Abstract
We present here a study of a eukaryotic trans-prenylsynthase from the malaria pathogen Plasmodium vivax. Based on the results of biochemical assays and contrary to previous indications, this enzyme catalyzes the production of geranylgeranyl pyrophosphate (GGPP) rather than farnesyl pyrophosphate (FPP). Structural analysis shows that the product length is constrained by a hydrophobic cavity formed primarily by a set of residues from the same subunit as the product as well as at least one other from the dimeric partner. Furthermore, Plasmodium GGPP synthase (GGPPS) can bind nitrogen-containing bisphosphonates (N-BPs) strongly with the energetically favorable cooperation of three Mg2+, resulting in inhibition by this class of compounds at IC50 concentrations below 100 nm. In contrast, human and yeast GGPPSs do not accommodate a third magnesium atom in the same manner, resulting in their insusceptibility to N-BPs. This differentiation is in part attributable to a deviation in a conserved motif known as the second aspartate-rich motif: whereas the aspartates at the start and end of the five-residue motif in FFPP synthases and P. vivax GGPPSs both participate in the coordination of the third Mg2+, an asparagine is featured as the last residue in human and yeast GGPPSs, resulting in a different manner of interaction with nitrogen-containing ligands.
Keywords: Enzymes/Inhibitors, Organisms/Parasite, Isoprenoid, Protein Structure, Protozoan, Plasmodium Parasites, Amino-bisphosphonates, Malaria, Structural Biology
Introduction
Isoprenoids make up the largest family of natural products and drugs derived from the five-carbon isoprene subunit. As precursors to steroids and sterols, they are utilized by prokaryotes and eukaryotes alike to modify proteins, such as G proteins, kinases, and phosphatases, and facilitate their localization to membranes. The production of isoprenoids begins with the synthesis of isopentenyl pyrophosphate (IPP)2 and dimethylallyl pyrophosphate (DMAPP) via either the mevalonate or nonmevalonate pathway. In most organisms, the first functionally significant product is farnesyl pyrophosphate (FPP). Condensation of FPP with another IPP molecule generates geranylgeranyl pyrophosphate (GGPP), which in turn begets successively longer isoprenoids as products of similar reactions constituting a multistep biopolymerization process. Chemists have designed numerous pyrophosphate analogs, including bisphosphonates, which feature substitution of oxygen by carbon as a phosphate linker. Nitrogen-containing bisphosphonates (N-BPs) have specifically been developed for bone diseases, targeting farnesyl pyrophosphate synthase (FPPS) (1–3), the producer of FPP with low nanomolar IC50 values in biochemical assays (3). These compounds are generally less effective against human and Saccharomyces cerevisiae geranylgeranyl pyrophosphate synthase (GGPPS), with high nanomolar or low micromolar IC50 values (4–6). On the other hand, alkylbisphosphonates (A-BPs) are capable of inhibiting the ability of GGPPS to convert FPP to GGPP at mid-nanomolar IC50 values (6). Crystallographic structures of human FPPS as well as human and yeast GGPPS, including complexes with products, substrates, and/or various inhibitors bound have led to descriptions of mechanisms of enzymatic action and inhibition (3, 6–8). One noteworthy observation is the number of magnesium atoms found to coordinate N-BPs in these structures, consistently three in FPPS and two in GGPPS (6).
Because isoprenoids are universally essential, bisphosphonates with and without nitrogen have also been tested as potential drugs against protozoan pathogens. Infection by trypanosomatids can be arrested by N-BPs at IC50 concentrations from low micromolar to nanomolar (9–11). In contrast, apicomplexan parasites have shown nonuniform susceptibility. For example, risedronate (RIS) and zoledronate (ZOL) can both be effective in impairing development of Toxoplasma gondii, Cryptosporidium parvum and Eimeria histolytica (10, 12), whereas Plasmodium falciparum cultures are less sensitive to N-BPs in the tests reported to date but can be controlled effectively by a number of A-BPs (4, 10, 13, 14). Recently, lipophilic bisphosphonates have been found to be effective in inhibiting liver stage infection in a mouse model (15).
Biochemical and structural studies on some of the protozoan enzymatic targets of bisphosphonates are also available to corroborate the parasitic disease models. For example, Trypanosoma cruzi FPPS is susceptible to nanomolar inhibition by RIS, with the mechanism elucidated by structural evidence (11). The T. gondii FPPS, also sensitive to inhibition by N-BPs, produces FPP along with a low level of GGPP when dosed with a high concentration of FPP (16); however, this bifunctional behavior is upstaged by a C. parvum nonspecific polyprenyl pyrophosphate synthase (CpNPPPS) that can synthesize a great range of products from C20 to C40 and can be inhibited by N-BPs at IC50 concentrations below 100 nm (12). Structural analysis identified the deviation in the chain length-determining (CLD) region of the Cryptosporidium enzyme critical in accommodating large products.
According to the PlasmoDB data base (17), there is only one Plasmodium enzyme homologous in sequence to human FPPS and GGPPS. The Plasmodium vivax ortholog can be inhibited by N-BPs and A-BPs (15, 18). Here, we present the findings of an investigation of the same P. vivax protein, focusing on enzymatic function and using crystallography to explain the mechanism of inhibition by bisphosphonates.
EXPERIMENTAL PROCEDURES
Reagents
All inhibitors used in this study, namely RIS, ZOL, and ibandronate (IBAN) were provided by Procter & Gamble Pharmaceuticals (Cincinnati, OH). The P. vivax Salvador I cDNA was a generous gift from Prof. Liwang Cui of Penn State University.
Cloning, Expression, and Purification of Plasmodium PVX_092040
Full-length P. vivax protein encoded by PVX_092040 (17) with an N-terminal His6 tag and TEV cleavage site (MGSSHHHHHHSSGRENLYFQ*G) was cloned from a P. vivax Salvador I cDNA library, expressed, and purified according to protocols described previously (19).
Enzymatic Characterization of PvGGPPS: Activity, Inhibition, and Product Studies
The activity of PvGGPPS3 was measured by a 14C radioactivity assay using the method of Reed and Rilling with some modifications (20). For the kinetic analysis, 35 nm purified recombinant protein was incubated for 3 min in 100 μl of buffer containing 50 mm Tris, pH 7.7, 2 mm MgCl2, 1 mm TCEP, and 5 μg/ml BSA. The concentrations of FPP and IPP (14C-IPP, 400 kBq/μmol) were typically 0.05–20 μm. The remainder of the assay was completed as previously described in Ref. 8. For inhibition experiments, 10 μl of 10× N-BP was added to the enzyme in a total volume of 80 μl. After a 10-min preincubation (to allow for any slow binding inhibition), 20 μl of a substrate mixture containing FPP and IPP (14C-IPP, 400 kBq/μmol) was added to start the reaction and giving a 10 μm final concentration for each substrate. Assays were carried out at 37 °C and subsequently terminated, extracted, and analyzed, as before (3). Preincubation of enzyme and inhibitor experiments showed tight binding characteristics, therefore data were evaluated as detailed previously (3). Reaction products were analyzed using thin layer chromatography (8). Silica Gel 60 TLC plates (Merck) spotted with reaction mixture were developed with propan-2-ol, ammonia, and water in a ratio of 9:3:1. Samples were identified by comparison with known standards and were visualized by staining with iodine vapor.
Crystallization, Data Collection, Structure Solution, and Refinement
PvGGPPS was crystallized in its apo form by mixing 1.5 μl of protein (at 16.5 mg/ml concentration in a buffer of 10 mm HEPES, pH 7.5, 500 mm NaCl) with 1.5 μl of reservoir solution consisting of 22% PEG 3350, 200 mm Li2SO4, 100 mm Tris, pH 8.5, in a hanging drop vapor diffusion setup with >350 μl of reservoir solution at 18 °C in VDXm plates (Hampton Research). For ZOL-added crystals, 1.5 μl of protein at 11.2 mg/ml containing an additional 10 mm ZOL, 10 mm IPP, and 10 mm MgCl2 was mixed with 1.5 μl of reservoir solution (20% PEG 3350, 200 mm Li2SO4, 100 mm Tris, 8.5 at 18 °C) and incubated in hanging drops over 350 μl of reservoir solution. For GGPP-added crystals, 1.5 μl of protein (at 12.1 mg/ml in 10 mm HEPES, pH 7.5, 500 mm NaCl containing 1 mm GGPP and 2 mm MgCl2) was mixed with 1.5 μl of reservoir solution consisting of 25% PEG 3350, 200 mm (NH4)2SO4, 100 mm Tris, pH 8.5, and incubated in hanging drops over 350 μl of reservoir solution at 18 °C. In each case crystals appeared overnight. Crystals were flash cooled in N2 (l). Data for apo and GGPP-bound crystals were collected on a Rigaku FR-E Superbright generator equipped with a Rigaku HTC image plate detector. The data for ZOL-incubated crystals were collected at the Cornell High Energy Synchrotron Source (CHESS), beam F1. Complete datasets were collected using 0.5° oscillations, and a total of 180° were collected. Native crystals diffracted to a maximum of 2.1 Å. The GGPP and ZOL crystals both diffracted to 2.5 Å, respectively. The native structure solution was obtained by molecular replacement using the program PHASER and Protein Data Bank (PDB) ID code 1UBV (Gallus gallus FPPS) as a model. Both ligand structures were subsequently solved using the original native PDB model as reference in PHASER. Initial maps from the molecular replacement for both sets showed obvious density for the ligands. All datasets and models were refined using REFMAC5 and the CCP4 package of programs. A test set consisting of 2% of the reflections was set aside for native data and 5% was set aside for both sets of ligand data. The final models have good geometry and no outliers in the Ramachandran analysis.
RESULTS
Plasmodium Prenylsynthases
Using the BLAST (21) utility served by the Web-based PlasmoDB database (17), the P. falciparum and P. vivax genes PF11_0295 and PVX_092040 were found to encode proteins 35–36% identical in sequence to human FPPS (HsFPPS) and 21–22% identical to human GGPPS (HsGGPPS). Similarly, they share a higher degree of sequence identity with FPPS than GGPPS from S. cerevisiae. Furthermore, the two Plasmodium orthologs are >70% identical to each other and approximately 30% identical to Trypanosoma and Toxoplasma FPPS as well as CpNPPPS, but less similar in sequence to GGPPS from E. histolytica.
In supplemental Fig. S1, the alignment of sequences of FPPS and/or GGPPS from humans, yeas,t and above-mentioned protozoan parasites are depicted. There is an insert of residues found specifically in the parasite prenylsynthases, including the kinetoplastids and apicomplexans, which is particularly long in the Toxoplasma enzyme and highly conserved between the two Plasmodium enzymes. Three of the critical sequence motifs identified in previous work (3, 8, 22), namely the first aspartate-rich motif (FARM), the second aspartate-rich motif (SARM) and the FPPS CLD region are also highlighted in supplemental Fig. S1. Previous studies indicate that (23, 24) two specific residues forming the CLD and starting 5 positions upstream of FARM limit the product length of FPPS to C15. These residues are both bulky in a specific FPPS and both small in GGPPS from humans, yeast, and Entamoeba. On the other hand, the reportedly bifunctional Toxoplasma FPPS (16) features a relatively small residue in cysteine and a large one in phenylalanine. Interestingly, both Plasmodium enzymes also have a similarly mixed pair in this position. Also distinctive is the asparagine in the fifth position in the SARM within HsGGPPS and ScGGPPS, where aspartate is present in all the other enzymes.
Previous analysis of structures of FPPS and (3, 6) have shown that the aspartates in FARM and SARM directly or indirectly (via water molecules) coordinate Mg2+ to interact with phosphate groups in substrates and inhibitors. In the Plasmodium enzymes as well as all FPPSs, an aspartate occupies the fifth position of SARM, where both human and yeast GGPPS feature asparagine.
Protein Expression and Activity Assays
We successfully expressed the P. vivax enzyme using an Escherichia coli-based expression platform proven to be effective for a significant number of apicomplexan proteins (19) but with two residues deviant from the sequence of PVX_092040 as published in PlasmoDB and NCBI (accession XP_001615401): T113M, N206D. This discrepancy may be attributed to either cloning mutation or genome sequencing error but is not part of any functional motif.
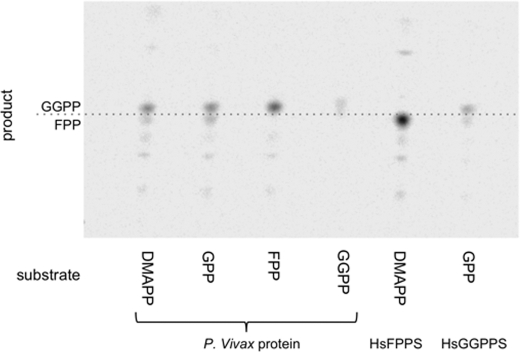
The activity of the recombinant protein was assayed using four potential allylic substrates, DMAPP, GPP, FPP, and GGPP. As indicated in Table 1 and Fig. 1, the enzyme catalyzed the conversion of all but GGPP. Among the productive substrates, the highest kcat value of 0.8 s−1 was obtained for FPP, with kcat = ∼0.5 s−1 for DMAPP and GPP (Table 1). These rate constants are comparable with the values of 0.42 and 0.204 s−1 obtained, respectively, for HsFPPS (3) and HsGGPPS (8). All three productive substrates had Km values in the low micromolar range (from 1.9 to 7.3 μm), as did IPP (8.4 μm), as found also for human (8) and yeast GGPPS (7). Judging kcat/Km ratios in Table 1, the efficiency of the Plasmodium enzyme is lowest with FPP as the substrate (however, the differences vis-à-vis DMAPP and IPP are not significant) and roughly in the same range as that of human GGPPS (8). Most significantly, TLC analysis (Fig. 1) revealed GGPP to be the final product for each of the three allylic substrates. Based on this, we propose that, at least under the conditions tested, the Plasmodium enzyme is a GGPPS (traces of intermediate products were present due to premature stoppage of the reaction as also in product analysis of human FPPS and GGPPS assays) (3, 8). Such a finding is intriguing given that this is the only Plasmodium enzyme homologous to both HsFPPS and HsGGPPS.
TABLE 1.
Kinetic parameters of PvGGPPS
Average values ± S.D. were obtained from multiple repetitions (n > 5) of the 14C radioactivity assay described in Ref. 20.
| Substrate | Apparent Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|
| μm | nmol/min per nmol | s−1 | μm−1s−1 | |
| IPP | 8.4 ± 1.6 | |||
| DMAPP | 3.2 ± 1.7 | 32.1 ± 1.7 | 0.5 | 0.2 |
| GPP | 1.9 ± 0.2 | 29.1 ± 1.0 | 0.5 | 0.3 |
| FPP | 7.3 ± 0.7 | 47.5 ± 1.6 | 0.8 | 0.1 |
| GGPP |
FIGURE 1.
Thin layer chromatography of products of P. vivax enzyme, human FPPS and GGPPS. The first four lanes on the left show 14C incorporation by products of the P. vivax enzyme from using GPP, DMAPP, FPP, and GGPP as substrates. The last two lanes on the right show 14C incorporation by products of HsFPPS and HsGGPPS, included in the experiment as a control. Comparison with HsGGPPS lanes shows clearly that the Plasmodium enzyme produced GGPP with DMAPP, GPP, or FPP as allylic substrate. No product is seen in the fourth lane, representing the use of GGPP as the substrate, indicating that the product size is limited to C20. Both the human and Plasmodium proteins produced a trace amount of FPP when GPP was the substrate, as did the Plasmodium protein using DMAPP, all due to premature stoppage of the reaction.
PvGGPPS Is Potently Inhibited by N-BPs
It has been previously reported that the PvGGPPS could be inhibited at submicromolar concentrations by bisphosphonates with and without nitrogen (18) (although the enzyme was assumed to be a FPPS in the study). The effectiveness of N-BPs is particularly interesting because these compounds have been found to be potent against FPPSs in general but ineffective on HsGGPPS (6). Accordingly, we chose three representative N-BPs, namely RIS, ZOL, and IBAN, to test against PvGGPPS under the same conditions as they were tested against HsFPPS (3), HsGGPPS (8), and CpNPPPS (12). Our experiments showed that all three compounds were more effective inhibitors by an order of magnitude than reported previously (18), with IC50 values of 47.5 nm for RIS, 43.3 nm for ZOL, and 61.9 nm for IBAN and Ki values also in the low nanomolar range (Table 2).
TABLE 2.
Inhibition of PvGGPPS by representative N-BPs (n > 5)
Average values ± S.D. were obtained from multiple repetitions (n > 5) of the 14C radioactivity assay described in Ref. 20.
| N-BP | IC50 | Kiapp | Ki |
|---|---|---|---|
| nm | nm | nm | |
| RIS | 47.5 ± 4.4 | 29.7 ± 4.0 | 12.4 ± 1.7 |
| ZOL | 43.3 ± 3.3 | 25.8 ± 2.9 | 10.7 ± 1.2 |
| IBAN | 61.9 ± 4.1 | 38.3 ± 4.5 | 16.0 ± 1.9 |
To the best of our knowledge, PvGGPPS is the only GGPPS studied to date that can be potently inhibited by N-BPs. We turned to structural biology to study this distinctive prenylsynthase further and determine the mechanism of inhibition by N-BPs.
Crystal Structures of PvGGPPS in Apo- and Ligand-bound Forms
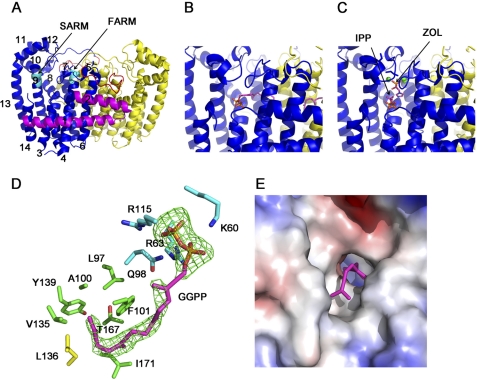
Structures of unliganded PvGGPPS (PDB ID code 3MAV; 2.1 Å; Fig. 2A), PvGGPPS/GGPP (PDB ID code 3PH7; 2.5 Å; Fig. 2B), and PvGGPPS/ZOL/IPP (PDB ID code 3LDW; 2.5 Å; Fig. 2C) were determined by means of x-ray crystallography, with the crystal parameters as well as processing and refinement data summarized in Table 3. Similar to FPPS structures but in contrast to the hexameric HsGGPPS (8), this protein is a homodimer of α-helices connected by loops of varying lengths (based on results of size exclusion chromatography; data not shown). In the crystals for all three structures, two dimers are in the asymmetric unit. In each subunit, the N-terminal two-helix hairpin (Fig. 2A) is curiously reminiscent of the FPPS fold (3, 25, 26) but distinct from both the single helix in the human GGPPS subunit (8) and the collinear pair of helices in ScGGPPS (7). The remaining helices of the PvGGPPS subunit, specifically α3–α14, form a cage-like arrangement common to both FPPS and other GGPPS to enclose a bicameral active site for housing the homoallylic and allylic substrates, as well as the eventual product. At the ends of helices α4 and α9, respectively, are the FARM and SARM, which contribute to binding of the allylic and homoallylic substrate in all prenylsynthases, e.g. Refs. 3, 7, 8, 12, 25, 26.
FIGURE 2.
PvGGPPS structures (all structures herein were rendered using PyMOL from Delano Scientific, Palo Alto, CA. A, PvGGPPS apo-dimer (PDB ID code 3MAV). The helices of the blue subunit are numbered. The N-terminal helical hairpin, consisting of α1 and α2, is highlighted in magenta. The FARM and SARM motifs are in cyan. The parasite-specific inserts are in red. B, active site of PvGGPPS occupied by product GGPP (PDB ID code 3PH7). C, active site of PvGGPPS occupied by IPP, ZOL, and magnesium ions (PDB ID code 3LDW). D, PvGGPPS active site. Close-up view shows the side chains around the two ends of GGPP: the key residues (carbon backbone in cyan) within hydrogen bonding distance of the pyrophosphate end and those forming a hydrophobic cavity (carbon backbone in green except for yellow Leu136 from the other subunit) anchoring the aliphatic end. An Fo − Fc omit map contoured at 2σ shows the electron density (in green) resulting from refinement of the complex without GGPP. A stereo figure of this active site is shown in supplemental Fig. S2. E, display of the electrostatic surface of a single subunit on the dimer interface side shows that, without the dimeric partner, the active site is wide open and may allow escape of the allylic substrate or intermediate product.
TABLE 3.
Data collection, phasing, and refinement statistics
| Data collection | PvGGPPS-apo | PvGGPPS+ZOL+IPP | PvGGPPS+GGPP |
|---|---|---|---|
| Space group | P21 | P212121 | P212121 |
| Cell dimensions | |||
| a (Å) | 84.00 | 111.27 | 107.10 |
| b (Å) | 116.46 | 109.70 | 108.99 |
| c (Å) | 92.43 | 139.53 | 141.35 |
| α° | 90.00 | 90.00 | 90.00 |
| β° | 116.01 | 90.00 | 90.00 |
| γ° | 90.00 | 90.00 | 90.00 |
| Wavelength | 1.5418 | 0.9179 | 1.5418 |
| Resolution | 50-2.1 | 25-2.5 | 50-2.5 |
| Measured reflections | 438,085 | 658,734 | 422,691 |
| Unique reflections | 92,263 | 60,963 | 57,809 |
| Rsym | 0.054 (0.564) | 0.115 (0.657) | 0.051 (0.759) |
| I/σ (I) | 35.4 (2.52) | 13.03 (2.03) | 39.83 (2.78) |
| Completeness (%) | 99.1 (96.6) | 100 (100) | 100 (100) |
| Redundancy | 4.7 (4.4) | 7.3 (7.2) | 7.3 (7.3) |
| PDB ID code | 3MAV | 3LDW | 3PH7 |
| Refinement | |||
| Resolution | 20.99-2.1 | 25-2.5 | 40-2.5 |
| No. of reflections | 92,136 | 60,724 | 57,715 |
| Test set | 1,850 | 3,026 | 2,901 |
| Rwork/Rfree | 0.229/0.262 | 0.243/0.285 | 0.237/0.273 |
| No. of atoms | 11,037 | 12,338 | 10,876 |
| Mean B factor | 46.29 | 34.21 | 68.77 |
| Ramachandran favored | 98 | 98.42 | 97.45 |
| Ramachandran disallowed | 0 | 0 | 0 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.011 | 0.01 | 0.01 |
| Bond angles (°) | 0.92 | 1.33 | 1.07 |
Enzyme-Product Complex
In the enzyme-product complex, GGPP is positioned with its pyrophosphate moiety in the homoallylic IPP chamber, forming hydrogen bonds with Lys60, Arg63, Gln98, and Arg115 (Fig. 2D). Overall, this highly hydrophilic region, where IPP is anchored before the condensation reaction, is conserved among FPPSs and GGPPSs. The middle portion of the aliphatic chain of GGPP is held in a channel within each subunit of PvGGPPS that is hydrophobic on the bottom and hydrophilic at the top. Interestingly, the CLD pair of phenylalanines in HsFPPS is aligned with Ala100 and Phe101 in PvGGPPS, where they form the wall of a hydrophobic cavity along with Leu97, Val135, Tyr139, Thr167, and Ile171 (Fig. 2D). In the same figure, we can see that helix 6 from the dimer partner, specifically Leu136, is complicit in closing this cavity. Without the presence of partner protein, as illustrated in the surface rendering of a single molecule of PvGGPPS in Fig. 2E, the hydrophobic end of the active site is wide open. This is distinctive from the yeast GGPPS structure (PDB ID code 2E8V), where each subunit provides complete self-containment of the GGPP pocket (7).
Enzyme-Inhibitor Complex
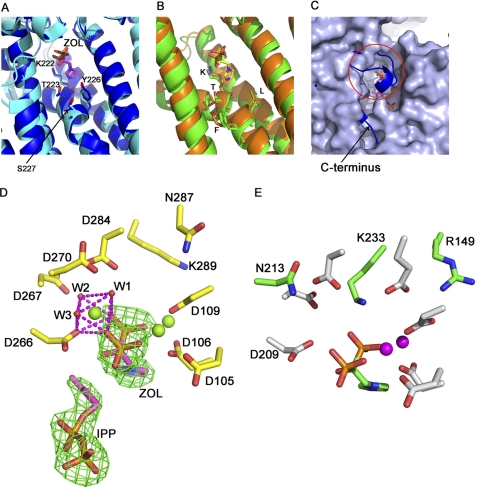
In the complex with ZOL and IPP (3LDW), the drug molecule interacts with the enzyme similarly to how N-BPs have been observed to bind FPPSs. First, the inhibitor is ensconced between the FARM- and SARM-containing helices via 3 Mg2+ atoms that interact specifically with the bisphosphonate moiety in a fashion similar to that found in the human FPPS·ZOL·IPP complex (3, 26). Furthermore, the nitrogen atom in the drug molecule heterocycle forms hydrogen bonds with the carbonyl oxygen on Lys222 and the Oγ on Thr223 (Fig. 3A). This N-Lys-Thr complex has also been previously described for HsFPPS (3, 26) and CpNPPPS (12) and is specifically responsible for the potent inhibition of both enzymes by N-BPs. More generally, the Lys-Thr pair is directly upstream of the kink on helix 8 and is conserved in all FPPSs as well as GGPPSs; however, variation between the two classes of prenylsynthases appears 4 positions downstream. Here, human and yeast GGPPSs feature a leucine and a phenylalanine (Leu173 and Phe174), creating a hydrophobic environment and counteracting to some degree the hydrogen bonding between nitrogen and the Lys-Thr pair (Fig. 3B). In contrast, as shown in Fig. 3A, both PvGGPPS and HsFPPS (and other FPPSs) feature a tyrosine (Tyr226) and a serine (Ser227) instead to create a comparatively polar environment. Although this effect distinguishes FPPSs and PvGGPPS from human and yeast GGPPSs, it does not fully explain the dramatically different levels of inhibition of these enzymes by N-BPs.
FIGURE 3.
ZOL binding. Both FPPSs and GGPPSs contain a duo of lysine and threonine just upstream of the kink on helix 8 capable of forming a strong bifurcated hydrogen bond with a heterocyclic ZOL (and similar N-BPs). In addition, these enzymes typically interact with the pyrophosphate moiety via Mg2+ and water molecules. A, in PvGGPPS (blue) and HsFPPS (cyan), as well as other FPPSs studied to date, there are also two residues downstream of the KT pair, namely tyrosine and serine, which form a polar region in place. Residues labeled are all from PvGGPPS. B, in most GGPPSs, including the human (orange) and yeast (green) enzymes, the N-Lys-Thr hydrogen-bonding complex is also conserved. In the vicinity of this interaction, there is a hydrophobic region formed by a Leu-Phe pair in place of the polar pair found in FPPS. The labeled residues are not numbered because they are the same in both human and yeast structures. C, surface rendering of apo-PvGGPPS superposed on schematic rendering (blue) of PvGGPPS with IPP and ZOL bound is shown. Both the C terminus and a loop connecting helices 9 and 10 (highlighted by a red circle) are stabilized by binding IPP. In particular, the stabilized loop blocks access to the active site and prevents competition with the drug molecule by the allylic substrate. D, PvGGPPS active site (3LDW) is shown. Three Mg2+ help bridge the binding of ZOL to FARM and SARM. On the SARM side, the third Mg2+ is coordinated by the phosphates on ZOL, water molecules W1, W2, and W3 as well as Asp266 in a fashion similar to that seen in FPPS (3). The positions of ZOL and IPP are verified by their Fo − Fc omit electron density maps contoured at 3σ. A stereo figure of this active site is shown in supplemental Fig. S3. E, in the ScGGPPS/ZOL structure, there are only two Mg2+ present, without a third metal ion to bridge SARM and ZOL. This may result from a change in the polar environment by the presence of the side chain of Lys233 (green), which is forced into this area in human and yeast GGPPS by Arg149 (green). Residue numbering is based on the scheme in Ref. 6. The substitution of asparagine (N213 in green) for aspartate in the last position of SARM, also found in human GGPPS, may be a factor as well.
In the apo structure, the C-terminal tail, specifically the last four residues (372FTGV375), is flexible, as is the loop following SARM and connecting helices 9 and 10 (with the segment 272FGDSKKTGKVG282 disordered). This leaves the “front” end of the active site wide open. In the ternary complex with ZOL and IPP, both regions are stabilized by ligand binding, resulting in a fully closed enzyme (Fig. 3C) and making the drug inaccessible to competition from the allylic substrate. A similar stabilization effect has been observed in HsFPPS (3, 26) where, however, the C-terminal tail is the primary segment blocking the active site once stabilized by the binding of IPP.
DISCUSSION
Both humans and yeast have distinctive enzymes to produce FPP and GGPP, whereas our study shows the single Plasmodium homolog to be a GGPP synthase. Three types of GGPPSs have been described in the literature (16). Based on our results, Plasmodium GGPPS deviates from all of them while sharing a few features with FPPS, namely (i) higher shared sequence identity with FPPSs than with GGPPSs; (ii) a similar fold, including a N-terminal hairpin seen previously only in FPPS structures; (iii) conservation of one bulky phenylalanine in the FPPS-CLD position typically featuring small residues in GGPPSs; (iv) aspartate in the last position of SARM rather than asparagine; (v) potent inhibition by N-BPs. To the best of our knowledge, no other GGPPS has been found to be inhibited by ZOL or RIS at the same level of potency.
As indicated in Table 1, the Plasmodium enzyme can use any one of DMAPP, GPP, and FPP readily as an allylic substrate; however, with this enzyme established by our results as a GGPPS, there is not another prenyl synthase available as a FPPS candidate in the Plasmodium genomes, based on sequence homology. Given that a methylerythritol phosphate-based Plasmodium isoprenoid biosynthetic pathway producing IPP has been firmly established (27), it is very likely PvGGPPS synthesizes GGPP in vivo with DMAPP and IPP as starting materials. On the other hand, previous studies have demonstrated the presence of active Plasmodium farnesyltransferase and geranylgeranyltransferase in the ring, trophozoite, and schizont stages (28, 29), suggesting ability to utilize both FPP and GGPP in these malaria pathogens. Significantly, in the same study, it has also been found that radiolabeled farnesol is elongated by Plasmodium parasites into geranylgeraniol before being used for protein prenylation (28), clearly supporting the presence of a GGPP synthase. On the other hand, PFT-I inhibitors have been found to be effective in inhibiting parasitic growth (28), which seems to suggest FPP to be essential. This invites questions about their source for this C15 product. At least three possibilities exist: (i) There is another Plasmodium enzyme producing FPP as a final product, one which is completely unique in its amino acid sequence so as to escape identification to date. (ii) FPP is imported from the human host. (iii) The Plasmodium enzyme in our study is in fact bifunctional in vivo, even though it produces only GGPP under the experimental conditions described herein. The last scenario is possible because FPP is a requisite intermediate product in the biosynthesis of GGPP and may be released under specific conditions by the enzyme without being turned into the final product via a currently unknown mechanism. Confirmation of this in future studies may be accompanied by elucidation of how the enzyme manipulates the substrates and products of three sequential steps of condensation.
Magnesium coordination is a ubiquitous mechanism of bisphosphonate binding in prenyl synthases and generally in binding of ligands with phosphate moieties, with the number of magnesium atoms varying among enzymes. In Ref. 6, it was first noted that the yeast GGPPS structures display a maximum of two Mg2+, whether the ligand is a substrate or inhibitor, in contrast to typical complement of three found in FPPSs. Interestingly, our structure PvGGPPS in complex with ZOL and IPP displays three Mg2+ involved in mediating the electrostatic interaction between the inhibitor and the FARM/SARM motifs, with two metal ions on the side of FARM and the remaining one on the other side facing SARM (Fig. 3D). Each Mg2+ is coordinated by 8 oxygen atoms contributed by the phosphate groups in the inhibitor, aspartates in the FARM and SARM, as well as water molecules. Specifically, the SARM-bridging Mg2+ is coordinated by ZOL, Asp266 (the first aspartate in the SARM), and 3 water molecules labeled W1, W2, and W3 in Fig. 3D. The W1 molecule forms hydrogen bonds with two aspartates, Asp270 and Asp284, with the former being the last aspartate in the SARM. In the yeast GGPPS structure with ZOL bound (PDB ID 2E91 and Fig. 3E), the two Mg2+ are both on the FARM side of the drug molecule, with the SARM-bridging metal ion absent. This is the case in almost all human and yeast GGPPS structures with 2 Mg2+/protein molecule. As noted above, both of these enzymes feature Asn as the last residue in their SARM, in contrast to Asp in FPPSs and PvGGPPS. At the time of writing this article, there are only two Saccharomyces GGPPS structures available in the PDB with three Mg2+ bound. In both cases, the ligand is an alkyl-BP. Likely due in part to the presence of Asn213, their SARM-bridging magnesium is coordinated differently, using 4 rather than 3 water molecules, with Asp209 (the first residue in the SARM motif) not participating directly. Notwithstanding the alternative coordination of the third metal ion, these A-BPs have been reported to be much more potent than ZOL in inhibiting yeast GGPPS (30). This supports the notion that the number of Mg2+ is an important factor in determining inhibitor potency.
Differences in the SARM motif may not be the only cause of deviation among prenylsynthases in the use of Mg2+ to coordinate phosphate groups. In human and yeast GGPPS, Arg149, which is not found in FPPSs or PvGGPPS, pushes Lys233 into the SARM region, numbering based on the scheme used for the yeast structure in (6). Together, the combination of a basic lysine and carboxamide-containing Asn213 in the yeast enzyme may change the solvent environment around the SARM region so as to prevent the coordination of a third Mg2+ in the manner seen in FPPSs and PvGGPPS.
Because Plasmodium cultures have been found to be much less sensitive to N-BPs than the kinetoplasts, Toxoplasma or Cryptosporidium (4, 10), our finding that Plasmodium GGPPS can be biochemically inhibited much more potently than previously believed is significant. This suggests the possibility that N-BPs may remain viable antimalarial candidates, if cell permeability problems described in the past (18) can be overcome with a next generation of compounds or transport enhancement. Furthermore, with an understanding of why N-BPs fail to affect HsGGPPS, it may be possible to modify existing drugs to permit a full complement of metal coordination and therefore improve the inhibition potency.
Finally, we believe that the structural and functional interpretations presented herein about the PvGGPPS enzyme are applicable to the P. falciparum ortholog. This is because the two proteins share 70% sequence identity, with nearly identical residues in key function-defining regions (supplemental Fig. S1).
Supplementary Material
Acknowledgments
We thank Helen Ren and Carmen Khuu for large scale expression of the PvGGPPS protein. The BL21(DE3)R3-pRARE2 strain of E. coli used in expressing the proteins came from Opher Gileadi of the Structural Genomics Consortium (SGC) at the University of Oxford. The cDNA for PvGGPPS was a generous gift from Prof. Liwang Cui of Pennsylvania State University.
This work was supported by the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Petrus and Augusta Hedlund's Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust (all to the Structural Genomics Consortium, registered charity number 1097737).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (codes 3MAV, 3PH7, and 3LDW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Residue numbering for PvGGPPS is based on the sequence of PVX_092040 in PlasmoDB and differs from that found in the PDB structure files.
- IPP
- isopentenyl pyrophosphate
- A-BP
- alkylbisphosphonate
- CLD
- chain length-determining
- CpNPPPS
- Cryptosporidium parvum nonspecific polyprenyl pyrophosphate synthase
- DMAPP
- dimethylallyl pyrophosphate
- FARM
- first aspartate-rich motif
- FPP
- farnesyl pyrophosphate
- FPPS
- farnesyl pyrophosphate synthase
- GGPP
- geranylgeranyl pyrophosphate
- GGPPS
- geranylgeranyl pyrophosphate synthase
- HsFPPS
- human FPPS
- HsGGPPS
- human GGPPS
- IBAN
- ibandronate
- N-BP
- nitrogen-containing bisphosphate
- PDB
- Protein Data Bank
- PvGGPPS
- Plasmodium vivax GGPPS
- RIS
- risedronte
- SARM
- second aspartate-rich motif
- ScGGPPS
- Saccharomyces cerevisiae GGPPS
- TCEP
- 1,2,3-tris(2-cyanoethoxy)propane
- ZOL
- zoledronate.
REFERENCES
- 1. Dunford J. E., Kwaasi A. A., Rogers M. J., Barnett B. L., Ebetino F. H., Russell R. G., Oppermann U., Kavanagh K. L. (2008) J. Med. Chem. 51, 2187–2195 [DOI] [PubMed] [Google Scholar]
- 2. Dunford J. E., Thompson K., Coxon F. P., Luckman S. P., Hahn F. M., Poulter C. D., Ebetino F. H., Rogers M. J. (2001) J. Pharmacol. Exp. Ther. 296, 235–242 [PubMed] [Google Scholar]
- 3. Kavanagh K. L., Guo K., Dunford J. E., Wu X., Knapp S., Ebetino F. H., Rogers M. J., Russell R. G., Oppermann U. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7829–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ghosh S., Chan J. M., Lea C. R., Meints G. A., Lewis J. C., Tovian Z. S., Flessner R. M., Loftus T. C., Bruchhaus I., Kendrick H., Croft S. L., Kemp R. G., Kobayashi S., Nozaki T., Oldfield E. (2004) J. Med. Chem. 47, 175–187 [DOI] [PubMed] [Google Scholar]
- 5. Szabo C. M., Matsumura Y., Fukura S., Martin M. B., Sanders J. M., Sengupta S., Cieslak J. A., Loftus T. C., Lea C. R., Lee H. J., Koohang A., Coates R. M., Sagami H., Oldfield E. (2002) J. Med. Chem. 45, 2185–2196 [DOI] [PubMed] [Google Scholar]
- 6. Guo R. T., Cao R., Liang P. H., Ko T. P., Chang T. H., Hudock M. P., Jeng W. Y., Chen C. K., Zhang Y., Song Y., Kuo C. J., Yin F., Oldfield E., Wang A. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10022–10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang T. H., Guo R. T., Ko T. P., Wang A. H., Liang P. H. (2006) J. Biol. Chem. 281, 14991–15000 [DOI] [PubMed] [Google Scholar]
- 8. Kavanagh K. L., Dunford J. E., Bunkoczi G., Russell R. G., Oppermann U. (2006) J. Biol. Chem. 281, 22004–22012 [DOI] [PubMed] [Google Scholar]
- 9. Montalvetti A., Bailey B. N., Martin M. B., Severin G. W., Oldfield E., Docampo R. (2001) J. Biol. Chem. 276, 33930–33937 [DOI] [PubMed] [Google Scholar]
- 10. Martin M. B., Grimley J. S., Lewis J. C., Heath H. T., 3rd, Bailey B. N., Kendrick H., Yardley V., Caldera A., Lira R., Urbina J. A., Moreno S. N., Docampo R., Croft S. L., Oldfield E. (2001) J. Med. Chem. 44, 909–916 [DOI] [PubMed] [Google Scholar]
- 11. Gabelli S. B., McLellan J. S., Montalvetti A., Oldfield E., Docampo R., Amzel L. M. (2006) Proteins 62, 80–88 [DOI] [PubMed] [Google Scholar]
- 12. Artz J. D., Dunford J. E., Arrowood M. J., Dong A., Chruszcz M., Kavanagh K. L., Minor W., Russell R. G., Ebetino F. H., Oppermann U., Hui R. (2008) Chem. Biol. 15, 1296–1306 [DOI] [PubMed] [Google Scholar]
- 13. Ling Y., Sahota G., Odeh S., Chan J. M., Araujo F. G., Moreno S. N., Oldfield E. (2005) J. Med. Chem. 48, 3130–3140 [DOI] [PubMed] [Google Scholar]
- 14. Moreno B., Bailey B. N., Luo S., Martin M. B., Kuhlenschmidt M., Moreno S. N., Docampo R., Oldfield E. (2001) Biochem. Biophys. Res. Commun. 284, 632–637 [DOI] [PubMed] [Google Scholar]
- 15. Singh A. P., Zhang Y., No J. H., Docampo R., Nussenzweig V., Oldfield E. (2010) Antimicrob. Agents Chemother. 54, 2987–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ling Y., Li Z. H., Miranda K., Oldfield E., Moreno S. N. (2007) J. Biol. Chem. 282, 30804–30816 [DOI] [PubMed] [Google Scholar]
- 17. Bahl A., Brunk B., Crabtree J., Fraunholz M. J., Gajria B., Grant G. R., Ginsburg H., Gupta D., Kissinger J. C., Labo P., Li L., Mailman M. D., Milgram A. J., Pearson D. S., Roos D. S., Schug J., Stoeckert C. J., Jr., Whetzel P. (2003) Nucleic Acids Res. 31, 212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mukkamala D., No J. H., Cass L. M., Chang T. K., Oldfield E. (2008) J. Med. Chem. 51, 7827–7833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vedadi M., Lew J., Artz J., Amani M., Zhao Y., Dong A., Wasney G. A., Gao M., Hills T., Brokx S., Qiu W., Sharma S., Diassiti A., Alam Z., Melone M., Mulichak A., Wernimont A., Bray J., Loppnau P., Plotnikova O., Newberry K., Sundararajan E., Houston S., Walker J., Tempel W., Bochkarev A., Kozieradzki I., Edwards A., Arrowsmith C., Roos D., Kain K., Hui R. (2007) Mol. Biochem. Parasitol. 151, 100–110 [DOI] [PubMed] [Google Scholar]
- 20. Reed B. C., Rilling H. C. (1975) Biochemistry 14, 50–54 [DOI] [PubMed] [Google Scholar]
- 21. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 22. Chen A., Kroon P. A., Poulter C. D. (1994) Protein Sci. 3, 600–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ohnuma S., Narita K., Nakazawa T., Ishida C., Takeuchi Y., Ohto C., Nishino T. (1996) J. Biol. Chem. 271, 30748–30754 [DOI] [PubMed] [Google Scholar]
- 24. Tarshis L. C., Proteau P. J., Kellogg B. A., Sacchettini J. C., Poulter C. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15018–15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tarshis L. C., Yan M., Poulter C. D., Sacchettini J. C. (1994) Biochemistry 33, 10871–10877 [DOI] [PubMed] [Google Scholar]
- 26. Rondeau J. M., Bitsch F., Bourgier E., Geiser M., Hemmig R., Kroemer M., Lehmann S., Ramage P., Rieffel S., Strauss A., Green J. R., Jahnke W. (2006) Chem. Med. Chem. 1, 267–273 [DOI] [PubMed] [Google Scholar]
- 27. Clastre M., Goubard A., Prel A., Mincheva Z., Viaud-Massuart M. C., Bout D., Rideau M., Velge-Roussel F., Laurent F. (2007) Exp. Parasitol. 116, 375–384 [DOI] [PubMed] [Google Scholar]
- 28. Chakrabarti D., Da Silva T., Barger J., Paquette S., Patel H., Patterson S., Allen C. M. (2002) J. Biol. Chem. 277, 42066–42073 [DOI] [PubMed] [Google Scholar]
- 29. Chakrabarti D., Azam T., DelVecchio C., Qiu L., Park Y. I., Allen C. M. (1998) Mol. Biochem. Parasitol. 94, 175–184 [DOI] [PubMed] [Google Scholar]
- 30. Chen K.-M., Hudock M. P., Zhang Y., Guo R. T., Cao R., No J. H., Liang P. H., Ko T. P., Chang T. H., Chang S. C., Song Y., Axelson J., Kumar A., Wang A. H., Oldfield E. (2008) J. Med. Chem. 51, 5594–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.