, RARE-KO (E−), and RARγ-KO (γ−). ESCs were cultured ± 1 μm RA for 1, 8, and 24 h, as indicated. The color scale of the log2 enrichment is indicated at the top of the figure. D, gene locations (blue tones) and CpG Islands (green tones) are indicated schematically. E, log2-transformed average (triplicate) mRNA microarray expression signal is represented for each gene in the locus as a grayscale level. The log2 averages is log2 transformed, normalized signal from the microarray. Each gray level is the average log2 transformed, normalized microarray signal for biological triplicate experiments. The expression scale is in the right-hand portion. F, quantitative analyses of the Hoxb1 and Hoxb5 regions shown in D as gray boxes. The -fold change in enrichment in acH3 (1st subpanel), H3K4me3 (2nd subpanel), and H3K27me3 (3rd subpanel) is shown for the Hoxb1 locus (solid lines) and the Hoxb5 locus (dashed lines) and each of the ESC lines as follows: WT (black), RARE-KO (green), and RARγ-KO (blue).
Like the Hoxa cluster, the Hoxb cluster has a bivalent character in WT ESCs, and RA induces a rapid reorganization of this domain, with focal placement of activating marks (acH3 and H3K4me3) preceding a more gradual and diffuse removal of H3K27me3 (Fig. 3, A–C, WT). Coincidently, RA triggers transcription of proximal Hoxb cluster genes (Hoxb1 to b7) within 1 h of RA exposure (Fig. 3E, WT). All of these transcriptional and epigenomic responses are severely blunted in RARγ-KO ESCs during the first 24 h after RA addition, suggesting that RA-mediated activation of the Hoxb cluster is largely dependent on RARγ at early time points (Fig. 3, A–C, and E, γ−) and that RARα and RARβ cannot compensate for the loss of RARγ. However, a surprising degree of cross-regulation between the Hoxa and Hoxb clusters was also evident.
Activation of the entire Hoxb cluster was attenuated in the RARE-KO ESCs (Fig. 3E, E−), and placement of activating histone modifications was blunted (Fig. 3, A and B, E−). However, RA-induced removal of the H3K27me3 mark proceeded normally across the Hoxb cluster genes in RARE-KO ESCs, even in the absence of normal transcriptional activation (Fig. 3C, WT, E−). To highlight the magnitude of epigenetic changes seen in the heat map (Fig. 3, A–C), we have shown the quantitative analysis for the acH3, H3K4me3, and H3K27me3 marks at the indicated regions in the Hoxa1 and Hoxa4 genes (Fig. 3F).These data indicate that RA signaling through RARγ bound to the Hoxb1 3′-RARE is not sufficient to trigger Hoxb cluster activation despite appropriate removal of the H3K27me3 mark. Although cross-regulation of Hoxa1 and Hoxb1 is well recognized during development, we show a major dependence of the Hoxb cluster on normal Hoxa cluster activation. This broad interdependence is initiated at the 3′-Hoxa1 RARE and subsequently reinforced by direct RARγ signaling within the 3′-Hoxb1 RARE and indirectly by both proximal Hoxa cluster genes (predominantly Hoxa1) and possibly other RARγ target genes (79, 80).
RARγ Is Required for RA-induced Transcription of Cyp26a1
The Cyp26a1 gene encodes an enzyme that metabolizes RA (81, 82) and is transcriptionally activated by RA through two closely localized RAREs in the proximal promoter (31, 32, 81–83). We examined the transcriptional activation of Cyp26a1 and its chromatin marks in response to RA in WT, RARE-KO, and RARγ-KO cells (Fig. 4, A–E).
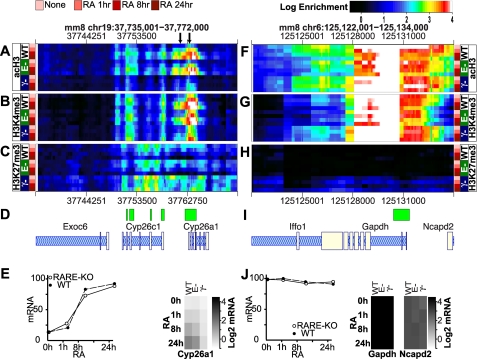
FIGURE 4.
Epigenetic landscape of Cyp26a1 and Gapdh after RA treatment. The loci of an RA target gene, Cyp26a1 (A–C) and Gapdh (F and G), are shown. The genomic coordinates of the loci are indicated at the top of the panels, and the location of the Cyp26a1 RAREs are indicated with arrows. ChIP-chip data acH3 (A and F), H3K4me3 (B and G), and H3K27me3 (C and H) are presented as heatmaps with rows representing ChIP-chip data sets (replicate means), columns indicating genomic loci, and color representing log2-transformed ChIP enrichment. ChIP-chip was performed for three ESC lines as follows: WT, RARE-KO (E−), and RARγ-KO (γ−). ESCs were cultured ± RA for 1, 8, and 24 h, as indicated. The color scale of the log2 enrichment is indicated at the top of the figure. D and I, gene locations (blue tones) and CpG Islands (green tones) are indicated schematically. E and J, line graph (left panel) shows transcript abundance quantified by quantitative RT-PCR normalized to the 36B4 reference control (mean ± S.E.). The grayscale image (right panel) shows the log2-transformed average (triplicate) mRNA microarray expression signal represented for Cyp26a1 (E) or Gapdh (J). The expression scale is in the right-hand portion of the panel.
In contrast to what we found at the Hox genes, RA-induced transcription of Cyp26a1 was robust in both WT and RARE-KO ESCs (Fig. 4E, WT, E−). RA exposure led to near normal epigenomic activation of the Cyp26a1 bivalent domain in these ESC lines. The activation marks (acH3, H3K4me3) rapidly increased and spread laterally, and the H3K27me3 mark was erased, initially from the RAREs and then across the promoter and proximal Cyp26a1 gene body (Fig. 4, A–C, WT, E−). In contrast, we found minimal histone acetylation and a comparatively small increase in H3K4me3 following RA treatment of RARγ-KO ESCs (Fig. 4E, γ−). As we saw at the Hox genes, the H3K27me3 mark was retained across this gene in the RARγ-KO line (Fig. 4C, γ−), and transcriptional activation of Cyp26a1 was virtually absent (Fig. 4E, γ−). Thus, RARγ is essential for RA-induced Cyp26a1 activation. Although other RA signal transducers, presumably other RARs, can affect some biochemical responses at this locus, only signaling mediated by RARγ leads to transcriptional activation.
As a control, we also examined a constitutively active gene, Gapdh (Fig. 4F), in the three ESC lines. Gapdh transcript levels were not responsive to RA treatment in any of the lines (Fig. 4J, and the epigenomic configuration of Gapdh was highly similar among the WT, RARE-KO, and RARγ-KO cell lines (Fig. 4, F–H). Thus, deletion of RARγ or of the 3′-Hoxa1 RARE causes RA-dependent and locus-specific abnormalities of the epigenome and gene transcription.
RARγ Is Bound to RAREs and RNA Polymerase II Is Bound to Proximal Promoters
We identified a central dependence on RARγ for the long range epigenomic reorganization of several RA-responsive genes in ESCs. To further investigate the mechanism of this dependence, we performed two-step ChIP (31, 32) to determine whether RARγ is present at the RAREs 3′ of the Hoxa1 and Hoxb1 genes and within the Cyp26a1 promoter. In WT ESCs, we identified RARγ at these RAREs prior to RA, with further recruitment after RA addition (Fig. 5, A, B, and D). We did not identify RARγ at the proximal promoter of either Hoxa1 or Hoxb1 (Fig. 5, C and E) or at a negative control region downstream of Hoxb1 (Fig. 5F). As expected, we detected no RARγ bound to any of the sites tested in the RARγ-KO ESCs (Fig. 5, A–F, filled squares).
FIGURE 5.
RARγ and RNA pol II binding to the Hoxa1, Hoxb1, and Cyp26a1 genes. A–F, RARγ ChIP was performed following dual cross-linking with disuccinimidyl glutarate and then formaldehyde. G–L, pol II Ser(P)-5 ChIP was performed following formaldehyde cross-linking. For all panels, WT, RARE-KO, and RARγ-KO ESCs were cultured ± 1 μm RA for 0, 1, or 8 h prior to cross-linking. ChIP DNA was quantified by quantitative PCR and is reported as percent enrichment of input (filled symbols, solid lines). Also shown are results using IgG as a negative control (open symbols, dotted lines). As an additional negative control we show enrichment at an intergenic region ∼18 kb 3′ of Hoxb1 (F and L). The loci analyzed are shown at the top of the figure. Each experiment was repeated 2–5 times, and data are reported as mean ± S.E.
We next examined the recruitment of RNA polymerase II (pol II) to Hoxa1, Hoxb1, and Cyp26a1 utilizing a monospecific antibody that recognizes the engaged/paused form of pol II phosphorylated at serine 5 (Ser(P)-5) (84). We did not identify pol II at the Hoxa1 or Hoxb1 3′-RARE prior to RA addition in any of the ESC lines (Fig. 5, H and J). Pol II was bound to the proximal promoters of these genes in both WT and mutant ESCs, but only in WT did RA induce further recruitment of pol II to these loci (Fig. 5, I and K). Thus, RA exposure increases the likelihood of RARγ binding to RAREs and leads to activation and recruitment of pol II to the proximal promoter regions of Hoxa1 and Hoxb1. Importantly, pol II levels increased normally at the Cyp26a1 proximal promoter in RARE-KO ESCs, indicating that the defects in this line are not common to all RA target genes (Fig. 5G). These results reinforce the primacy of RA-RARγ signaling at the 3′-Hoxa1 RARE for normal transcriptional and epigenetic activation of the Hoxa and Hoxb loci (Figs. 2 and 3).
RA-induced Placement of H3K4me3 Is Not Explained by Ash2l Recruitment
Histone lysine methyltransferases that can place the H3K4me3 mark reside in several complexes (MLL complexes) that share core components as follows: Ash2l, Rbbp5, Wdr5, and Dpy-30 (85). We next assessed whether changes in the H3K4me3 mark across the Hox clusters result from altered recruitment of MLL complexes in the mutant ESC lines. We used standard ChIP assays to probe the occupancy of a common MLL complex component, Ash2l, at the Hoxa1 and Hoxb1 3′-RAREs and proximal promoters, and at the Cyp26a1 proximal promoter. We found Ash2l associated with all of these loci prior to the addition of RA in WT, RARE-KO, and RARγ-KO ESCs (Fig. 6). (The Hoxa1 RARE cannot be assessed in the RARE-KO cells because it has been knocked out.) Basal occupation of these loci by the MLL complex is expected in ESCs because the product, H3K4me3, is also present (Figs. 2B, 3B, and 4B). Following RA exposure, Ash2l was further recruited to these loci in WT cells, but the level was slightly lower in the mutant ESC lines. Ash2l recruitment did not correlate with transcriptional activation or with Ser-5 phosphorylation of pol II at these loci (e.g. Figs. 4E, 5, G–K, and 6), and the magnitude of the change was modest (≤2-fold) when compared with the degree of H3K4me3 placement at these loci following RA exposure (Figs. 2A, 3A, and 4A). These results suggest that RA-induced transcription requires not just locus-specific recruitment of the MLL complex but also activation of some complex component or interacting partners.
FIGURE 6.
Recruitment of Ash2l to the Hoxa1, Hoxb1, and Cyp26a1 genes. A–E, Ash2L ChIP was performed on WT, RARE-KO, and RARγ-KO ESCs cultured ± 1 μm RA for 0, 1, or 8 h prior to cross-linking. ChIP DNA was quantified by quantitative PCR and is reported as percent enrichment of input (solid symbols, solid lines). Also shown are results using IgG as a negative control (open symbols, dashed lines). The loci analyzed are shown at the top of the figure. Each experiment was repeated 2–5 times, and data are reported as mean ± S.E.
DISCUSSION
We combined genetic tools with epigenomic technologies to investigate the spatiotemporal mechanisms of RA-induced Hox gene activation in ESCs. We identified RARγ as the predominant mediator of RA signaling in ESCs in the context of transcriptional activation of the Hoxa and Hoxb chromosomal clusters at early time points after RA addition. RARγ is required to trigger broad epigenomic reorganization of the Hoxa and Hoxb chromosomal clusters and for gene-specific removal of the H3K27me3 mark. We found rapid intra-cluster reorganization follows RA exposure, with transcription and the placement of activating marks preceding the removal of the polycomb-repressive mark, H3K27me3, by several hours. These results suggest that H3K27me3 erasure is temporally uncoupled from MLL complex activities and the coincident transcriptional induction. Our data also revealed a striking degree of inter-cluster coordination between the Hoxa and Hoxb clusters. This coordination results from cross-regulation of the proximal Hoxa and Hoxb cluster genes triggered by RARγ localized to the Hoxa1 3′-RARE. Thus, the 3′-Hoxa1 RARE is a critical regulatory element necessary for RA-RARγ-mediated activation of both the Hoxa1 and Hoxb1 clusters.
We found that large bivalent chromatin domains encompass the clustered Hox genes in self-renewing ESCs, as also demonstrated by other groups (52). Assembly of these regional domains did not require basal occupation of RAREs by RARγ because the bivalent character of these clusters was maintained in our mutant ESC lines (Figs. 2 and 3 and supplemental Figs. S1 and S2). It may be that these bivalent domains are self-assembling with their organization linked to the underlying sequence characteristics of the loci (52, 64). Placement of the activating marks (acH3 and H3K4me3) was highly periodic and tightly correlated with the transcription start site of cluster genes. This periodicity suggests a topological organization of the cluster with the activating marks collected by an intranuclear hub such as a “transcription factory” (86, 87). Although we detected no basal transcription from the clustered Hox genes, it is clear that these genes are maintained in a “poised” state so that they may be transcribed when triggered by developmental cues.
RA is an essential developmental signaling molecule that affects body plan organization through its regulation of Hox genes. We found that RA induces synchronous activation of the proximal Hoxa and Hoxb cluster genes in WT ESCs (Fig. 2E and Fig. 3E). Coincident with transcriptional activation, the epigenomic configuration of these clusters is rapidly remodeled, with the earliest changes occurring near the TSSs marked by H3K4me3 and acH3 (Figs. 2, A and B, and 3, A and B, WT). H3K4me3 can serve as an anchor for post-initiation and splicing factors, and the induced placement of this mark may provide a link to co-transcriptional RNA processing (88). Elegant studies of ESC nuclear organization have previously shown that Hoxb1 rapidly relocates to a transcription factory in response to RA (89). Transcription factories allow for shared enhancer activity and the concentration of regulatory proteins necessary for localized spreading of chromatin modifications (86, 90). In this light, the association of clustered Hox genes with transcription factories (either by de novo assembly or recruitment of preformed factories) may facilitate coordinated cluster regulation (87). In many ways, the synchronous spread of acH3 and H3K4me3 surrounding genes with RA-induced transcription appears to be a linear representation of the Hox cluster topology. It is tempting to speculate that transcription factories processing the proximal Hoxa and Hoxb clusters are managed by RARγ seated at the Hoxa1 3′-RARE because deletion of either the receptor or its this binding site diminishes or abolishes RA induction of both the Hoxa and Hoxb cluster genes (Figs. 2 and 3, γ−, E−).
Dynamic co-localization of actively transcribed genes appears to be the rule rather than the exception (91). Active transcription factories typically harbor the elongating form of pol II phosphorylated on Ser-2 (Ser(P)-2) (87, 91). However, certain developmental genes residing within PRC1-positive bivalent domains (64) are instead associated with poised transcription factories containing pol II Ser(P)-5 in place of pol II Ser(P)-2 (92, 93). Surprisingly, RNA synthesis does occur in poised transcription factories, but pol II elongation is ineffectively coupled to nascent RNA maturation, and transcription is nonproductive. Strikingly, induction of such developmental genes is not associated with major changes in pol II occupancy but is instead linked to removal of the PRC1/Ring1-mediated H2A ubiquitination, at least at later time points (92, 93). We found pol II Ser(P)-5 preloaded onto the promoters of Hoxa1 and Hoxb1 in self-renewing ESCs, suggesting that these genes are basally associated with poised transcription factories (Fig. 5). RA triggered only modest further recruitment of the polymerase to these promoters implicating other mechanisms in the rapid transcriptional induction of these genes (Figs. 2E and 3E).
Epigenomic reorganization of the proximal Hox clusters quickly follows RA exposure. The transition from a bivalent configuration to a mature pattern is largely complete by 24 h, but the kinetics of activation are much more rapid than the pace of H3K27me3 erasure (Figs. 2 and 3). Prior studies have also shown that tri-methylation of H3K4 by a MLL2/3 complex precedes H3K27me3 demethylation by the JmjC domain protein UTX, but these studies evaluated the Hox genes following relatively long (18–72 h) exposure to RA (73, 94). Our studies extend these prior reports by providing mechanistic insights from the early kinetics of Hox gene activation in ESCs. Although the action of UTX may be required for full transcriptional activation of Hox genes by RA at late time points (94), removal of H3K27me3 is not required to initiate transcriptional induction (Figs. 2, C and E, and 3, C and E). Furthermore, H3K27me3 erasure does not appear to be sufficient for normal transcriptional induction because transcription is severely depressed in the RARE-KO ESC line despite the tempo of demethylation being preserved (Fig. 2, C and E, E−). Thus, if RA-triggered induction of the proximal Hox clusters requires removal of H2Aub1 to release the poised pol II Ser(P)-5, it occurs too rapidly to depend upon shedding of the PRC1 complexes responsible for placing the H2Aub1 mark (93). Perhaps then, RA recruits to the Hox genes an H2A deubiquitinase such as 2A-DUB through its interactions with the RAR/RXR co-activator p300/PCAF (31, 32, 95).
Although we and others have shown that RA stimulates RARs to shed co-repressors and recruit co-activator molecules to specific RAREs (31, 32), recruitment alone does not fully explain Hox activation in ESCs. Like pol II Ser(P)-5 within a poised transcription factory, histone acetyltransferases and MLL complexes are preassembled at the Hox loci, yet their activity is insufficient to initiate productive transcription (Figs. 2, 3, 5, and 6). Similarly, UTX associates with the Hoxb1 locus in self-renewing ESCs at a time the locus is broadly marked by H3K27me3; thus, UTX activity must also be constrained prior to RA addition (94). Neither RARγ nor the Hoxa1 3′-RARE is necessary to assemble these factors, and further recruitment of these complexes to the Hox loci following RA exposure is modest compared with the pace and extent of transcriptional activation. Our data clearly show the primacy of RARγ as the mediator of early Hox activation by RA, but they do not explain how the Hox genes are so rapidly transcriptionally activated. Perhaps even minor changes in a specific factor or histone mark trigger a cascade of events leading to effective transcription. We speculate that RA induces an RARγ-dependent activity that unharnesses the pre-assembled complexes and permits transcription to initiate.
Our data support a model in which RARγ functions in ESCs as a critical regulator of the Hoxa and Hoxb clusters through interactions with distinct enhancer elements. We show that these Hox clusters fail to undergo normal transcription and epigenomic patterning following RA if either RARγ or its binding site 3′ of Hoxa1 is missing. The kinetics of these events suggest that activation of poised transcription does not wait for erasure of PRC2 marks. These results invite the discovery of the initial arbiter of RARγ-induced Hox activation.
Supplementary Material
Acknowledgments
We thank Dr. Alex Langston and Elana Wilen for some initial experiments, Christopher Kelly for editorial assistance, and the Gudas and Scandura laboratory members for helpful discussions. We thank Dr. Pierre Chambon for RARγ and RARβ knock-out mice and Dr. Jeffrey Dilworth for the Ash2l antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA043796 (to L. J. G.). This work was also supported by Cornell Belfer Family startup funds (to J. M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S3, Table S1, and additional references.
- RA
- all-trans-retinoic acid
- ESC
- embryonic stem cell
- H3K4me3
- trimethylation of lysine 4 of histone 3
- H3K27me3
- trimethylation of lysine 27 of histone H3
- acH3
- acetylation of H3K9/K14
- MLL
- mixed lineage leukemia
- PRC
- polycomb repressive complex
- pol II
- RNA polymerase II
- RAR
- retinoic acid receptor
- RARE
- retinoic acid response element.
REFERENCES
- 1. Boncinelli E., Simeone A., Acampora D., Mavilio F. (1991) Trends Genet. 7, 329–334 [DOI] [PubMed] [Google Scholar]
- 2. Langston A. W., Gudas L. J. (1994) Curr. Opin. Genet. Dev. 4, 550–555 [DOI] [PubMed] [Google Scholar]
- 3. Lemons D., McGinnis W. (2006) Science 313, 1918–1922 [DOI] [PubMed] [Google Scholar]
- 4. Duboule D. (2007) Development 134, 2549–2560 [DOI] [PubMed] [Google Scholar]
- 5. Duboule D., Morata G. (1994) Trends Genet. 10, 358–364 [DOI] [PubMed] [Google Scholar]
- 6. Garcia-Fernàndez J. (2005) Nat. Rev. Genet. 6, 881–892 [DOI] [PubMed] [Google Scholar]
- 7. Argiropoulos B., Humphries R. K. (2007) Oncogene 26, 6766–6776 [DOI] [PubMed] [Google Scholar]
- 8. Pfeifer G. P., Rauch T. A. (2009) Semin. Cancer Biol. 19, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaspar N., Marshall L., Perryman L., Bax D. A., Little S. E., Viana-Pereira M., Sharp S. Y., Vassal G., Pearson A., Reis R. M., Hargrave D., Workman P., Jones C. (2010) Cancer Res. 70, 9243–9252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ke X. S., Qu Y., Rostad K., Li W. C., Lin B., Halvorsen O. J., Haukaas S. A., Jonassen I., Petersen K., Goldfinger N., Rotter V., Akslen L. A., Oyan A. M., Kalland K. H. (2009) PLoS One 4, e4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Norris J. D., Chang C. Y., Wittmann B. M., Kunder R. S., Cui H., Fan D., Joseph J. D., McDonnell D. P. (2009) Mol. Cell 36, 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shah N., Sukumar S. (2010) Nat. Rev. Cancer 10, 361–371 [DOI] [PubMed] [Google Scholar]
- 13. Lengerke C., Schmitt S., Bowman T. V., Jang I. H., Maouche-Chretien L., McKinney-Freeman S., Davidson A. J., Hammerschmidt M., Rentzsch F., Green J. B., Zon L. I., Daley G. Q. (2008) Cell Stem Cell 2, 72–82 [DOI] [PubMed] [Google Scholar]
- 14. Pearson J. C., Lemons D., McGinnis W. (2005) Nat. Rev. Genet. 6, 893–904 [DOI] [PubMed] [Google Scholar]
- 15. Soshnikova N., Duboule D. (2009) Science 324, 1320–1323 [DOI] [PubMed] [Google Scholar]
- 16. Dupé V., Davenne M., Brocard J., Dollé P., Mark M., Dierich A., Chambon P., Rijli F. M. (1997) Development 124, 399–410 [DOI] [PubMed] [Google Scholar]
- 17. Frasch M., Chen X., Lufkin T. (1995) Development 121, 957–974 [DOI] [PubMed] [Google Scholar]
- 18. Gavalas A., Studer M., Lumsden A., Rijli F. M., Krumlauf R., Chambon P. (1998) Development 125, 1123–1136 [DOI] [PubMed] [Google Scholar]
- 19. Huang D., Chen S. W., Gudas L. J. (2002) Dev. Dyn. 223, 353–370 [DOI] [PubMed] [Google Scholar]
- 20. Langston A. W., Gudas L. J. (1992) Mech. Dev. 38, 217–227 [DOI] [PubMed] [Google Scholar]
- 21. Langston A. W., Thompson J. R., Gudas L. J. (1997) J. Biol. Chem. 272, 2167–2175 [DOI] [PubMed] [Google Scholar]
- 22. Mainguy G., In der Rieden P. M., Berezikov E., Woltering J. M., Plasterk R. H., Durston A. J. (2003) Trends Genet. 19, 476–479 [DOI] [PubMed] [Google Scholar]
- 23. Marshall H., Studer M., Pöpperl H., Aparicio S., Kuroiwa A., Brenner S., Krumlauf R. (1994) Nature 370, 567–571 [DOI] [PubMed] [Google Scholar]
- 24. Means A. L., Gudas L. J. (1995) Annu. Rev. Biochem. 64, 201–233 [DOI] [PubMed] [Google Scholar]
- 25. Oosterveen T., van Vliet P., Deschamps J., Meijlink F. (2003) J. Biol. Chem. 278, 24103–24107 [DOI] [PubMed] [Google Scholar]
- 26. Pöpperl H., Featherstone M. S. (1993) Mol. Cell. Biol. 13, 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roelen B. A., de Graaff W., Forlani S., Deschamps J. (2002) Mech. Dev. 119, 81–90 [DOI] [PubMed] [Google Scholar]
- 28. Thompson J. R., Chen S. W., Ho L., Langston A. W., Gudas L. J. (1998) Dev. Dyn. 211, 97–108 [DOI] [PubMed] [Google Scholar]
- 29. Huang D., Chen S. W., Langston A. W., Gudas L. J. (1998) Development 125, 3235–3246 [DOI] [PubMed] [Google Scholar]
- 30. Delacroix L., Moutier E., Altobelli G., Legras S., Poch O., Choukrallah M. A., Bertin I., Jost B., Davidson I. (2010) Mol. Cell. Biol. 30, 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gillespie R. F., Gudas L. J. (2007) J. Mol. Biol. 372, 298–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gillespie R. F., Gudas L. J. (2007) J. Biol. Chem. 282, 33421–33434 [DOI] [PubMed] [Google Scholar]
- 33. Mongan N. P., Gudas L. J. (2007) Differentiation 75, 853–870 [DOI] [PubMed] [Google Scholar]
- 34. Zhang M., Kim H. J., Marshall H., Gendron-Maguire M., Lucas D. A., Baron A., Gudas L. J., Gridley T., Krumlauf R., Grippo J. F. (1994) Development 120, 2431–2442 [DOI] [PubMed] [Google Scholar]
- 35. Tümpel S., Cambronero F., Ferretti E., Blasi F., Wiedemann L. M., Krumlauf R. (2007) Dev. Biol. 302, 646–660 [DOI] [PubMed] [Google Scholar]
- 36. Gavalas A., Ruhrberg C., Livet J., Henderson C. E., Krumlauf R. (2003) Development 130, 5663–5679 [DOI] [PubMed] [Google Scholar]
- 37. Barrow J. R., Stadler H. S., Capecchi M. R. (2000) Development 127, 933–944 [DOI] [PubMed] [Google Scholar]
- 38. Studer M., Gavalas A., Marshall H., Ariza-McNaughton L., Rijli F. M., Chambon P., Krumlauf R. (1998) Development 125, 1025–1036 [DOI] [PubMed] [Google Scholar]
- 39. Tvrdik P., Capecchi M. R. (2006) Dev. Cell 11, 239–250 [DOI] [PubMed] [Google Scholar]
- 40. Atkinson S. P., Koch C. M., Clelland G. K., Willcox S., Fowler J. C., Stewart R., Lako M., Dunham I., Armstrong L. (2008) Stem Cells 26, 1174–1185 [DOI] [PubMed] [Google Scholar]
- 41. Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 42. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 43. Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., Ye Z., Lee L. K., Stuart R. K., Ching C. W., Ching K. A., Antosiewicz-Bourget J. E., Liu H., Zhang X., Green R. D., Lobanenkov V. V., Stewart R., Thomson J. A., Crawford G. E., Kellis M., Ren B. (2009) Nature 459, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim T. H., Barrera L. O., Zheng M., Qu C., Singer M. A., Richmond T. A., Wu Y., Green R. D., Ren B. (2005) Nature 436, 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. (2007) Cell 130, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 47. Cao R., Zhang Y. (2004) Mol. Cell 15, 57–67 [DOI] [PubMed] [Google Scholar]
- 48. Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. (2001) Nature 410, 116–120 [DOI] [PubMed] [Google Scholar]
- 49. Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., Helin K. (2004) EMBO J. 23, 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., Jenuwein T. (2001) Cell 107, 323–337 [DOI] [PubMed] [Google Scholar]
- 51. Schlesinger Y., Straussman R., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B. E., Bergman Y., Simon I., Cedar H. (2007) Nat. Genet. 39, 232–236 [DOI] [PubMed] [Google Scholar]
- 52. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 53. Giadrossi S., Dvorkina M., Fisher A. G. (2007) Curr. Opin. Genet. Dev. 17, 132–138 [DOI] [PubMed] [Google Scholar]
- 54. Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G. A., Stewart R., Thomson J. A. (2007) Cell Stem Cell 1, 299–312 [DOI] [PubMed] [Google Scholar]
- 56. Martinez-Ceballos E., Chambon P., Gudas L. J. (2005) J. Biol. Chem. 280, 16484–16498 [DOI] [PubMed] [Google Scholar]
- 57. Martinez-Ceballos E., Gudas L. J. (2008) J Neurosci. Res. 86, 2809–2819 [DOI] [PubMed] [Google Scholar]
- 58. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 59. Bracken A. P., Dietrich N., Pasini D., Hansen K. H., Helin K. (2006) Genes Dev. 20, 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pasini D., Bracken A. P., Hansen J. B., Capillo M., Helin K. (2007) Mol. Cell. Biol. 27, 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ren X., Vincenz C., Kerppola T. K. (2008) Mol. Cell. Biol. 28, 2884–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., Koseki H., Fuchikami T., Abe K., Murray H. L., Zucker J. P., Yuan B., Bell G. W., Herbolsheimer E., Hannett N. M., Sun K., Odom D. T., Otte A. P., Volkert T. L., Bartel D. P., Melton D. A., Gifford D. K., Jaenisch R., Young R. A. (2006) Cell 125, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Suganuma T., Workman J. L. (2008) Cell 135, 604–607 [DOI] [PubMed] [Google Scholar]
- 64. Ku M., Koche R. P., Rheinbay E., Mendenhall E. M., Endoh M., Mikkelsen T. S., Presser A., Nusbaum C., Xie X., Chi A. S., Adli M., Kasif S., Ptaszek L. M., Cowan C. A., Lander E. S., Koseki H., Bernstein B. E. (2008) PLoS Genet. 4, e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sakamoto Y., Watanabe S., Ichimura T., Kawasuji M., Koseki H., Baba H., Nakao M. (2007) J. Biol. Chem. 282, 16391–16400 [DOI] [PubMed] [Google Scholar]
- 66. Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., Brugmann S. A., Goodnough L. H., Helms J. A., Farnham P. J., Segal E., Chang H. Y. (2007) Cell 129, 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sessa L., Breiling A., Lavorgna G., Silvestri L., Casari G., Orlando V. (2007) RNA 13, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hong S., Cho Y. W., Yu L. R., Yu H., Veenstra T. D., Ge K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jepsen K., Solum D., Zhou T., McEvilly R. J., Kim H. J., Glass C. K., Hermanson O., Rosenfeld M. G. (2007) Nature 450, 415–419 [DOI] [PubMed] [Google Scholar]
- 70. Jung J., Mysliwiec M. R., Lee Y. (2005) Dev. Dyn. 232, 21–32 [DOI] [PubMed] [Google Scholar]
- 71. Klose R. J., Kallin E. M., Zhang Y. (2006) Nat. Rev. Genet. 7, 715–727 [DOI] [PubMed] [Google Scholar]
- 72. Lan F., Bayliss P. E., Rinn J. L., Whetstine J. R., Wang J. K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., Roberts T. M., Chang H. Y., Shi Y. (2007) Nature 449, 689–694 [DOI] [PubMed] [Google Scholar]
- 73. Lee M. G., Villa R., Trojer P., Norman J., Yan K. P., Reinberg D., Di Croce L., Shiekhattar R. (2007) Science 318, 447–450 [DOI] [PubMed] [Google Scholar]
- 74. Lee N., Zhang J., Klose R. J., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2007) Nat. Struct. Mol. Biol. 14, 341–343 [DOI] [PubMed] [Google Scholar]
- 75. Ghyselinck N. B., Dupé V., Dierich A., Messaddeq N., Garnier J. M., Rochette-Egly C., Chambon P., Mark M. (1997) Int. J. Dev. Biol. 41, 425–447 [PubMed] [Google Scholar]
- 76. Lohnes D., Kastner P., Dierich A., Mark M., LeMeur M., Chambon P. (1993) Cell 73, 643–658 [DOI] [PubMed] [Google Scholar]
- 77. Kawase E., Suemori H., Takahashi N., Okazaki K., Hashimoto K., Nakatsuji N. (1994) Int. J. Dev. Biol. 38, 385–390 [PubMed] [Google Scholar]
- 78. Chen A. C., Gudas L. J. (1996) J. Biol. Chem. 271, 14971–14980 [DOI] [PubMed] [Google Scholar]
- 79. Su D., Gudas L. J. (2008) Exp. Hematol. 36, 624–641 [DOI] [PubMed] [Google Scholar]
- 80. Su D., Gudas L. J. (2008) Biochem. Pharmacol. 75, 1129–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Loudig O., Babichuk C., White J., Abu-Abed S., Mueller C., Petkovich M. (2000) Mol. Endocrinol. 14, 1483–1497 [DOI] [PubMed] [Google Scholar]
- 82. Loudig O., Maclean G. A., Dore N. L., Luu L., Petkovich M. (2005) Biochem. J. 392, 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dilworth F. J., Chambon P. (2001) Oncogene 20, 3047–3054 [DOI] [PubMed] [Google Scholar]
- 84. Fuda N. J., Ardehali M. B., Lis J. T. (2009) Nature 461, 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cho Y. W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G. R., Copeland T. D., Kalkum M., Ge K. (2007) J. Biol. Chem. 282, 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kosak S. T., Groudine M. (2004) Genes Dev. 18, 1371–1384 [DOI] [PubMed] [Google Scholar]
- 87. Sutherland H., Bickmore W. A. (2009) Nat. Rev. Genet. 10, 457–466 [DOI] [PubMed] [Google Scholar]
- 88. Sims R. J., 3rd., Millhouse S., Chen C. F., Lewis B. A., Erdjument-Bromage H., Tempst P., Manley J. L., Reinberg D. (2007) Mol. Cell 28, 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chambeyron S., Bickmore W. A. (2004) Genes Dev. 18, 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Spilianakis C. G., Lalioti M. D., Town T., Lee G. R., Flavell R. A. (2005) Nature 435, 637–645 [DOI] [PubMed] [Google Scholar]
- 91. Osborne C. S., Chakalova L., Brown K. E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J. A., Lopes S., Reik W., Fraser P. (2004) Nat. Genet. 36, 1065–1071 [DOI] [PubMed] [Google Scholar]
- 92. Brookes E., Pombo A. (2009) EMBO Rep. 10, 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Stock J. K., Giadrossi S., Casanova M., Brookes E., Vidal M., Koseki H., Brockdorff N., Fisher A. G., Pombo A. (2007) Nat. Cell Biol. 9, 1428–1435 [DOI] [PubMed] [Google Scholar]
- 94. Agger K., Cloos P. A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A. E., Helin K. (2007) Nature 449, 731–734 [DOI] [PubMed] [Google Scholar]
- 95. Zhu P., Zhou W., Wang J., Puc J., Ohgi K. A., Erdjument-Bromage H., Tempst P., Glass C. K., Rosenfeld M. G. (2007) Mol. Cell 27, 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.